Dissection of Genetic Basis Underpinning Kernel Weight-Related Traits in Common Wheat
Abstract
1. Introduction
2. Results
2.1. Phenotypic Statistics
2.2. Linkage Map Construction
2.3. QTL Analyses of Three Grain Related-Traits
2.4. Co-Located Loci for All Three Traits
2.5. Co-Located Loci for Two Yield Related-Traits
2.6. The QTL Only Mapped for One Trait Effect
2.7. Factor ANOVA Analysis between Two Co-Located Loci for Three Traits
3. Discussion
3.1. Phenotypic Variation Caused by Environments
3.2. Comparison of Stable QTL for Grain-Related Traits with Previous Studies
4. Materials and Methods
4.1. Plant Materials
4.2. Field Trials and Phenotype Evaluation
4.3. DNA Extraction and Genotyping
4.4. Genetic Linkage Maps Construction and QTL Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Hawkesford, M.J.; Araus, J.-L.; Park, R.; Calderini, D.; Miralles, D.; Shen, T.; Zhang, J.; Parry, M.A.J. Prospects of doubling global wheat yields. Food Energy Secur. 2013, 2, 34–48. [Google Scholar] [CrossRef]
- Alexander, W.L.; Smith, E.L.; Dhanasobhan, C. A comparison of yield and yield component selection in winter wheat. Euphytica 1984, 33, 953–961. [Google Scholar] [CrossRef]
- Simmonds, J.; Scott, P.; Leverington-Waite, M.; Turner, A.S.; Brinton, J.; Korzun, V.; Snape, J.; Uauy, C. Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.). BMC Plant. Biol. 2014, 14, 191. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, D.; Meng, Z.; Xu, K.; Yan, J.; Xia, X.; Cao, S.; Tian, Y.; He, Z.; Zhang, Y. QTL mapping for grain yield-related traits in bread wheat via SNP-based selective genotyping. Theor. Appl. Genet. 2020, 133, 857–872. [Google Scholar] [CrossRef]
- Prashant, R.; Kadoo, N.; Desale, C.; Kore, P.; Dhaliwal, H.S.; Chhuneja, P.; Gupta, V. Kernel morphometric traits in hexaploid wheat (Triticum aestivum L.) are modulated by intricate QTL × QTL and genotype × environment interactions. J. Cereal Sci. 2012, 56, 432–439. [Google Scholar] [CrossRef]
- Lizana, X.C.; Riegel, R.; Gomez, L.D.; Herrera, J.; Isla, A.; McQueen-Mason, S.J.; Calderini, D.F. Expansins expression is associated with grain size dynamics in wheat (Triticum aestivum L.). J. Exp. Bot 2010, 61, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Mayes, S.; Sparkes, D.L. Carpel size, grain filling, and morphology determine individual grain weight in wheat. J. Exp. Bot 2015, 66, 6715–6730. [Google Scholar] [CrossRef]
- Fahy, B.; Siddiqui, H.; David, L.C.; Powers, S.J.; Borrill, P.; Uauy, C.; Smith, A.M. Final grain weight is not limited by the activity of key starch-synthesising enzymes during grain filling in wheat. J. Exp. Bot 2018, 69, 5461–5475. [Google Scholar] [CrossRef] [PubMed]
- Bhusal, N.; Sarial, A.K.; Sharma, P.; Sareen, S. Mapping QTLs for grain yield components in wheat under heat stress. PLoS ONE 2017, 12, e0189594. [Google Scholar] [CrossRef]
- Liu, B.; Asseng, S.; Liu, L.; Tang, L.; Cao, W.; Zhu, Y. Testing the responses of four wheat crop models to heat stress at anthesis and grain filling. Glob. Chang. Biol. 2016, 22, 1890–1903. [Google Scholar] [CrossRef]
- Wang, X.; Hou, L.; Lu, Y.; Wu, B.; Gong, X.; Liu, M.; Wang, J.; Sun, Q.; Vierling, E.; Xu, S. Metabolic adaptation of wheat grain contributes to a stable filling rate under heat stress. J. Exp. Bot 2018, 69, 5531–5545. [Google Scholar] [CrossRef]
- Al-Sheikh Ahmed, S.; Zhang, J.; Ma, W.; Dell, B. Contributions of TaSUTs to grain weight in wheat under drought. Plant. Mol. Biol. 2018, 98, 333–347. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Pozniak, C.J. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Xu, D.; Wen, W.; Fu, L.; Li, F.; Li, J.; Xie, L.; Xia, X.; Ni, Z.; He, Z.; Cao, S. Genetic dissection of a major QTL for kernel weight spanning the Rht-B1 locus in bread wheat. Theor. Appl. Genet. 2019, 132, 3191–3200. [Google Scholar] [CrossRef]
- He, X.; Lillemo, M.; Shi, J.; Wu, J.; Bjørnstad, Å.; Belova, T.; Dreisigacker, S.; Duveiller, E.; Singh, P. QTL characterization of fusarium head blight resistance in CIMMYT bread wheat line Soru#1. PLoS ONE 2016, 11, e0158052. [Google Scholar]
- Liu, J.; He, Z.; Wu, L.; Bai, B.; Wen, W.; Xie, C.; Xia, X. Genome-wide linkage mapping of QTL for black point reaction in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2179–2190. [Google Scholar] [CrossRef]
- Edae, E.A.; Rouse, M.N. Bulked segregant analysis RNA-seq (BSR-Seq) validated a stem resistance locus in Aegilops umbellulata, a wild relative of wheat. PLoS ONE 2019, 14, e0215492. [Google Scholar] [CrossRef]
- Shi, F.; Tibbits, J.; Pasam, R.K.; Kay, P.; Wong, D.; Petkowski, J.; Forrest, K.L.; Hayes, B.J.; Akhunova, A.; Davies, J.; et al. Exome sequence genotype imputation in globally diverse hexaploid wheat accessions. Theor. Appl. Genet. 2017, 130, 1393–1404. [Google Scholar] [CrossRef]
- Chen, Z.; Cheng, X.; Chai, L.; Wang, Z.; Bian, R.; Li, J.; Zhao, A.; Xin, M.; Guo, W.; Hu, Z.; et al. Dissection of genetic factors underlying grain size and fine mapping of QTgw.cau-7D in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 149–162. [Google Scholar] [CrossRef]
- Echeverry-Solarte, M.; Kumar, A.; Kianian, S.; Simsek, S.; Alamri, M.S.; Mantovani, E.E.; McClean, P.E.; Deckard, E.L.; Elias, E.; Schatz, B.; et al. New QTL alleles for quality-related traits in spring wheat revealed by RIL population derived from supernumerary × non-supernumerary spikelet genotypes. Theor. Appl. Genet. 2015, 128, 893–912. [Google Scholar] [CrossRef]
- Garcia, M.; Eckermann, P.; Haefele, S.; Satija, S.; Sznajder, B.; Timmins, A.; Baumann, U.; Wolters, P.; Mather, D.E.; Fleury, D. Genome-wide association mapping of grain yield in a diverse collection of spring wheat (Triticum aestivum L.) evaluated in southern Australia. PLoS ONE 2019, 14, e0211730. [Google Scholar] [CrossRef]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef]
- Guan, P.; Lu, L.; Jia, L.; Kabir, M.R.; Zhang, J.; Lan, T.; Zhao, Y.; Xin, M.; Hu, Z.; Yao, Y.; et al. Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front. Plant. Sci. 2018, 9, 529. [Google Scholar] [CrossRef]
- Guan, P.; Shen, X.; Mu, Q.; Wang, Y.; Wang, X.; Chen, Y.; Zhao, Y.; Chen, X.; Zhao, A.; Mao, W.; et al. Dissection and validation of a QTL cluster linked to Rht-B1 locus controlling grain weight in common wheat (Triticum aestivum L.) using near-isogenic lines. Theor. Appl. Genet. 2020, 133, 2639–2653. [Google Scholar] [CrossRef]
- Kumar, A.; Mantovani, E.E.; Simsek, S.; Jain, S.; Elias, E.M.; Mergoum, M. Genome wide genetic dissection of wheat quality and yield related traits and their relationship with grain shape and size traits in an elite × non-adapted bread wheat cross. PLoS ONE 2019, 14, e0221826. [Google Scholar] [CrossRef]
- Kumari, S.; Jaiswal, V.; Mishra, V.K.; Paliwal, R.; Balyan, H.S.; Gupta, P.K. QTL mapping for some grain traits in bread wheat (Triticum aestivum L.). Physiol Mol. Biol. Plants 2018, 24, 909–920. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; He, Z.; Liu, J.; Jin, H.; Cao, S.; Geng, H.; Yan, J.; Zhang, P.; Wan, Y.; et al. Genome-wide linkage mapping of yield-related traits in three Chinese bread wheat populations using high-density SNP markers. Theor. Appl. Genet. 2018, 131, 1903–1924. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; Liu, J.; Zhang, Y.; Cao, S.; He, Z.; Rasheed, A.; Jin, H.; Zhang, C.; Yan, J.; et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant. Biol. 2019, 19, 168. [Google Scholar] [CrossRef]
- Li, X.; Xu, X.; Liu, W.; Li, X.; Yang, X.; Ru, Z.; Li, L. Dissection of Superior Alleles for Yield-Related Traits and Their Distribution in Important Cultivars of Wheat by Association Mapping. Front. Plant. Sci. 2020, 11, 175. [Google Scholar] [CrossRef]
- Liu, T.; Wu, L.; Gan, X.; Chen, W.; Liu, B.; Fedak, G.; Cao, W.; Chi, D.; Liu, D.; Zhang, H.; et al. Mapping Quantitative Trait Loci for 1000-Grain Weight in a Double Haploid Population of Common Wheat. Int. J. Mol. Sci. 2020, 21, 3960. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P.; McIntyre, C.L.; Mathews, K.L.; Jalal Kamali, M.R.; Mossad, M.; Feltaous, Y.; Tahir, I.S.; Chatrath, R.; Ogbonnaya, F.; et al. QTL for yield and associated traits in the Seri/Babax population grown across several environments in Mexico, in the West Asia, North Africa, and South Asia regions. Theor. Appl. Genet. 2013, 126, 971–984. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, H.; Li, S.; Zou, Y.; Li, T.; Liu, J.; Ding, P.; Mu, Y.; Tang, H.; Deng, M.; et al. Identification of quantitative trait loci for kernel traits in a wheat cultivar Chuannong16. BMC Genet. 2019, 20, 77. [Google Scholar] [CrossRef]
- Su, Q.; Zhang, X.; Zhang, W.; Zhang, N.; Song, L.; Liu, L.; Xue, X.; Liu, G.; Liu, J.; Meng, D.; et al. QTL Detection for Kernel Size and Weight in Bread Wheat (Triticum aestivum L.) Using a High-Density SNP and SSR-Based Linkage Map. Front. Plant. Sci. 2018, 9, 1484. [Google Scholar] [CrossRef]
- Tura, H.; Edwards, J.; Gahlaut, V.; Garcia, M.; Sznajder, B.; Baumann, U.; Shahinnia, F.; Reynolds, M.; Langridge, P.; Balyan, H.S.; et al. QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments. Theor. Appl. Genet. 2020, 133, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.X.; Hai, L.; Zhang, X.Y.; You, G.X.; Yan, C.S.; Xiao, S.H. QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai x Yu8679. Theor. Appl. Genet. 2009, 118, 313–325. [Google Scholar] [CrossRef]
- Xin, F.; Zhu, T.; Wei, S.; Han, Y.; Zhao, Y.; Zhang, D.; Ma, L.; Ding, Q. QTL Mapping of Kernel Traits and Validation of a Major QTL for Kernel Length-Width Ratio Using SNP and Bulked Segregant Analysis in Wheat. Sci. Rep. 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Liang, F.; Xu, H.; Zhang, X.; Zhai, H.; Sun, Q.; Ni, Z. Identification of QTL for grain size and shape on the D genome of natural and synthetic allohexaploid wheats with near-identical AABB genomes. Front. Plant. Sci. 2017, 8, 1705. [Google Scholar] [CrossRef]
- Zanke, C.D.; Ling, J.; Plieske, J.; Kollers, S.; Ebmeyer, E.; Korzun, V.; Argillier, O.; Stiewe, G.; Hinze, M.; Neumann, F.; et al. Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Front. Plant. Sci. 2015, 6, 644. [Google Scholar] [CrossRef] [PubMed]
- Brinton, J.; Simmonds, J.; Minter, F.; Leverington-Waite, M.; Snape, J.; Uauy, C. Increased pericarp cell length underlies a major quantitative trait locus for grain weight in hexaploid wheat. New Phytol. 2017, 215, 1026–1038. [Google Scholar] [CrossRef]
- Hu, M.J.; Zhang, H.P.; Liu, K.; Cao, J.J.; Wang, S.X.; Jiang, H.; Wu, Z.Y.; Lu, J.; Zhu, X.F.; Xia, X.C.; et al. Cloning and Characterization of TaTGW-7A Gene Associated with Grain Weight in Wheat via SLAF-seq-BSA. Front. Plant. Sci. 2016, 7, 1902. [Google Scholar] [CrossRef]
- Zhai, H.; Feng, Z.; Du, X.; Song, Y.; Liu, X.; Qi, Z.; Song, L.; Li, J.; Li, L.; Peng, H.; et al. A novel allele of TaGW2-A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2018, 131, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Di, N.; Mu, Q.; Shen, X.; Wang, Y.; Wang, X.; Yu, K.; Song, W.; Chen, Y.; Xin, M.; et al. Use of near-isogenic lines to precisely map and validate a major QTL for grain weight on chromosome 4AL in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 2367–2379. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Xu, D.; Hanif, M.; Xia, X.; He, Z. Genetic architecture underpinning yield component traits in wheat. Theor. Appl. Genet. 2020, 133, 1811–1823. [Google Scholar] [CrossRef] [PubMed]
- IWGSC. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef]
- Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping:Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop. J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Mccouch, S.; Cho, Y.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Mccouch, S.; Cho, Y.; Paul, E.; Morishima, H. Report on QTL nomenclature. Rice Genet. Newsl. 1997, 14, 11–13. [Google Scholar]
- Ogbonnaya, F.C.; Rasheed, A.; Okechukwu, E.C.; Jighly, A.; Makdis, F.; Wuletaw, T.; Hagras, A.; Uguru, M.I.; Agbo, C.U. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor. Appl. Genet. 2017, 130, 1819–1835. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Guanter, L.; Guan, K.; You, L.; Huete, A.; Ju, W.; Zhang, Y. Satellite sun-induced chlorophyll fluorescence detects early response of winter wheat to heat stress in the Indian Indo-Gangetic Plains. Glob. Chang. Biol. 2018, 24, 4023–4037. [Google Scholar] [CrossRef]
- Sukumaran, S.; Reynolds, M.P.; Sansaloni, C. Genome-wide association analyses identify QTL hotspots for yield and component traits in durum wheat grown under yield potential, drought, and heat stress environments. Front. Plant. Sci. 2018, 9, 81. [Google Scholar] [CrossRef]
- Balla, K.; Karsai, I.; Bónis, P.; Kiss, T.; Berki, Z.; Horváth, Á.; Mayer, M.; Bencze, S.; Veisz, O. Heat stress responses in a large set of winter wheat cultivars (Triticum aestivum L.) depend on the timing and duration of stress. PLoS ONE 2019, 14, e0222639. [Google Scholar] [CrossRef] [PubMed]
- Valluru, R.; Reynolds, M.P.; Davies, W.J.; Sukumaran, S. Phenotypic and genome-wide association analysis of spike ethylene in diverse wheat genotypes under heat stress. New Phytol. 2017, 214, 271–283. [Google Scholar] [CrossRef]
- Chen, W.; Sun, D.; Yan, X.; Li, R.; Wang, S.; Shi, Y.; Jing, R. QTL analysis of wheat kernel traits, and genetic effects of qKW-6A on kernel width. Euphytica 2019, 215, 11. [Google Scholar] [CrossRef]
- Deng, Z.; Cui, Y.; Han, Q.; Fang, W.; Li, J.; Tian, J. Discovery of consistent QTLs of wheat spike-related traits under nitrogen treatment at different development stages. Front. Plant. Sci. 2017, 8, 2120. [Google Scholar] [CrossRef]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the chinese wheat cross Zhou 8425B/Chinese Spring. Front. Plant. Sci. 2015, 6, 1099. [Google Scholar] [CrossRef]
- Li, X.; Xia, X.; Xiao, Y.; He, Z.; Wang, D.; Trethowan, R.; Wang, H.; Chen, X. QTL mapping for plant height and yield components in common wheat under water-limited and full irrigation environments. Crop. Pasture Sci. 2015, 66, 660–670. [Google Scholar] [CrossRef]
- Zhang, J.; Dell, B.; Biddulph, B.; Drake-Brockman, F.; Walker, E.; Khan, N.; Wong, D.; Hayden, M.; Appels, R. Wild-type alleles of Rht-B1 and Rht-D1 as independent determinants of thousand-grain weight and kernel number per spike in wheat. Mol. Breed. 2013, 32, 771–783. [Google Scholar] [CrossRef]
- Dixon, L.E.; Greenwood, J.R.; Bencivenga, S.; Zhang, P.; Cockram, J.; Mellers, G.; Ramm, K.; Cavanagh, C.; Swain, S.M.; Boden, S.A. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum L.). Plant. Cell 2018, 30, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Jia, L.; Lu, L.; Qin, D.; Zhang, J.; Guan, P.; Ni, Z.; Yao, Y.; Sun, Q.; Peng, H. Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor. Appl. Genet. 2014, 127, 2415–2432. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, Y.; Wu, Q.; Chen, Y.; Zhang, P.; Zhang, Y.; Hu, W.; Wang, X.; Zhao, H.; Dong, L.; et al. Molecular characterization of a novel TaGL3–5A allele and its association with grain length in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 1799–1814. [Google Scholar] [CrossRef]
- Gegas, V.C.; Nazari, A.; Griffiths, S.; Simmonds, J.; Fish, L.; Orford, S.; Sayers, L.; Doonan, J.H.; Snape, J.W. A genetic framework for grain size and shape variation in wheat. Plant. Cell 2010, 22, 1046–1056. [Google Scholar] [CrossRef]
- Sajjad, M.; Ma, X.; Khan, S.H.; Shoaib, M.; Song, Y.; Yang, W.; Zhang, A.; Liu, D. TaFlo2-A1, an ortholog of rice Flo2, is associated with thousand grain weight in bread wheat (Triticum aestivum L.). BMC Plant. Biol 2017, 17, 164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ren, Y.; Yan, Y.; Fu, J.; Geng, H. Effect of Allelic Variations in TaGS-D1 and TaFlo2-A1 on Thousand Kernel Weight of Wheat. J. Triticeae Crop. 2020, 40, 533–546. [Google Scholar]
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant. Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef]
- Golan, G.; Ayalon, I.; Perry, A.; Zimran, G.; Ade-Ajayi, T.; Mosquna, A.; Distelfeld, A.; Peleg, Z. GNI-A1 mediates trade-off between grain number and grain weight in tetraploid wheat. Theor. Appl. Genet. 2019, 132, 2353–2365. [Google Scholar] [CrossRef]
- Sukumaran, S.; Lopes, M.; Dreisigacker, S.; Reynolds, M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor. Appl. Genet. 2018, 131, 985–998. [Google Scholar] [CrossRef]
- Griffiths, S.; Wingen, L.; Pietragalla, J.; Garcia, G.; Hasan, A.; Miralles, D.; Calderini, D.F.; Ankleshwaria, J.B.; Waite, M.L.; Simmonds, J.; et al. Genetic dissection of grain size and grain number trade-offs in CIMMYT wheat germplasm. PLoS ONE 2015, 10, e0118847. [Google Scholar] [CrossRef]
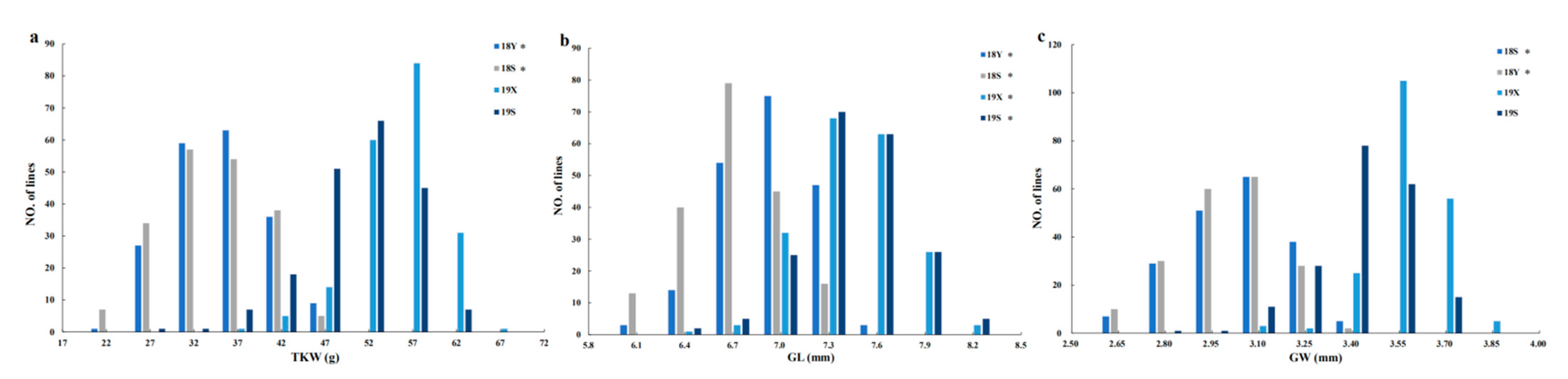
| Trait a | Environment b | HN5290 | 06Dn23 | Signi. d | 194 RILs Population | ||
|---|---|---|---|---|---|---|---|
| Mean ± SD c | Mean ± SD | Mean ± SD | Range | CV e (%) | |||
| TKW (g) | 18Y | 25.91 ± 1.89 | 46.29 ± 7.25 | *** | 37.62 ± 5.45 | 20.99–49.70 | 14.49 |
| 18S | 30.00 ± 3.96 | 51.46 ± 4.67 | *** | 36.96 ± 5.85 | 17.59–49.71 | 15.83 | |
| 19X | 45.78 ±1.22 | 70.79 ±2.12 | *** | 57.81 ± 4.45 | 40.28–66.97 | 7.70 | |
| 19S | 41.66 ± 2.72 | 59.44 ± 3.16 | *** | 53.06 ± 5.61 | 31.96–64.89 | 10.57 | |
| GL (mm) | 18Y | 6.08 ± 0.06 | 7.52 ± 0.18 | *** | 7.10 ± 0.29 | 6.34–7.84 | 4.02 |
| 18S | 6.30 ± 0.13 | 7.73 ± 0.20 | *** | 6.85 ± 0.31 | 5.92–7.53 | 4.51 | |
| 19X | 6.67 ± 0.07 | 8.33 ± 0.08 | *** | 7.59 ± 0.30 | 6.80–8.42 | 3.90 | |
| 19S | 6.64 ± 0.12 | 8.11 ± 0.14 | *** | 7.60 ± 0.30 | 6.60–8.45 | 3.97 | |
| GW (mm) | 18Y | 2.79 ± 0.05 | 3.37 ± 0.15 | *** | 3.11 ± 0.16 | 2.56–3.53 | 5.30 |
| 18S | 2.88 ± 0.12 | 3.44 ± 0.11 | *** | 3.09 ± 0.16 | 2.57–3.41 | 5.19 | |
| 19X | 3.39 ± 0.04 | 3.89 ± 0.06 | *** | 3.65 ± 0.11 | 3.23–3.93 | 3.03 | |
| 19S | 3.23 ± 0.10 | 3.63 ± 0.07 | *** | 3.50 ± 0.15 | 2.89–3.82 | 4.17 | |
| Source of Variation | df | Mean Square | ||
|---|---|---|---|---|
| TKW | GL | GW | ||
| Environments | 3 | 43,584.04 *** | 52.32 *** | 31.11 *** |
| Lines | 193 | 179.50 *** | 0.62 *** | 0.13 *** |
| Lines * Environments | 579 | 16.44 *** | 0.03 | 0.02 |
| Error | 768 | 6.82 | 0.01 | 0.01 |
| Heritability | 0.80 | 0.89 | 0.75 | |
| Environment | TKW18Y a | TKW18S b | TKW19X c | TKW19S | GL d 18Y | GL18S | GL19X | GL19S | GW e 18S | GW18Y | GW19X |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TKW18S | 0.80 ** | ||||||||||
| TKW19X | 0.71 ** | 0.69 ** | |||||||||
| TKW19S | 0.70 ** | 0.67 ** | 0.77 ** | ||||||||
| GL18Y | 0.66 ** | 0.61 ** | 0.66 ** | 0.61 ** | |||||||
| GL18S | 0.59 ** | 0.71 ** | 0.66 ** | 0.61 ** | 0.87 ** | ||||||
| GL19X | 0.47 ** | 0.49 ** | 0.71 ** | 0.51 ** | 0.83 ** | 0.82 ** | |||||
| GL19S | 0.42 ** | 0.47 ** | 0.59 ** | 0.65 ** | 0.81 ** | 0.78 ** | 0.81 ** | ||||
| GW18S | 0.76 ** | 0.94 ** | 0.62 ** | 0.61 ** | 0.46 ** | 0.57 ** | 0.32 ** | 0.32 ** | |||
| GW18Y | 0.92 ** | 0.71 ** | 0.62 ** | 0.60 ** | 0.44 ** | 0.39 ** | 0.25 ** | 0.21 ** | 0.78 ** | ||
| GW19X | 0.57 ** | 0.54 ** | 0.82 ** | 0.62 ** | 0.33 ** | 0.35 ** | 0.28 ** | 0.25 ** | 0.62 ** | 0.65 ** | |
| GW19S | 0.62 ** | 0.57 ** | 0.63 ** | 0.88 ** | 0.38 ** | 0.40 ** | 0.22 ** | 0.34 ** | 0.61 ** | 0.64 ** | 0.69 ** |
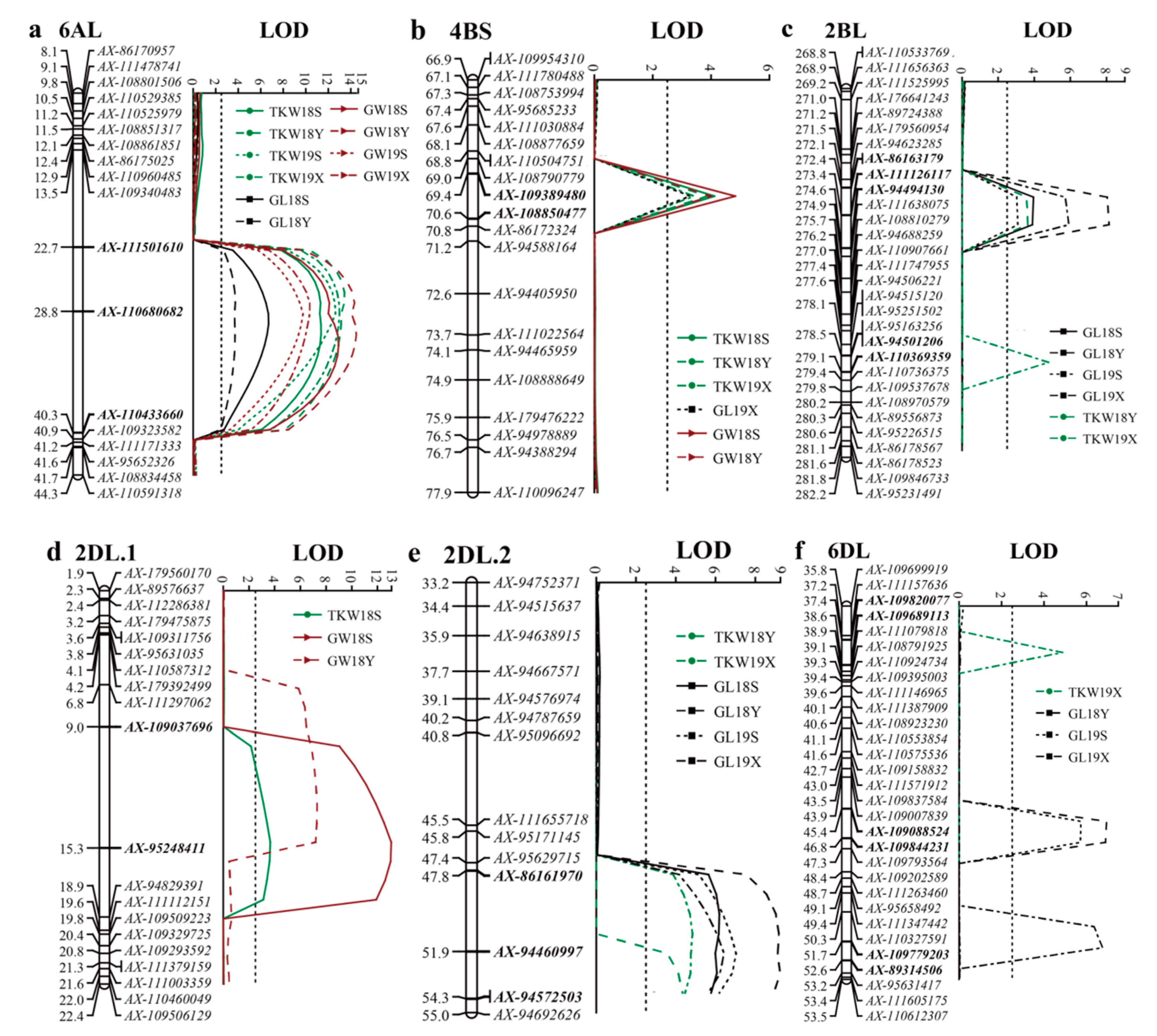
| Traits | QTL | Environment | Position | Left Marker | Right Marker | LOD a | PVE (%) b | Add c |
|---|---|---|---|---|---|---|---|---|
| TKW | QTkw.haaf-2BL | 18Y | 274 | AX-111126117 | AX-94494130 | 3.6 | 8.5 | −1.60 |
| 19X | 279 | AX-94501206 | AX-110369359 | 4.8 | 13.5 | −1.47 | ||
| QTkw.haaf-2DL.1 | 18S | 15 | AX-109037696 | AX-95248411 | 3.7 | 4.6 | −1.21 | |
| QTkw.haaf-2DL.2 | 18Y | 54 | AX-94460997 | AX-94572503 | 4.3 | 9.8 | −1.73 | |
| 19X | 51 | AX-86161970 | AX-94460997 | 4.8 | 10.7 | −1.51 | ||
| QTkw.haaf-4BS | 18S | 70 | AX-109389480 | AX-108850477 | 4.1 | 10.2 | −1.79 | |
| 18Y | 70 | AX-109389480 | AX-108850477 | 3.9 | 9.5 | −1.62 | ||
| 19X | 70 | AX-109389480 | AX-108850477 | 3.4 | 8.4 | −1.25 | ||
| QTkw.haaf-5AL | 18S | 187 | AX-86165895 | AX-110976589 | 5.1 | 6.5 | 1.47 | |
| 19S | 186 | AX-109966154 | AX-86165895 | 4.9 | 5.9 | 1.41 | ||
| 19X | 187 | AX-86165895 | AX-110976589 | 9.4 | 10.7 | 1.47 | ||
| QTkw.haaf-6AL | 18S | 31 | AX-110680682 | AX-110433660 | 11.4 | 17.2 | −2.40 | |
| 18Y | 27 | AX-111501610 | AX-110680682 | 13.4 | 24.8 | −2.58 | ||
| 19S | 29 | AX-111501610 | AX-110680682 | 12.6 | 16.6 | −2.39 | ||
| 19X | 29 | AX-111501610 | AX-110680682 | 13.0 | 15.4 | −1.78 | ||
| QTkw.haaf-6DL | 19X | 38 | AX-109820077 | AX-109689113 | 4.9 | 5.3 | −1.03 | |
| GL | QGl.haaf-1BS.1 | 18S | 99 | AX-94535608 | AX-86174948 | 24.9 | 5.3 | 0.26 |
| 18Y | 99 | AX-94535608 | AX-86174948 | 21.5 | 4.3 | 0.22 | ||
| 19S | 99 | AX-94535608 | AX-86174948 | 30.7 | 6.8 | 0.29 | ||
| 19X | 99 | AX-94535608 | AX-86174948 | 24.5 | 5.3 | 0.25 | ||
| QGl.haaf-1BS.2 | 18S | 103 | AX-179477422 | AX-179476084 | 35.0 | 8.5 | −0.33 | |
| 18Y | 103 | AX-179477422 | AX-179476084 | 30.3 | 6.9 | −0.28 | ||
| 19S | 103 | AX-179477422 | AX-179476084 | 42.9 | 11.3 | −0.37 | ||
| 19X | 103 | AX-179477422 | AX-179476084 | 35.5 | 8.9 | −0.32 | ||
| QGl.haaf-2AL | 18S | 16 | AX-108902945 | AX-111489408 | 5.3 | 7.3 | −0.08 | |
| 18Y | 18 | AX-108902945 | AX-111489408 | 5.5 | 6.3 | −0.07 | ||
| 19S | 17 | AX-108902945 | AX-111489408 | 5.3 | 1.8 | −0.07 | ||
| 19X | 16 | AX-108902945 | AX-111489408 | 5.3 | 5.6 | −0.06 | ||
| QGl.haaf-2BL | 18S | 273 | AX-86163179 | AX-111126117 | 4.0 | 11.9 | −0.09 | |
| 18Y | 274 | AX-111126117 | AX-94494130 | 8.1 | 17.8 | −0.12 | ||
| 19S | 274 | AX-111126117 | AX-94494130 | 3.1 | 9.7 | −0.08 | ||
| 19X | 274 | AX-111126117 | AX-94494130 | 5.9 | 16.5 | −0.10 | ||
| QGl.haaf-2DS | 18Y | 1 | AX-111112187 | AX-179558004 | 3.8 | 3.8 | 0.05 | |
| 19S | 1 | AX-111112187 | AX-179558004 | 6.4 | 6.4 | 0.08 | ||
| 19X | 7 | AX-109634352 | AX-110507164 | 6.2 | 5.4 | 0.07 | ||
| QGl.haaf-2DL.2 | 18S | 50 | AX-86161970 | AX-94460997 | 6.2 | 13.6 | −0.12 | |
| 18Y | 53 | AX-94460997 | AX-94572503 | 9.2 | 19.8 | −0.13 | ||
| 19S | 52 | AX-94460997 | AX-94572503 | 7.0 | 15.4 | −0.12 | ||
| 19X | 52 | AX-94460997 | AX-94572503 | 6.4 | 17.6 | −0.10 | ||
| QGl.haaf-4BS | 19X | 70 | AX-109389480 | AX-108850477 | 3.3 | 8.8 | −0.08 | |
| QGl.haaf-6AL | 18S | 30 | AX-110680682 | AX-110433660 | 6.7 | 8.8 | −0.09 | |
| 18Y | 28 | AX-111501610 | AX-110680682 | 3.7 | 4.1 | −0.05 | ||
| QGl.haaf-6DL | 18Y | 46 | AX-109088524 | AX-109844231 | 6.9 | 7.3 | −0.07 | |
| 19S | 46 | AX-109088524 | AX-109844231 | 5.7 | 5.8 | −0.08 | ||
| 19X | 52 | AX-109779203 | AX-89314506 | 6.8 | 5.9 | −0.07 | ||
| GW | QGw.haaf-2AS | 18S | 15 | AX-110478994 | AX-111530828 | 4.7 | 10.5 | −0.05 |
| 18Y | 15 | AX-110478994 | AX-111530828 | 3.9 | 8.9 | −0.05 | ||
| 19S | 15 | AX-110478994 | AX-111530828 | 6.5 | 14.3 | −0.06 | ||
| 19X | 15 | AX-110478994 | AX-111530828 | 5.3 | 11.7 | −0.04 | ||
| QGw.haaf-2DL.1 | 18S | 15 | AX-109037696 | AX-95248411 | 13.1 | 13.8 | −0.06 | |
| 18Y | 13 | AX-109037696 | AX-95248411 | 7.3 | 10.4 | −0.05 | ||
| QGw.haaf-4BS | 18S | 70 | AX-109389480 | AX-108850477 | 4.8 | 11.2 | −0.05 | |
| 18Y | 70 | AX-109389480 | AX-108850477 | 4.0 | 9.3 | −0.05 | ||
| QGw.haaf-6AL | 18S | 32 | AX-110680682 | AX-110433660 | 12.9 | 15.7 | −0.07 | |
| 18Y | 31 | AX-110680682 | AX-110433660 | 14.5 | 24.4 | −0.08 | ||
| 19S | 29 | AX-111501610 | AX-110680682 | 9.8 | 19.3 | −0.06 | ||
| 19X | 28 | AX-111501610 | AX-110680682 | 10.4 | 15.9 | −0.05 |
| Source | df | Type III SS a | Mean Square | F Value | p > F b | Variation (%) |
|---|---|---|---|---|---|---|
| Year | 2 | 54,492.3 | 27,246.1 | 1338.1 | <0.0001 | 80.9 |
| QTkw.haaf-4BS | 3 | 1079.9 | 534.0 | 26.5 | <0.0001 | 3.0 |
| QTkw.haaf-6AL | 3 | 1533.6 | 766.8 | 37.7 | <0.0001 | 4.4 |
| QTkw.haaf-4BS& QTkw.haaf-6AL | 3 | 48.6 | 16.2 | 0.8 | 0.4966 | 0 |
| Year | 1 | 6.1 | 6.1 | 80.7 | <0.0001 | 23.6 |
| QGL.haaf-4BS | 2 | 2.0 | 1.0 | 13.5 | <0.0001 | 9.0 |
| QGL.haaf-6AL | 2 | 1.3 | 0.6 | 8.4 | 0.0003 | 9.7 |
| QGL.haaf-4BS& QGL.haaf-6AL | 2 | 0.1 | 0.1 | 0.2 | 0.9271 | 0 |
| Year | 1 | 0.1 | 0.1 | 2.1 | 0.1441 | 0.4 |
| QGW.haaf-4BS | 2 | 0.8 | 0.4 | 20.8 | <0.0001 | 17.4 |
| QGW.haaf-6AL | 2 | 0.7 | 0.3 | 16.7 | <0.0001 | 17.7 |
| QGW.haaf-4BS& QGW.haaf-6AL | 2 | 0.1 | 0.1 | 1.0 | 0.4233 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wang, L.; Meng, Y.; Hao, Y.; Xu, H.; Hao, M.; Lan, S.; Zhang, Y.; Lv, L.; Zhang, K.; et al. Dissection of Genetic Basis Underpinning Kernel Weight-Related Traits in Common Wheat. Plants 2021, 10, 713. https://doi.org/10.3390/plants10040713
Li S, Wang L, Meng Y, Hao Y, Xu H, Hao M, Lan S, Zhang Y, Lv L, Zhang K, et al. Dissection of Genetic Basis Underpinning Kernel Weight-Related Traits in Common Wheat. Plants. 2021; 10(4):713. https://doi.org/10.3390/plants10040713
Chicago/Turabian StyleLi, Shunda, Liang Wang, Yaning Meng, Yuanfeng Hao, Hongxin Xu, Min Hao, Suque Lan, Yingjun Zhang, Liangjie Lv, Kai Zhang, and et al. 2021. "Dissection of Genetic Basis Underpinning Kernel Weight-Related Traits in Common Wheat" Plants 10, no. 4: 713. https://doi.org/10.3390/plants10040713
APA StyleLi, S., Wang, L., Meng, Y., Hao, Y., Xu, H., Hao, M., Lan, S., Zhang, Y., Lv, L., Zhang, K., Peng, X., Lan, C., Li, X., & Zhang, Y. (2021). Dissection of Genetic Basis Underpinning Kernel Weight-Related Traits in Common Wheat. Plants, 10(4), 713. https://doi.org/10.3390/plants10040713







