Exploring the End-Use Quality Potential of a Collection of Spanish Bread Wheat Landraces
Abstract
1. Introduction
2. Results
2.1. Genotypic Characterization of Bread Wheat Landraces
2.1.1. Puroindoline Genotyping
2.1.2. HMW-Gs Subunits Characterization
2.2. Evaluation of End-Use Quality-Related Traits
2.2.1. Landrace Performances in Comparison to Reference Varieties
2.2.2. Year and Genetic Structure in Relation to GPC and SDSS Values of Landraces
2.3. HMW-Gs Influence on Gluten Strength
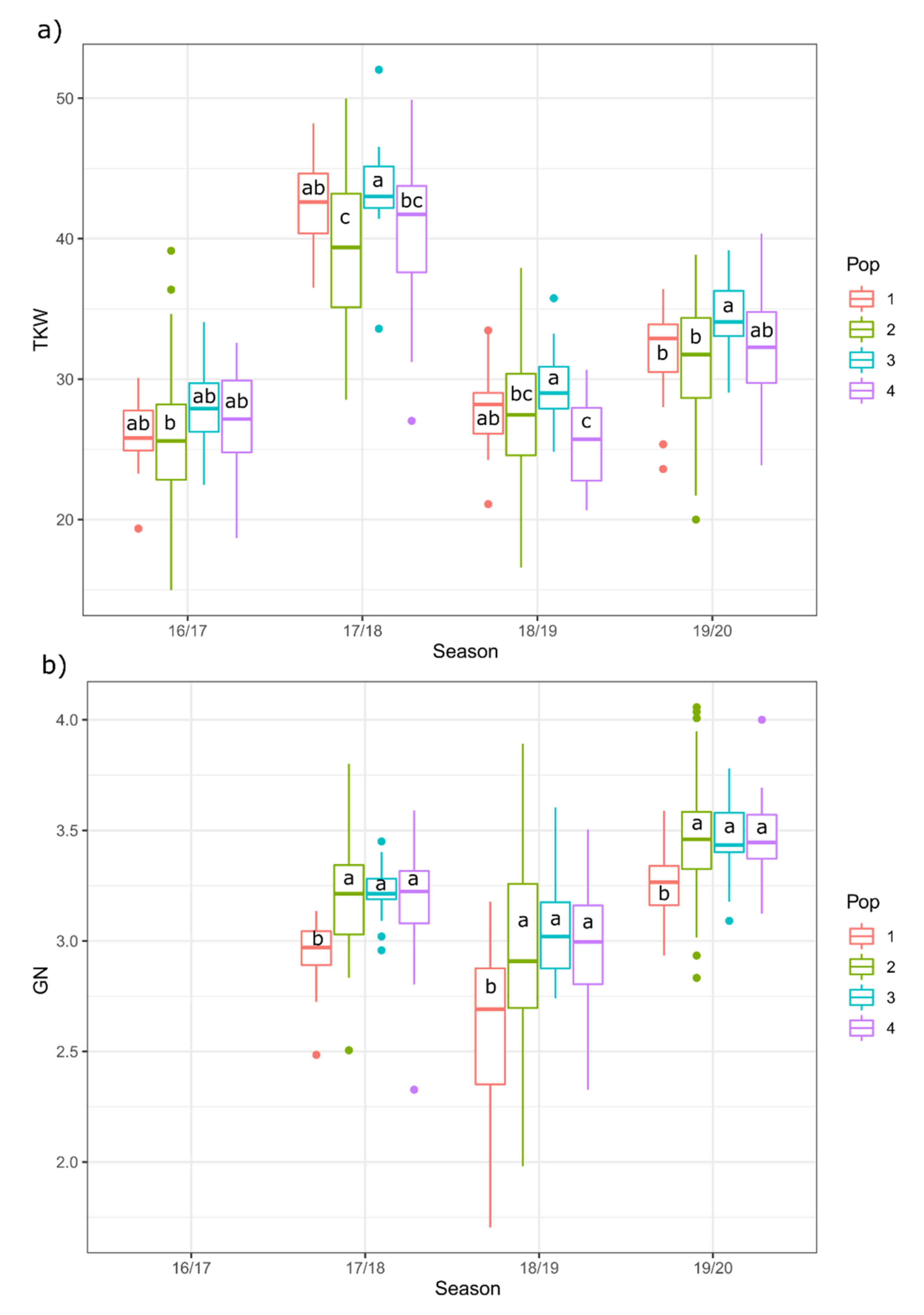
2.4. Evaluation of Yield Components (TKW and GN)
2.4.1. Landrace Performances in Comparison to Modern Varieties
2.4.2. Year and Genetic Structure Influence in TKW and GN
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Isolation and Genotyping
4.3. Grain Quality and Yieldparameters
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations FAOSTAT. Available online: http://www.fao:faostat/es/#data/QC. (accessed on 1 February 2021).
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F.; Olsen, O. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.S. Deepening the wheat gene pool. J. Crop Prod. 1997, 1, 1–25. [Google Scholar] [CrossRef]
- Ficiciyan, A.; Loos, J.; Sievers-Glotzbach, S.; Tscharntke, T. More than yield: Ecosystem services of traditional versus modern crop varieties revisited. Sustainability 2018, 10, 2834. [Google Scholar] [CrossRef]
- Alipour, H.; Bihamta, M.R.; Mohammadi, V.; Peyghambari, S.A.; Bai, G.; Zhang, G. Genotyping-by-sequencing (GBS) revealed molecular genetic diversity of Iranian wheat landraces and cultivars. Front. Plant. Sci. 2017, 8, 1293. [Google Scholar] [CrossRef] [PubMed]
- Nazco, R.; Villegas, D.; Ammar, K.; Pena, R.J.; Moragues, M.; Royo, C. Can Mediterranean durum wheat landraces contribute to improved grain quality attributes in modern cultivars? Euphytica 2012, 185, 1–17. [Google Scholar] [CrossRef]
- Wingen, L.U.; Orford, S.; Goram, R.; Leverington-Waite, M.; Bilham, L.; Patsiou, T.S.; Ambrose, M.; Dicks, J.; Griffiths, S. Establishing the AE Watkins landrace cultivar collection as a resource for systematic gene discovery in bread wheat. Theor. Appl. Genet. 2014, 127, 1831–1842. [Google Scholar] [CrossRef]
- Jordan, K.W.; Wang, S.; Lun, Y.; Gardiner, L.; MacLachlan, R.; Hucl, P.; Wiebe, K.; Wong, D.; Forrest, K.L.; Sharpe, A.G. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Sehgal, D.; Vikram, P.; Sansaloni, C.P.; Ortiz, C.; Saint Pierre, C.; Payne, T.; Ellis, M.; Amri, A.; Petroli, C.D.; Wenzl, P. Exploring and mobilizing the gene bank biodiversity for wheat improvement. PLoS ONE 2015, 10, e0132112. [Google Scholar] [CrossRef] [PubMed]
- Riaz, A.; Hathorn, A.; Dinglasan, E.; Ziems, L.; Richard, C.; Singh, D.; Mitrofanova, O.; Afanasenko, O.; Aitken, E.; Godwin, I. Into the vault of the Vavilov wheats: Old diversity for new alleles. Genet. Resour. Crop Evol. 2017, 64, 531–544. [Google Scholar] [CrossRef]
- Asseng, S.; Martre, P.; Maiorano, A.; Rötter, R.P.; O’Leary, G.J.; Fitzgerald, G.J.; Girousse, C.; Motzo, R.; Giunta, F.; Babar, M.A. Climate change impact and adaptation for wheat protein. Global Chang. Biol. 2019, 25, 155–173. [Google Scholar] [CrossRef]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef]
- Brancourt-Hulmel, M.; Doussinault, G.; Lecomte, C.; Bérard, P.; Le Buanec, B.; Trottet, M. Genetic improvement of agronomic traits of winter wheat cultivars released in France from 1946 to 1992. Crop Sci. 2003, 43, 37–45. [Google Scholar] [CrossRef]
- De Vita, P.; Matteu, L.; Mastrangelo, A.M.; Di Fonzo, N.; Cattivelli, L. Effects of breeding activity on durum wheat traits breed in Italy during the 20th century. Ital. J. Agron. 2007, 2, 451–462. [Google Scholar] [CrossRef]
- Payne, P.I.; Nightingale, M.A.; Krattiger, A.F.; Holt, L.M. The relationship between HMW glutenin subunit composition and the bread-making quality of British-grown wheat varieties. J. Sci. Food Agric. 1987, 40, 51–65. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G.; Tatham, A.S. High molecular weight subunits of wheat glutenin. J. Cereal Sci. 1992, 15, 105–120. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G.; Lafiandra, D. Genetics of Wheat Gluten Proteins. Adv. Genet. 2003, 49, 111–184. [Google Scholar] [CrossRef] [PubMed]
- Payne, P.I.; Holt, L.M.; Law, C.N. Structural and genetical studies on the high-molecular-weight subunits of wheat glutenin. Theor. Appl. Genet. 1981, 60, 229–236. [Google Scholar] [CrossRef]
- Pogna, N.E.; Borghi, B.; Mellini, F.; Peruffo, A.D.B.; Nash, R.J. Electrophoresis of gliadins for estimating the genetic purity in bread wheat seed production. Genet. Agr. 1986, 40, 201–212. [Google Scholar]
- Branlard, G.; Pierre, J.; Rousset, M. Selection indices for quality evaluation in wheat breeding. Theor. Appl. Genet. 1992, 84, 57–64. [Google Scholar] [CrossRef]
- Branlard, G.; Dardevet, M. Diversity of grain protein and bread wheat quality: II. Correlation between high molecular weight subunits of glutenin and flour quality characteristics. J. Cereal Sci. 1985, 3, 345–354. [Google Scholar] [CrossRef]
- Pirozi, M.R.; Margiotta, B.; Lafiandra, D.; MacRitchie, F. Composition of polymeric proteins and bread-making quality of wheat lines with allelic HMW-GS differing in number of cysteines. J. Cereal Sci. 2008, 48, 117–122. [Google Scholar] [CrossRef]
- Martin, J.M.; Frohberg, R.C.; Morris, C.F.; Talbert, L.E.; Giroux, M.J. Milling and bread baking traits associated with puroindoline sequence type in hard red spring wheat. Crop Sci. 2001, 41, 228–234. [Google Scholar] [CrossRef]
- Morris, C.F.; DeMacon, V.L.; Giroux, M.J. Wheat grain hardness among chromosome 5D homozygous recombinant substitution lines using different methods of measurement. Cereal Chem. 1999, 76, 249–254. [Google Scholar] [CrossRef]
- Morris, C.F.; Rose, S.P. Wheat. In Cereal Grain Quality; Springer: Berlin/Heidelberg, Germany, 1996; pp. 3–54. [Google Scholar]
- Morris, C.F. Puroindolines: The molecular genetic basis of wheat grain hardness. Plant Mol. Biol. 2002, 48, 633–647. [Google Scholar] [CrossRef]
- Bhave, M.; Morris, C.F. Molecular genetics of puroindolines and related genes: Allelic diversity in wheat and other grasses. Plant Mol. Biol. 2008, 66, 205–219. [Google Scholar] [CrossRef]
- Ma, X.; Sajjad, M.; Wang, J.; Yang, W.; Sun, J.; Li, X.; Zhang, A.; Liu, D. Diversity, distribution of Puroindoline genes and their effect on kernel hardness in a diverse panel of Chinese wheat germplasm. BMC Plant Biol. 2017, 17, 158. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Arora, S.; Singh, K.; Garg, M. Puroindoline allelic diversity in Indian wheat germplasm and identification of new allelic variants. Breed. Sci. 2015, 65, 319–326. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Sun, J.; Liu, D.; Yang, W.; Wang, D.; Tong, Y.; Zhang, A. Analysis of Pina and Pinb alleles in the micro-core collections of Chinese wheat germplasm by Ecotilling and identification of a novel Pinb allele. J. Cereal Sci. 2008, 48, 836–842. [Google Scholar] [CrossRef]
- Lillemo, M.; Chen, F.; Xia, X.; William, M.; Peña, R.J.; Trethowan, R.; He, Z. Puroindoline grain hardness alleles in CIMMYT bread wheat germplasm. J. Cereal Sci. 2006, 44, 86–92. [Google Scholar] [CrossRef]
- Ayala, M.; Guzmán, C.; Alvarez, J.B.; Peña, R.J. Characterization of genetic diversity of puroindoline genes in Mexican wheat landraces. Euphytica 2013, 190, 53–63. [Google Scholar] [CrossRef]
- Brown, A. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Rharrabti, Y.; Villegas, D.; Royo, C.; Martos-Núñez, V.; Del Moral, L.G. Durum wheat quality in Mediterranean environments: II. Influence of climatic variables and relationships between quality parameters. Field Crops Res. 2003, 80, 133–140. [Google Scholar] [CrossRef]
- Sehgal, D.; Autrique, E.; Singh, R.; Ellis, M.; Singh, S.; Dreisigacker, S. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Sukumaran, S.; Lopes, M.; Dreisigacker, S.; Reynolds, M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor. Appl. Genet. 2018, 131, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garcia, M.; Álvaro, F.; Martín-Sánchez, J.A.; Sillero, J.C.; Escribano, J.; Royo, C. Breeding effects on the genotype× environment interaction for yield of bread wheat grown in Spain during the 20th century. Field Crops Res. 2012, 126, 79–86. [Google Scholar] [CrossRef]
- Ruiz, M.; Giraldo, P.; Royo, C.; Villegas, D.; Aranzana, M.J.; Carrillo, J.M. Diversity and genetic structure of a collection of Spanish durum wheat landraces. Crop Sci. 2012, 52, 2262–2275. [Google Scholar] [CrossRef]
- Giraldo, P.; Royo, C.; González, M.; Carrillo, J.M.; Ruiz, M. Genetic diversity and association mapping for agromorphological and grain quality traits of a structured collection of durum wheat landraces including subsp. durum, turgidum and diccocon. PLoS ONE 2016, 11, e0166577. [Google Scholar] [CrossRef]
- Pascual, L.; Ruiz, M.; López-Fernández, M.; Pérez-Peña, H.; Benavente, E.; Vázquez, J.F.; Sansaloni, C.; Giraldo, P. Genomic analysis of Spanish wheat landraces reveals their variability and potential for breeding. BMC Genom. 2020, 21, 122. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, P.; Rodríguez-Quijano, M.; Simon, C.; Vázquez, J.F.; Carrillo, J.M. Allelic variation in HMW glutenins in Spanish wheat landraces and their relationship with bread quality. Span. J. Agric. Res. 2010, 8, 1012–1023. [Google Scholar] [CrossRef]
- Chacón, E.A.; Vázquez, F.J.; Giraldo, P.; Carrillo, J.M.; Benavente, E.; Rodríguez-Quijano, M. Allelic variation for prolamins in Spanish durum wheat landraces and its relationship with quality traits. Agronomy 2020, 10, 136. [Google Scholar] [CrossRef]
- Martos, V.; Royo, C.; Rharrabti, Y.; Del Moral, L.G. Using AFLPs to determine phylogenetic relationships and genetic erosion in durum wheat cultivars released in Italy and Spain throughout the 20th century. Field Crops Res. 2005, 91, 107–116. [Google Scholar] [CrossRef]
- Ayala, M.; Guzmán, C.; Peña, R.J.; Alvarez, J.B. Genetic diversity and molecular characterization of puroindoline genes (Pina-D1 and Pinb-D1) in bread wheat landraces from Andalusia (Southern Spain). J. Cereal Sci. 2016, 71, 61–65. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Appels, R.; Xia, X.C. Catalogue of gene symbols for wheat: 2013–2014. In Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan, 8–14 September 2013. [Google Scholar]
- García, A.G. Cultivos Herbáceos Extensivos; Mundi-Prensa Libros: Madrid, Spain, 1999; pp. 26–27. [Google Scholar]
- Gautier, M.; Aleman, M.; Guirao, A.; Marion, D.; Joudrier, P. Triticum aestivum puroindolines, two basic cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol. Biol. 1994, 25, 43–57. [Google Scholar] [CrossRef]
- Iftikhar, A.; Ali, I. Kernel softness in wheat is determined by starch granule bound Puroindoline proteins. J. Plant Biochem. Biotechnol. 2017, 26, 247–262. [Google Scholar] [CrossRef]
- Nadolska-Orczyk, A.; Gasparis, S.; Orczyk, W. The determinants of grain texture in cereals. J. Appl. Genet. 2009, 50, 185–197. [Google Scholar] [CrossRef]
- Giroux, M.J.; Morris, C.F. Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc. Natl. Acad. Sci USA 1998, 95, 6262–6266. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, H.; Li, X.; Dong, Z.; Zuo, A.; Shang, X.; Cui, D. Alveograph and Mixolab parameters associated with Puroindoline-D1 genes in Chinese winter wheats. J. Sci. Food Agric. 2013, 93, 2541–2548. [Google Scholar] [CrossRef]
- Eagles, H.A.; Cane, K.; Eastwood, R.F.; Hollamby, G.J.; Kuchel, H.; Martin, P.J.; Cornish, G.B. Contributions of glutenin and puroindoline genes to grain quality traits in southern Australian wheat breeding programs. Aust. J. Agric. Res. 2006, 57, 179–186. [Google Scholar] [CrossRef]
- Chang, C.; Zhang, H.; Xu, J.; Li, W.; Liu, G.; You, M.; Li, B. Identification of allelic variations of puroindoline genes controlling grain hardness in wheat using a modified denaturing PAGE. Euphytica 2006, 152, 225–234. [Google Scholar] [CrossRef]
- Przyborowski, M.; Gasparis, S.; Kała, M.; Orczyk, W.; Nadolska-Orczyk, A. The variability of puroindoline-encoding alleles and their influence on grain hardness in modern wheat cultivars cultivated in Poland, breeding lines and Polish old landraces (Triticum aestivum L.). Agronomy 2020, 10, 1075. [Google Scholar] [CrossRef]
- Lillemo, M.; Morris, C.F. A leucine to proline mutation in puroindoline b is frequently present in hard wheats from Northern Europe. Theor. Appl. Genet. 2000, 100, 1100–1107. [Google Scholar] [CrossRef]
- Huang, X.; Röder, M.S. Development of SNP assays for genotyping the Puroindoline b gene for grain hardness in wheat using pyrosequencing. J. Agric. Food Chem. 2005, 53, 2070–2075. [Google Scholar] [CrossRef]
- Rodríguez-Quijano, M.; Vázquez, J.F.; Carrillo, J.M. Variation of high molecular weight glutenin subunits in Spanish landraces of Triticum aestivum ssp. vulgare and ssp. spelta. J. Genet. Breed. 1990, 44, 121–126. [Google Scholar]
- Igrejas, G.; Carnide, V.; Guedes Pinto, H.; Branlard, G.; Gateau, I. Storage protein diversity within the old Portuguese bread wheat Barbela population [Triticum aestivum]. J. Genet. Breed. 1997. [Google Scholar]
- Fang, J.; Liu, Y.; Luo, J.; Wang, Y.; Shewry, P.R.; He, G. Allelic variation and genetic diversity of high molecular weight glutenin subunit in Chinese endemic wheats (Triticum aestivum L.). Euphytica 2009, 166, 177. [Google Scholar] [CrossRef]
- Li, Y.; Huang, C.; Sui, X.; Fan, Q.; Li, G.; Chu, X. Genetic variation of wheat glutenin subunits between landraces and varieties and their contributions to wheat quality improvement in China. Euphytica 2009, 169, 159. [Google Scholar] [CrossRef]
- Goel, S.; Yadav, M.; Singh, K.; Jaat, R.S.; Singh, N.K. Exploring diverse wheat germplasm for novel alleles in HMW-GS for bread quality improvement. J. Food Sci. Technol. 2018, 55, 3257–3262. [Google Scholar] [CrossRef]
- Dai, S.; Xu, D.; Yan, Y.; Wen, Z.; Zhang, J.; Chen, H.; Lu, Z.; Li, H.; Cong, H.; Wei, Y. Characterization of high-and low-molecular-weight glutenin subunits from Chinese Xinjiang wheat landraces and historical varieties. J. Food Sci. Technol. 2020, 1–13. [Google Scholar] [CrossRef]
- Maryami, Z.; Azimi, M.R.; Guzman, C.; Dreisigacker, S.; Najafian, G. Puroindoline (Pina-D1 and Pinb-D1) and waxy (Wx-1) genes in Iranian bread wheat (Triticum aestivum L.) landraces. Biotechnol. Biotechnol. Equip. 2020, 34, 1019–1027. [Google Scholar] [CrossRef]
- Caballero, L.; Martin, L.M.; Alvarez, J.B. Intra-and interpopulation diversity for HMW glutenin subunits in Spanish spelt wheat. Genet. Resour. Crop Evol. 2004, 51, 175–181. [Google Scholar] [CrossRef]
- Brites, C.; Bagulho, A.S.; Rodriguez-Quijano, M.; Carrillo, J.M. Effects of HMW glutenin subunits on some quality parameters of Portuguese landraces of Triticum aestivum ssp. vulgare. In Wheat Gluten, Proceedings of the 7th International Workshop Gluten 2000, Bristol, UK, 2–6 April 2000; Royal Society of Chemistry: London, UK, 2000; pp. 55–60. [Google Scholar]
- Payne, P.I.; Lawrence, G.J. Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1, and Glu-D1 which code for high-molecular-weight subunits of glutenin in hexaploid wheat. Cereal Res. Commun. 1983, 11, 29–35. [Google Scholar]
- Morgunov, A.I.; Pena, R.J.; Crossa, J.; Rajaram, S. Worldwide distribution of Glu-1 alleles in bread wheat. J. Genet. Breed. 1993, 47, 53–60. [Google Scholar]
- Tohver, M. High molecular weight (HMW) glutenin subunit composition of some Nordic and Middle European wheats. Genet. Resour. Crop Evol. 2007, 54, 67–81. [Google Scholar] [CrossRef]
- Morris, C.F.; Paszczynska, B.; Bettge, A.D.; King, G.E. A critical examination of the sodium dodecyl sulfate (SDS) sedimentation test for wheat meals. J. Sci. Food Agric. 2007, 87, 607–615. [Google Scholar] [CrossRef]
- Cornish, G.; Békés, F.; Eagles, H.; Payne, P. Prediction of dough properties for bread wheats. In Gliadin and Glutenin: The Unique Balance of Wheat Quality; American Association of Cereal Chemists International: Saint Paul, MN, USA, 2006. [Google Scholar]
- Espí, A.; Giraldo, P.; Rodriguez-Quijano, M.; Carrillo, J.M. A PCR-based method for discriminating between high molecular weight glutenin subunits Bx7 and Bx7* in Triticum aestivum L. Plant Breed. 2012, 131, 571–573. [Google Scholar] [CrossRef]
- Nucia, A.; Okoń, S.; Tomczyńska-Mleko, M. Characterization of HMW glutenin subunits in European spring common wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2019, 66, 579–588. [Google Scholar] [CrossRef]
- Brunori, A.; Galterio, G.; Zannettino, C.; Pogna, N.E. Bread-Making Quality indices in Triticum aestivum orogenies. Implications in breeding for better bread wheat. Plant Breed. 1989, 102, 222–231. [Google Scholar] [CrossRef]
- Joppa, L.R.; Du, C.; Hart, G.E.; Hareland, G.A. Mapping gene (s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 1997, 37, 1586–1589. [Google Scholar] [CrossRef]
- Prasad, M.; Varshney, R.K.; Kumar, A.; Balyan, H.S.; Sharma, P.C.; Edwards, K.J.; Dhaliwal, H.S.; Roy, J.K.; Gupta, P.K. A microsatellite marker associated with a QTL for grain protein content on chromosome arm 2DL of bread wheat. Theor. Appl. Genet. 1999, 99, 341–345. [Google Scholar] [CrossRef]
- Perretant, M.R.; Cadalen, T.; Charmet, G.; Sourdille, P.; Nicolas, P.; Boeuf, C.; Tixier, M.H.; Branlard, G.; Bernard, S. QTL analysis of bread-making quality in wheat using a doubled haploid population. Theor. Appl. Genet. 2000, 100, 1167–1175. [Google Scholar] [CrossRef]
- Zanetti, S.; Winzeler, M.; Feuillet, C.; Keller, B.; Messmer, M. Genetic analysis of bread-making quality in wheat and spelt. Plant Breed. 2001, 120, 13–19. [Google Scholar] [CrossRef]
- Bhullar, S.S.; Jenner, C.F. Differential responses to high temperatures of starch and nitrogen accumulation in the grain of four cultivars of wheat. Funct. Plant Biol. 1985, 12, 363–375. [Google Scholar] [CrossRef]
- Wardlaw, I.F.; Wrigley, C.W. Heat tolerance in temperate cereals: An overview. Funct. Plant Biol. 1994, 21, 695–703. [Google Scholar] [CrossRef]
- Daniel, C.; Triboi, E. Effects of temperature and nitrogen nutrition on the grain composition of winter wheat: Effects on gliadin content and composition. J. Cereal Sci. 2000, 32, 45–56. [Google Scholar] [CrossRef]
- López-Bellido, L.; Fuentes, M.; Castillo, J.E.; López-Garrido, F.J. Effects of tillage, crop rotation and nitrogen fertilization on wheat-grain quality grown under rainfed Mediterranean conditions. Field Crops Res. 1998, 57, 265–276. [Google Scholar] [CrossRef]
- Gürsoy, S.; Sessiz, A.; Malhi, S.S. Short-term effects of tillage and residue management following cotton on grain yield and quality of wheat. Field Crops Res. 2010, 119, 260–268. [Google Scholar] [CrossRef]
- Dotlačil, L.; Hermuth, J.; Stehno, Z.; Dvořáček, V.; Bradová, J.; Leišová, L. How can wheat landraces contribute to present breeding. Czech J. Genet. Plant Breed. 2010, 46, S70–S74. [Google Scholar] [CrossRef]
- Ruiz, M.; Zambrana, E.; Fite, R.; Sole, A.; Tenorio, J.L.; Benavente, E. Yield and quality performance of traditional and improved bread and durum wheat varieties under two conservation tillage systems. Sustainability 2019, 11, 4522. [Google Scholar] [CrossRef]
- Zeven, A.C. Landraces: A review of definitions and classifications. Euphytica 1998, 104, 127–139. [Google Scholar] [CrossRef]
- Kyzeridis, N.; Biesantz, A.; Limberg, P. Comparative trials with durum-wheat landraces and cultivars in different ecological environments in the Mediterranean region. J. Agron. Crop. Sci. 1995, 174, 133–144. [Google Scholar] [CrossRef]
- Carranza-Gallego, G.; Guzmán, G.I.; Garcia-Ruiz, R.; Gonzalez de Molina, M.; Aguilera, E. Addressing the role of landraces in the sustainability of Mediterranean agroecosystems. Sustainability 2019, 11, 6029. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P.; Manes, Y.; Singh, R.P.; Crossa, J.; Braun, H.J. Genetic yield gains and changes in associated traits of CIMMYT spring bread wheat in a “historic” set representing 30 years of breeding. Crop Sci. 2012, 52, 1123–1131. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Campbell, K.G.; Bergman, C.J.; Gualberto, D.G.; Anderson, J.A.; Giroux, M.J.; Hareland, G.; Fulcher, R.G.; Sorrells, M.E.; Finney, P.L. Quantitative trait loci associated with kernel traits in a soft× hard wheat cross. Crop Sci. 1999, 39, 1184–1195. [Google Scholar] [CrossRef]
- Royo, C.; Abaza, M.; Blanco, R.; del Moral, L.F.G. Triticale grain growth and morphometry as affected by drought stress, late sowing and simulated drought stress. Funct. Plant Biol. 2000, 27, 1051–1059. [Google Scholar] [CrossRef]
- Diacono, M.; Castrignanò, A.; Troccoli, A.; De Benedetto, D.; Basso, B.; Rubino, P. Spatial and temporal variability of wheat grain yield and quality in a Mediterranean environment: A multivariate geostatistical approach. Field Crops Res. 2012, 131, 49–62. [Google Scholar] [CrossRef]
- Austin, R.B.; Bingham, J.; Blackwell, R.D.; Evans, L.T.; Ford, M.A.; Morgan, C.L.; Taylor, M. Genetic improvements in winter wheat yields since 1900 and associated physiological changes. J. Agric. Sci. 1980, 94, 675–689. [Google Scholar] [CrossRef]
- Cerere Project. Available online: http://cere2020.eu/ptoject/ (accessed on 19 February 2021).
- Pascual, L.; Fernández, M.; Aparicio, N.; López-Fernández, M.; Fité, R.; Giraldo, P.; Ruiz, M. Development of a multipurpose core collection of bread wheat based on high-throughput genotyping data. Agronomy 2020, 10, 534. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Ribeiro, M.; Rodríguez-Quijano, M.; Giraldo, P.; Pinto, L.; Vázquez, J.F.; Carrillo, J.M.; Igrejas, G. Effect of allelic variation at glutenin and puroindoline loci on bread-making quality: Favorable combinations occur in less toxic varieties of wheat for celiac patients. Eur. Food Res. Technol. 2017, 243, 743–752. [Google Scholar] [CrossRef]
- Geneious; Biomatters Ltd.: Auckland, New Zealand, 2017.
- Singh, N.K.; Shepherd, K.W.; Cornish, G.B. A simplified SDS-page procedure for separating LMW subunits of glutenin. J Cereal Sci. 1991, 14, 203–208. [Google Scholar] [CrossRef]
- Payne, P.I.; Law, C.N.; Mudd, E.E. Control by homoeologous group 1 chromosomes of the high-molecular-weight subunits of glutenin, a major protein of wheat endosperm. Theor. Appl. Genet. 1980, 58, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Ragupathy, R.; Naeem, H.A.; Reimer, E.; Lukow, O.M.; Sapirstein, H.D.; Cloutier, S. Evolutionary origin of the segmental duplication encompassing the wheat GLU-B1 locus encoding the overexpressed Bx7 (Bx7 OE) high molecular weight glutenin subunit. Theor. Appl. Genet. 2008, 116, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Dick, J.W.; Quick, J.S. A Modified Screening Test for Rapid Estimation of Gluten Strength in Early-Generation Durum Wheat Breeding Lines. Cereal Chem. 1983, 60, 315–318. [Google Scholar]
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]



| Landraces | Reference Set | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pinA-D1 allele | a | a | a | a | a | b | a | a | a | b |
| pinB-D1 allele | a | b | d | new1 (ad) | new2 | a | a | b | d | a |
| Endosperm texture | soft | hard | hard | hard | n.d. | hard | soft | hard | hard | hard |
| N | 150 | 5 | 5 | 15 | 3 | 8 | 8 | 3 | 3 | 3 |
| % | 80.65 | 2.69 | 2.69 | 8.06 | 1.61 | 4.30 | 47.06 | 17.65 | 17.65 | 17.65 |
| Locus | Allele | HMW-GS | N | % |
|---|---|---|---|---|
| Glu-A1 | a | 1 | 31 | 16.40 |
| b | 2* | 101 | 53.44 | |
| c | null | 22 | 11.64 | |
| y | 2·· | 34 | 17.99 | |
| New1 | - | 1 | 0.53 | |
| Glu-B1 | a | 7 | 6 | 3.17 |
| d | 6+8 | 11 | 5.82 | |
| e | 20x+20y | 89 | 47.09 | |
| f | 13+16 | 30 | 15.87 | |
| aq | 32+33 | 6 | 3.17 | |
| 7*+9 | 7*+9 | 7 | 3.70 | |
| al | 7oe+8 | 3 | 1.59 | |
| am | 18 | 4 | 2.12 | |
| i | 17+18 | 3 | 1.59 | |
| u | 7*+8 | 26 | 13.76 | |
| New2 | - | 1 | 0.53 | |
| New3 | - | 1 | 0.53 | |
| New4 | - | 1 | 0.53 | |
| New5 | - | 1 | 0.53 | |
| Glu-D1 | a | 2+12 | 127 | 67.20 |
| c | 4+12 | 44 | 23.28 | |
| d | 5+10 | 8 | 4.23 | |
| h | 5+12 | 1 | 0.53 | |
| j | 2+12* | 2 | 1.06 | |
| l | 12 | 3 | 1.59 | |
| u | 2+10’ | 1 | 0.53 | |
| New6 | - | 2 | 1.06 | |
| New7 | - | 1 | 0.53 |
| Mean | Min | Max | sd | p-Value Ld/Ref # | |||
|---|---|---|---|---|---|---|---|
| GPC (%) | 2016/2017 | Ld | 17.59 | 13.97 | 20.96 | 1.24 | - |
| Ref | - | - | - | - | |||
| 2017/2018 | Ld | 11.51 | 8.97 | 15.44 | 1.16 | *** | |
| Ref | 9.28 | 7.84 | 10.86 | 0.76 | |||
| 2018/2019 | Ld | 17.05 | 13.41 | 20.33 | 1.19 | *** | |
| Ref | 15.30 | 12.96 | 16.96 | 1.15 | |||
| 2019/2020 | Ld | 14.47 | 11.23 | 18.78 | 1.60 | *** | |
| Ref | 13.23 | 11.77 | 15.05 | 0.95 | |||
| SDSS (mm) | 2016/2017 | Ld | 55.45 | 23.50 | 115.50 | 23.28 | - |
| Ref | - | - | - | - | |||
| 2017/2018 | Ld | 37.90 | 18 | 111 | 14.78 | *** | |
| Ref | 52.28 | 34 | 69 | 10.44 | |||
| 2018/2019 | Ld | 41.51 | 10 | 93.50 | 17.02 | *** | |
| Ref | 87.89 | 58.5 | 112 | 16.09 | |||
| 2019/2020 | Ld | 58.14 | 18.50 | 107.50 | 20.39 | *** | |
| Ref | 83.22 | 66 | 97.5 | 10.89 |
| Mean | Min | Max | sd | p-Value Ld/Ref # | |||
|---|---|---|---|---|---|---|---|
| TKW (g) | 2016/2017 | Ld | 26.20 | 14.98 | 39.14 | 3.99 | - |
| Ref | - | - | - | - | |||
| 2017/2018 | Ld | 40.24 | 27.03 | 52.03 | 5.01 | *** | |
| Ref | 35.11 | 25.73 | 43.71 | 5.13 | |||
| 2018/2019 | Ld | 27.19 | 16.59 | 37.92 | 3.84 | ns | |
| Ref | 27.42 | 22.37 | 33.31 | 3.09 | |||
| 2019/2020 | Ld | 31.92 | 20.00 | 40.36 | 4.02 | *** | |
| Ref | 29.84 | 22.08 | 35.60 | 3.87 | |||
| GN | 2016/2017 | Ld | - | - | - | - | - |
| Ref | - | - | - | - | |||
| 2017/2018 | Ld | 24.67 | 10.25 | 44.75 | 6.31 | *** | |
| Ref | 36.36 | 24.5 | 52.25 | 7.04 | |||
| 2018/2019 | Ld | 19.91 | 5.5 | 49.00 | 7.61 | *** | |
| Ref | 40.16 | 18 | 57.75 | 8.99 | |||
| 2019/2020 | Ld | 31.76 | 17.00 | 57.80 | 7.24 | *** | |
| Ref | 46.46 | 29.4 | 62.8 | 8.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Fernández, M.; Pascual, L.; Faci, I.; Fernández, M.; Ruiz, M.; Benavente, E.; Giraldo, P. Exploring the End-Use Quality Potential of a Collection of Spanish Bread Wheat Landraces. Plants 2021, 10, 620. https://doi.org/10.3390/plants10040620
López-Fernández M, Pascual L, Faci I, Fernández M, Ruiz M, Benavente E, Giraldo P. Exploring the End-Use Quality Potential of a Collection of Spanish Bread Wheat Landraces. Plants. 2021; 10(4):620. https://doi.org/10.3390/plants10040620
Chicago/Turabian StyleLópez-Fernández, Matilde, Laura Pascual, Isabel Faci, Mario Fernández, Magdalena Ruiz, Elena Benavente, and Patricia Giraldo. 2021. "Exploring the End-Use Quality Potential of a Collection of Spanish Bread Wheat Landraces" Plants 10, no. 4: 620. https://doi.org/10.3390/plants10040620
APA StyleLópez-Fernández, M., Pascual, L., Faci, I., Fernández, M., Ruiz, M., Benavente, E., & Giraldo, P. (2021). Exploring the End-Use Quality Potential of a Collection of Spanish Bread Wheat Landraces. Plants, 10(4), 620. https://doi.org/10.3390/plants10040620







