Prokaryotic Expression of Phosphoenolpyruvate Carboxylase Fragments from Peanut and Analysis of Osmotic Stress Tolerance of Recombinant Strains
Abstract
1. Introduction
2. Results
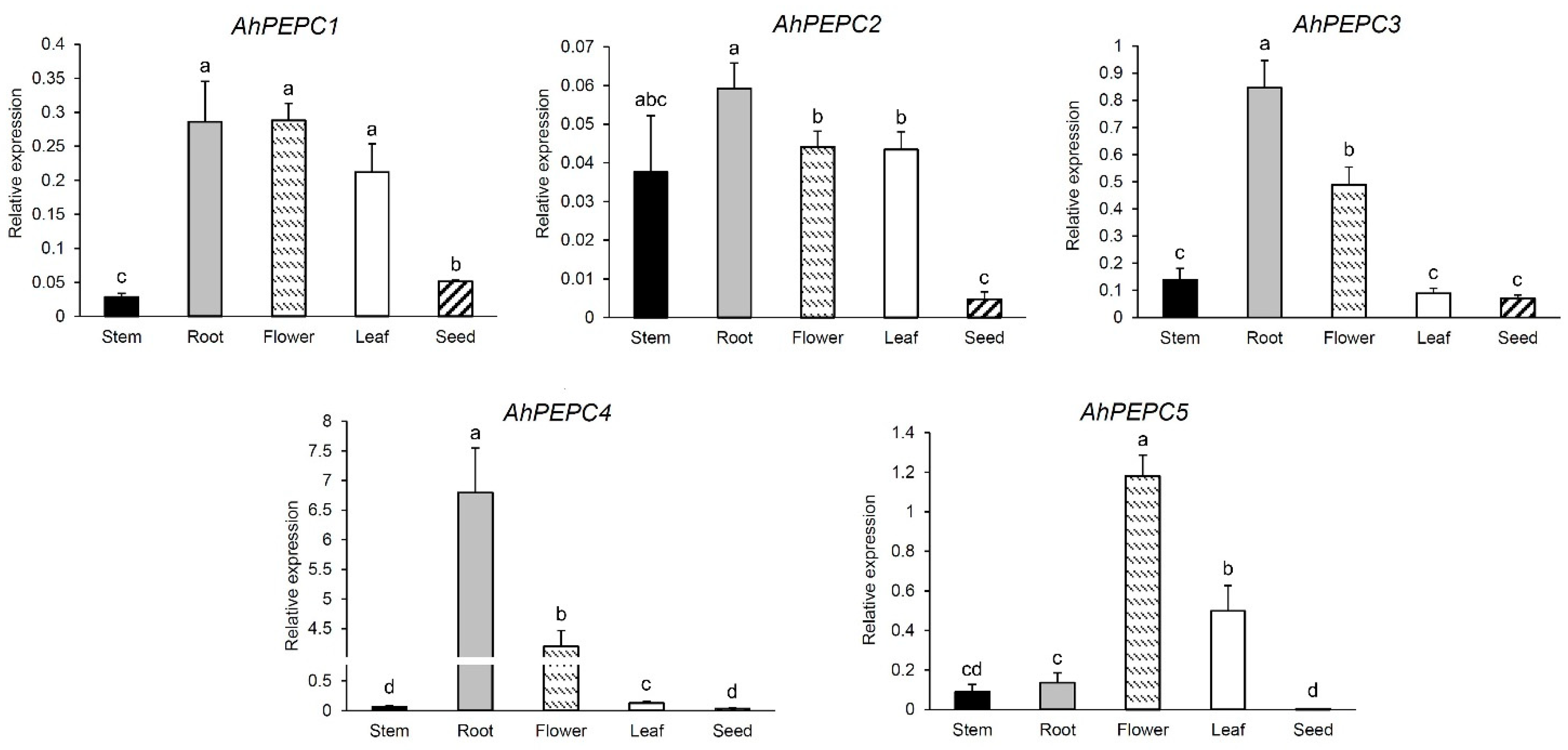
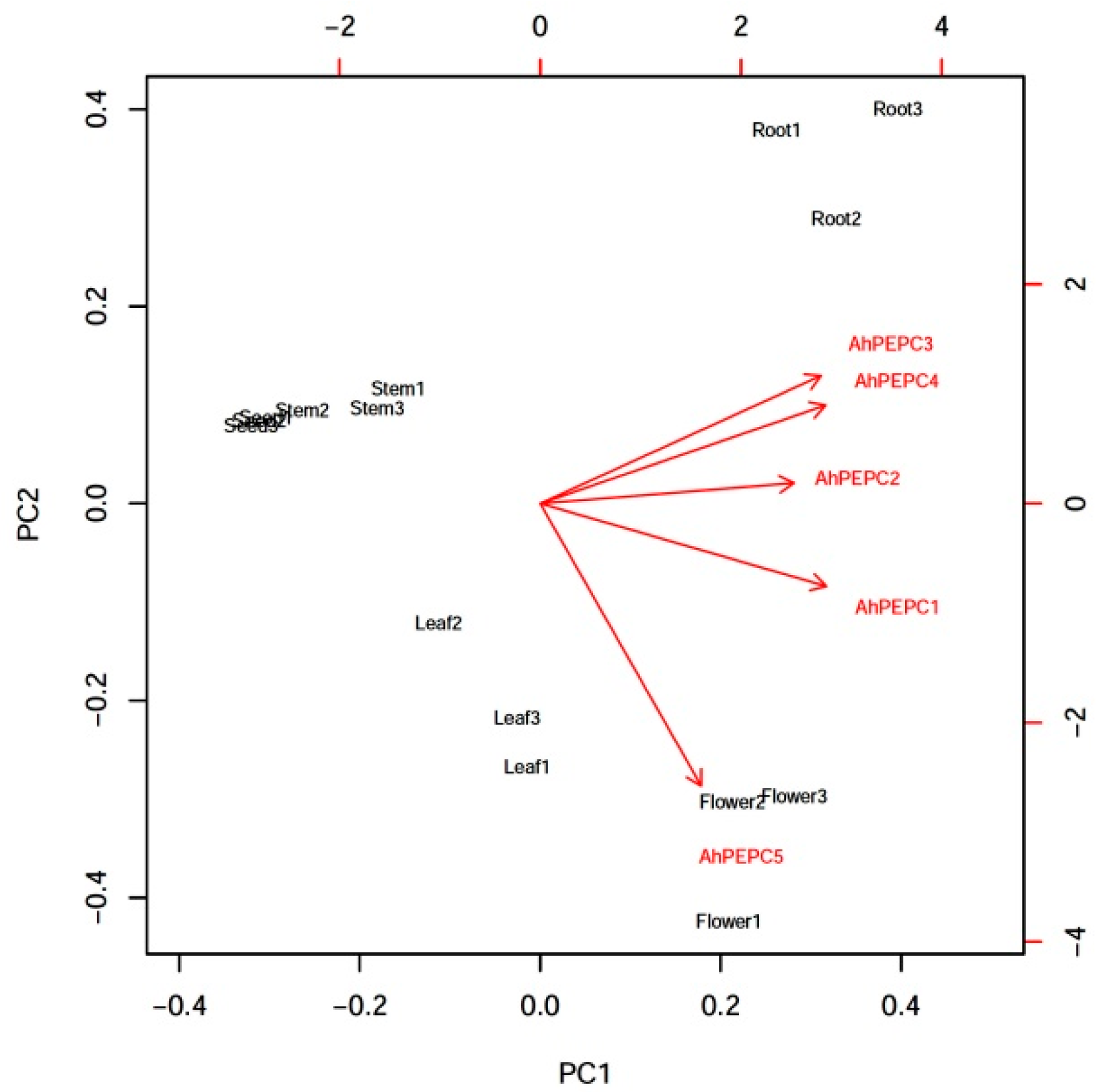
2.1. Expression Patterns of AhPEPCs in Peanut
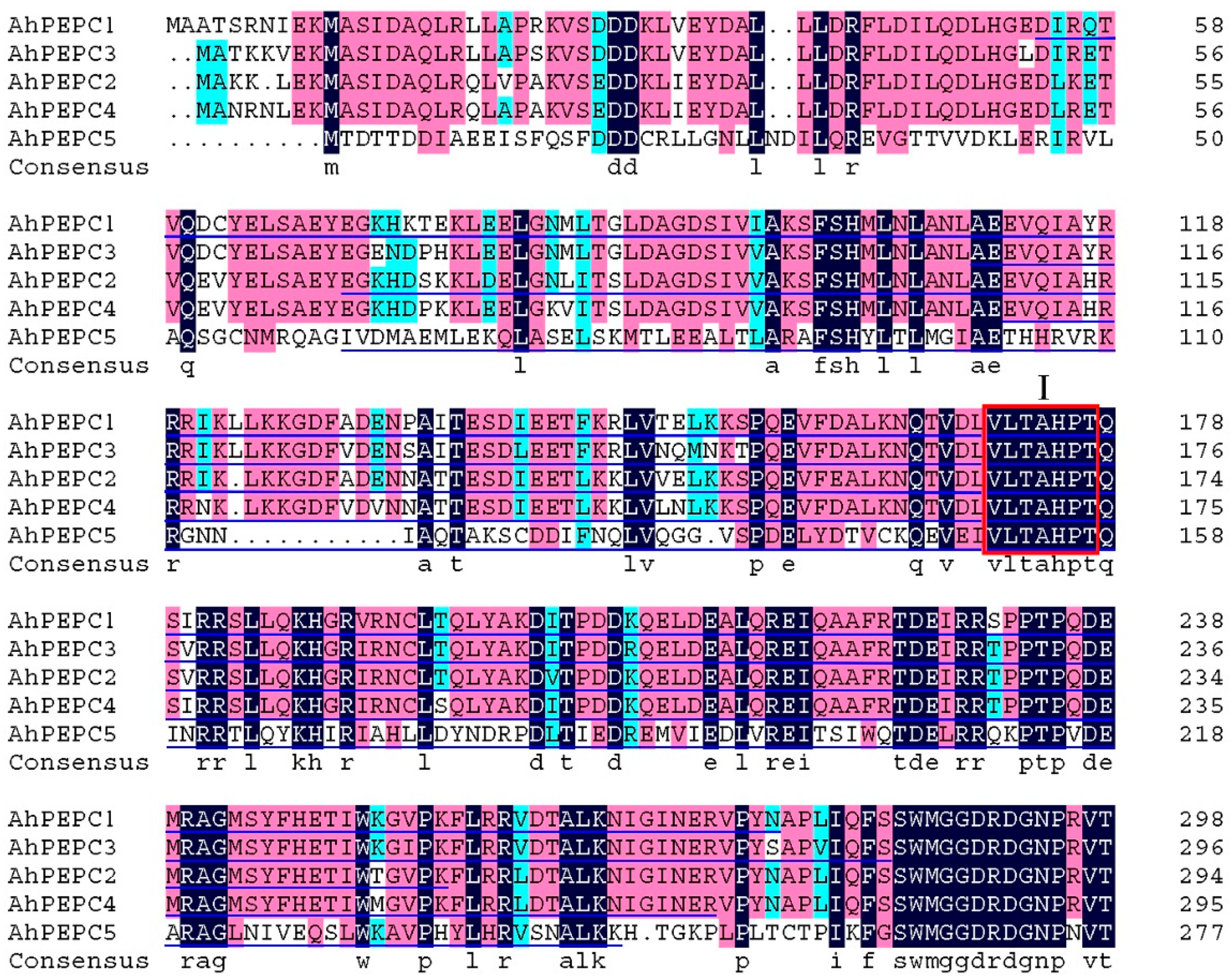
2.2. Cloning of AhPEPC cDNA Fragments and Construction of Prokaryotic Expression Vectors
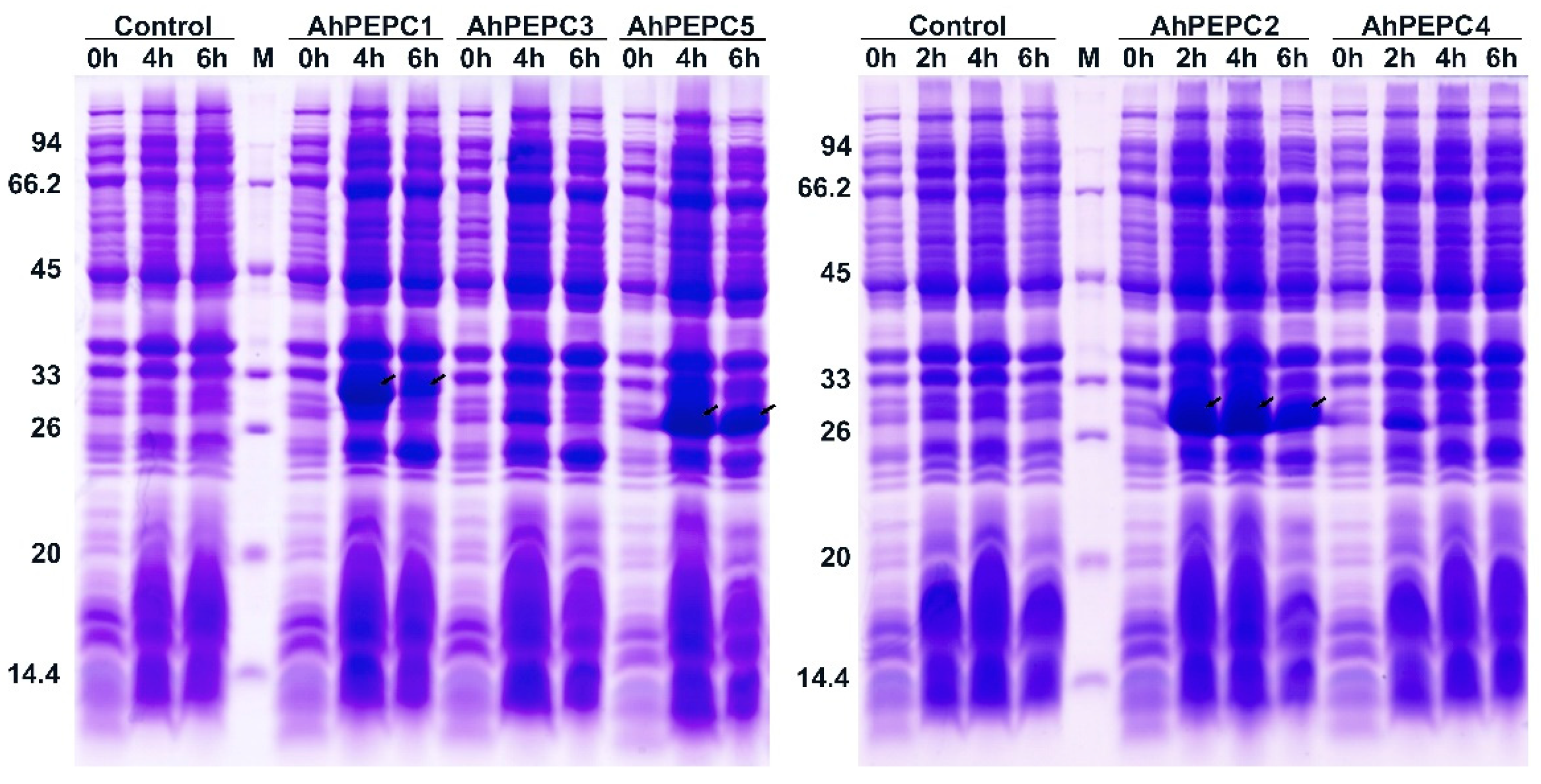
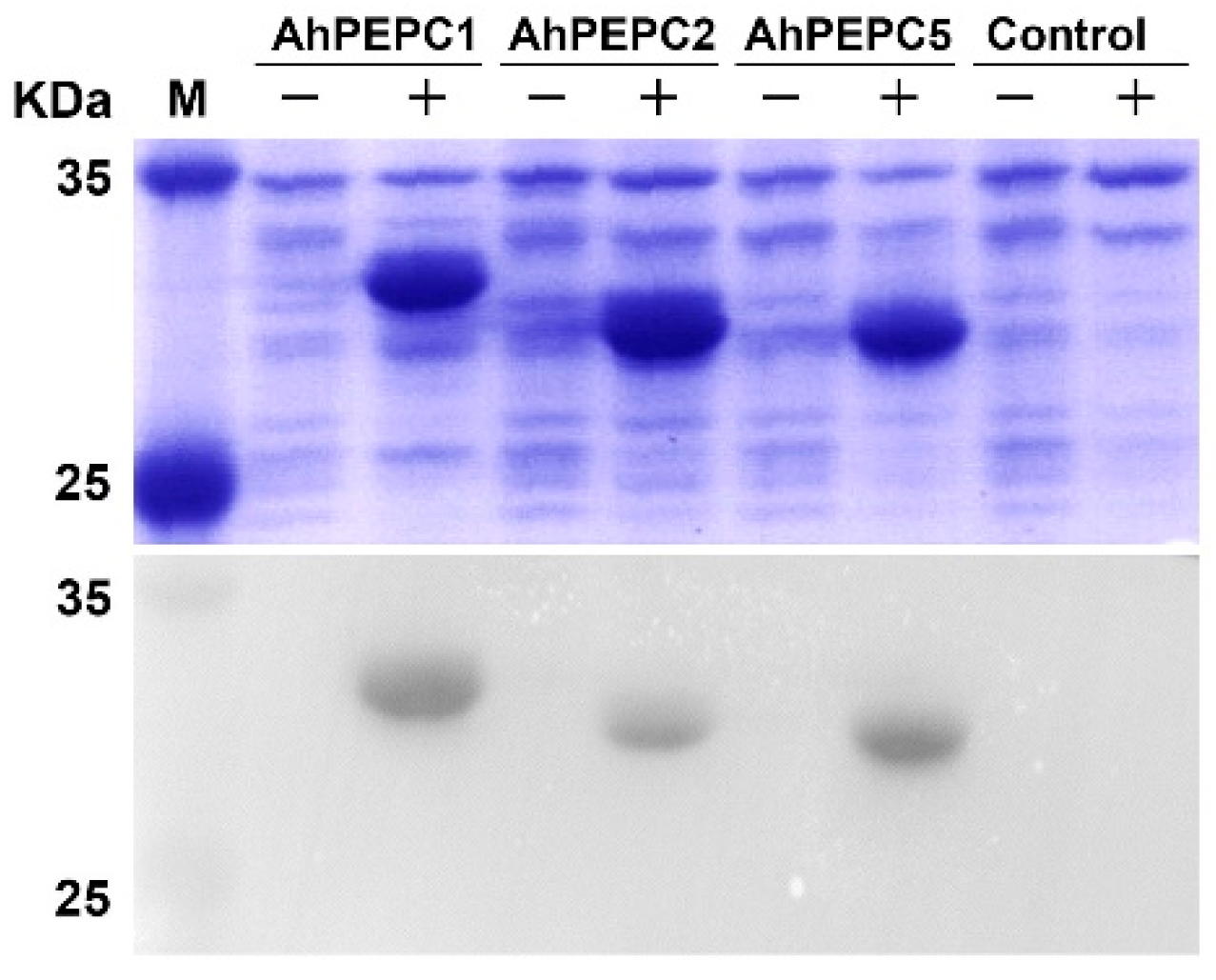
2.3. Expression of AhPEPC Fragments in E. coli
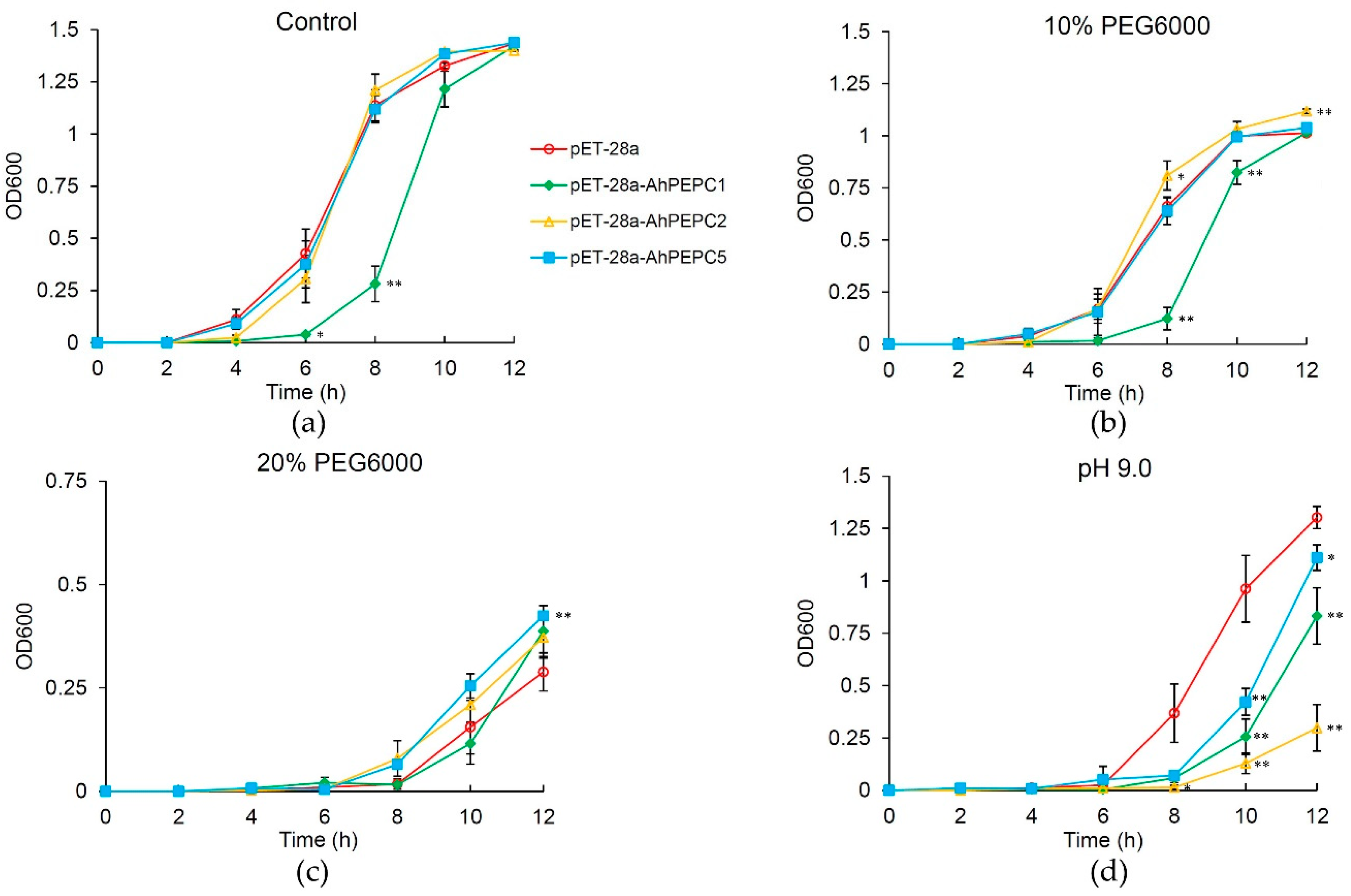
2.4. Effect of Osmotic Stress on the Growth of Recombinant Strains
2.5. Acid/base Tolerance Test of Recombinant Strains
3. Discussion
4. Materials and Methods
4.1. Plant Materials, E. coli Strains, and Plasmids
4.2. AhPEPC Fragments Cloning and Construction of Expression Vectors
4.3. Quantitative Gene Expression Analysis
4.4. Prokaryotic Expression and Western Blot Analysis
4.5. Growth and Stress Tolerance Assay
4.6. Statistical Analysis and PCA Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, M.; Tang, Y.; Zhang, J.; Yang, M.; Xu, Y. RNA interference-based suppression of phosphoenolpyruvate carboxylase results in susceptibility of rapeseed to osmotic stress. J. Integr. Plant Biol. 2010, 52, 585–592. [Google Scholar] [CrossRef]
- O’ Leary, B.; Park, J.; Plaxton, W.C. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): Recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 2011, 436, 15–34. [Google Scholar] [CrossRef]
- Sánchez, R.; Cejudo, F.J. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 2003, 132, 949–957. [Google Scholar] [CrossRef]
- Svensson, P.; Bläsing, O.E.; Westhoff, P. Evolution of C4 phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys. 2003, 414, 180–188. [Google Scholar] [CrossRef]
- Izui, K.; Matsumura, H.; Furumoto, T.; Kai, Y. Phosphoenolpyruvate carboxylase: A new era of structural biology. Annu. Rev. Plant Biol. 2004, 55, 69–84. [Google Scholar] [CrossRef] [PubMed]
- O’ Leary, B.; Fedosejevs, E.T.; Hill, A.T.; Bettridge, J.; Park, J.; Rao, S.K.; Leach, C.A.; Plaxton, W.C. Tissue-specific expression and post-translational modifications of plant- and bacterial-type phosphoenolpyruvate carboxylase isozymes of the castor oil plant, Ricinus communis L. J. Exp. Bot. 2011, 62, 5485–5495. [Google Scholar] [CrossRef]
- Igawa, T.; Fujiwara, M.; Tanaka, I.; Fukao, Y.; Yanagawa, Y. Characterization of bacterial-type phosphoenolpyruvate carboxylase expressed in male gametophyte of higher plants. BMC Plant Biol. 2010, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Waseem, M.; Ahmad, F. The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene 2019, 690, 11–20. [Google Scholar] [CrossRef]
- Miyao, M.; Fukayama, H. Metabolic consequences of overproduction of phosphoenolpyruvate carboxylase in C3 plants. Arch. Biochem. Biophys. 2003, 414, 197–203. [Google Scholar] [CrossRef]
- Sánchez, R.; Flores, A.; Cejudo, F.J. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 2006, 223, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Doubnerová Hysková, V.; Miedzińska, L.; Dobrá, J.; Vankova, R.; Ryšlavá, H. Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J. Plant Physiol. 2014, 171, 19–25. [Google Scholar] [CrossRef]
- Wang, N.; Zhong, X.; Cong, Y.; Wang, T.; Yang, S.; Li, Y.; Gai, J. Genome-wide analysis of phosphoenolpyruvate carboxylase gene family and their response to abiotic stresses in soybean. Sci. Rep. 2016, 6, 38448. [Google Scholar] [CrossRef]
- Pan, L.; Zhang, J.; Chen, N.; Chen, M.; Wang, M.; Wang, T.; Chi, X.; Yuan, M.; Wan, Y.; Yu, S.; et al. Molecular Characterization and Expression Profiling of the Phosphoenolpyruvate Carboxylase Genes in Peanut (Arachis hypogaea L.). J. Plant Physiol. 2017, 64, 576–587. [Google Scholar] [CrossRef]
- Liu, H.; Hong, Y.; Lu, Q.; Li, H.; Gu, J.; Ren, L.; Deng, L.; Zhou, B.; Chen, X.; Liang, X. Integrated Analysis of Comparative Lipidomics and Proteomics Reveals the Dynamic Changes of Lipid Molecular Species in High-Oleic Acid Peanut Seed. J. Agric. Food Chem. 2020, 68, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Pan, L.; Yang, Q.; Chen, M.; Zhang, H. Identification and Expression Analysis of the Phosphoenolpyruvate Carboxylase Gene Family in Peanut (Arachis hypogaea L.). Agric. Sci. 2010, 9, 477–487. [Google Scholar] [CrossRef]
- Pan, L.; Yang, Q.; Chi, X.; Chen, M.; Yang, Z.; Chen, N.; Wang, T.; Wang, M.; He, Y.; Yu, S. Functional analysis of the phosphoenolpyruvate carboxylase on the lipid accumulation of peanut (Arachis hypogaea L.) seeds. J. Integr. Agric. 2013, 12, 36–44. [Google Scholar] [CrossRef]
- Singh, J.; Reddy, G.M.; Agarwal, A.; Chandrasekhar, K.; Sopory, S.K.; Reddy, M.K.; Kaul, T. Molecular and structural analysis of C4-specific PEPC isoform from Pennisetum glaucum plays a role in stress adaptation. Gene 2012, 500, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Wang, L.; Lan, H. Cloning of PEPC-1 from a C4 halophyte Suaeda aralocaspica without Kranz anatomy and its recombinant enzymatic activity in responses to abiotic stresses. Enzyme Microb. Tech. 2016, 83, 57–67. [Google Scholar] [CrossRef]
- Yamamoto, N.; Sasou, A.; Saito, Y.; Sugimoto, T.; Masumura, T. Protein and gene expression characteristics of a rice phosphoenolpyruvate carboxylase Osppc3; its unique role for seed cell maturation. J. Cereal Sci. 2015, 64, 100–108. [Google Scholar] [CrossRef]
- Yamamoto, N.; Kubota, T.; Masumura, T.; Shiraishi, N.; Tanaka, K.; Sugimoto, T.; Oji, Y. Molecular cloning, gene expression and functional expression of a phosphoenolpyruvate carboxylase Osppc1 in developing rice seeds: Implication of involvement in nitrogen accumulation. Seed Sci. Res. 2014, 24, 23–36. [Google Scholar] [CrossRef]
- Yamamoto, N.; Masumura, T.; Yano, K.; Sugimoto, T. Pattern analysis suggests that phosphoenolpyruvate carboxylase in maturing soybean seeds promotes the accumulation of protein. Biosci. Biotechnol. Biochem. 2019, 83, 2238–2243. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Sugimoto, T.; Takano, T.; Sasou, A.; Morita, S.; Yano, K.; Masumura, T. The plant-type phosphoenolpyruvate carboxylase Gmppc2 is developmentally induced in immature soy seeds at the late maturation stage: A potential protein biomarker for seed chemical composition. Biosci. Biotechnol. Biochem. 2020, 84, 552–562. [Google Scholar] [CrossRef]
- Yamamoto, N.; Takano, T.; Masumura, T.; Sasou, A.; Morita, S.; Sugimoto, T.; Yano, K. Rapidly evolving phosphoenolpyruvate carboxylase Gmppc1 and Gmppc7 are highly expressed in the external seed coat of immature soybean seeds. Gene 2020, 762, 145015. [Google Scholar] [CrossRef]
- Andre, C.; Froehlich, J.E.; Moll, M.R.; Benning, C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2006–2022. [Google Scholar] [CrossRef] [PubMed]
- Junker, B.H.; Lonien, J.; Heady, L.E.; Rogers, A.; Schwender, J. Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 2007, 68, 2232–2242. [Google Scholar] [CrossRef]
- González, M.C.; Sánchez, R.; Cejudo, F.J. Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 2003, 216, 985–992. [Google Scholar] [CrossRef]
- García-Mauriño, S.; Monreal, J.A.; Alvarez, R.; Vidal, J.; Echevarríaa, C. Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: Independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 2003, 216, 648–655. [Google Scholar] [CrossRef]
- Carmo-Silva, A.E.; Bernardes da Silva, A.; Keys, A.J.; Parry, M.A.J.; Arrabaça, M.C. The activities of PEP carboxylase and the C4 acid decarboxylases are little changed by drought stress in three C4 grasses of different subtypes. Photosynth. Res. 2008, 97, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Xu, W.; Hu, L.; Wang, H.; Qi, X.; Fang, Y.; Hua, X. Drought tolerance and proteomics studies of transgenic wheat containing the maize C4 phosphoenolpyruvate carboxylase (PEPC) gene. Protoplasma 2016, 253, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Shi, D.; Jia, X.; Mi, H.; Huang, X.; He, P. Recombinant expression and functional analysis of a Chlamydomonas reinhardtii bacterial-type phosphoenolpyruvate carboxylase gene fragment. Biotechnol. Lett. 2014, 36, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Vidal, J.; Chollet, R.; Gadal, P.; Crétin, C. Phosphoenolpyruvate carboxylase: Structure, regulation and evolution. Plant Sci. 1994, 99, 111–124. [Google Scholar] [CrossRef]
- Terada, K.; Izui, K. Site-directed mutagenesis of the conserved histidine residue of phosphoenolpyruvate carboxylase. European Journal of Biochemistry. Eur. J. Biochem. 1991, 202, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Matsumura, H.; Inoue, T.; Terada, T.; Nagara, Y.; Yoshinaga, T.; Kihara, A.; Tsumura, K.; Izui, K. Three-dimensional structure of phosphoenolpyruvate carboxylase: A proposed mechanism for allosteric inhibition. Proc. Natl. Acad. Sci. USA 1999, 96, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Kai, Y.; Matsumura, H.; Izui, K. Phosphoenolpyruvate carboxylase: Three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 2003, 414, 170–179. [Google Scholar] [CrossRef]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef]
- Doubnerová, V.; Ryšlavá, H. What can enzymes of C4 photosynthesis do for C3 plants under stress? Plant Sci. 2011, 180, 575–583. [Google Scholar] [CrossRef]
- Park, S.; Lee, W.; Kim, H.; Pack, S.P.; Lee, J. Characterization of Phosphoenolpyruvate Carboxylase from Oceanimonas smirnovii in Escherichia coli. Appl. Biochem. Biotechnol. 2015, 177, 217–225. [Google Scholar] [CrossRef]
- López-Millán, A.F.; Morales, F.; Andaluz, S.; Gogorcena, Y.; Abadía, A.; De Las Rivas, J.; Abadía, J. Responses of sugar beet roots to iron deficiency. Changes in carbon assimilation and oxygen use. Plant Physiol. 2000, 124, 885–898. [Google Scholar] [CrossRef]
- Sakano, K. Revision of biochemical pH-stat: Involvement of alternative pathway metabolisms. Plant. Cell Physiol. 1998, 39, 467–473. [Google Scholar] [CrossRef]
- Wen, S.; Liu, H.; Li, X.; Chen, X.; Hong, Y.; Li, H.; Lu, Q.; Liang, X. TALEN-mediated targeted mutagenesis of fatty acid desaturase 2 (FAD2) in peanut (Arachis hypogaea L.) promotes the accumulation of oleic acid. Plant Mol. Biol. 2018, 97, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, J.; Feng, L.; Hong, Y.; Liu, Q.; Huang, X.; Li, Y. Prokaryotic Expression of Phosphoenolpyruvate Carboxylase Fragments from Peanut and Analysis of Osmotic Stress Tolerance of Recombinant Strains. Plants 2021, 10, 365. https://doi.org/10.3390/plants10020365
Tu J, Feng L, Hong Y, Liu Q, Huang X, Li Y. Prokaryotic Expression of Phosphoenolpyruvate Carboxylase Fragments from Peanut and Analysis of Osmotic Stress Tolerance of Recombinant Strains. Plants. 2021; 10(2):365. https://doi.org/10.3390/plants10020365
Chicago/Turabian StyleTu, Jiaqi, Lanlan Feng, Yanbin Hong, Qiuyun Liu, Xia Huang, and Yin Li. 2021. "Prokaryotic Expression of Phosphoenolpyruvate Carboxylase Fragments from Peanut and Analysis of Osmotic Stress Tolerance of Recombinant Strains" Plants 10, no. 2: 365. https://doi.org/10.3390/plants10020365
APA StyleTu, J., Feng, L., Hong, Y., Liu, Q., Huang, X., & Li, Y. (2021). Prokaryotic Expression of Phosphoenolpyruvate Carboxylase Fragments from Peanut and Analysis of Osmotic Stress Tolerance of Recombinant Strains. Plants, 10(2), 365. https://doi.org/10.3390/plants10020365






