NAC and MYB Families and Lignin Biosynthesis-Related Members Identification and Expression Analysis in Melilotus albus
Abstract
:1. Introduction
2. Results
2.1. Identification of NAC and MYB Genes
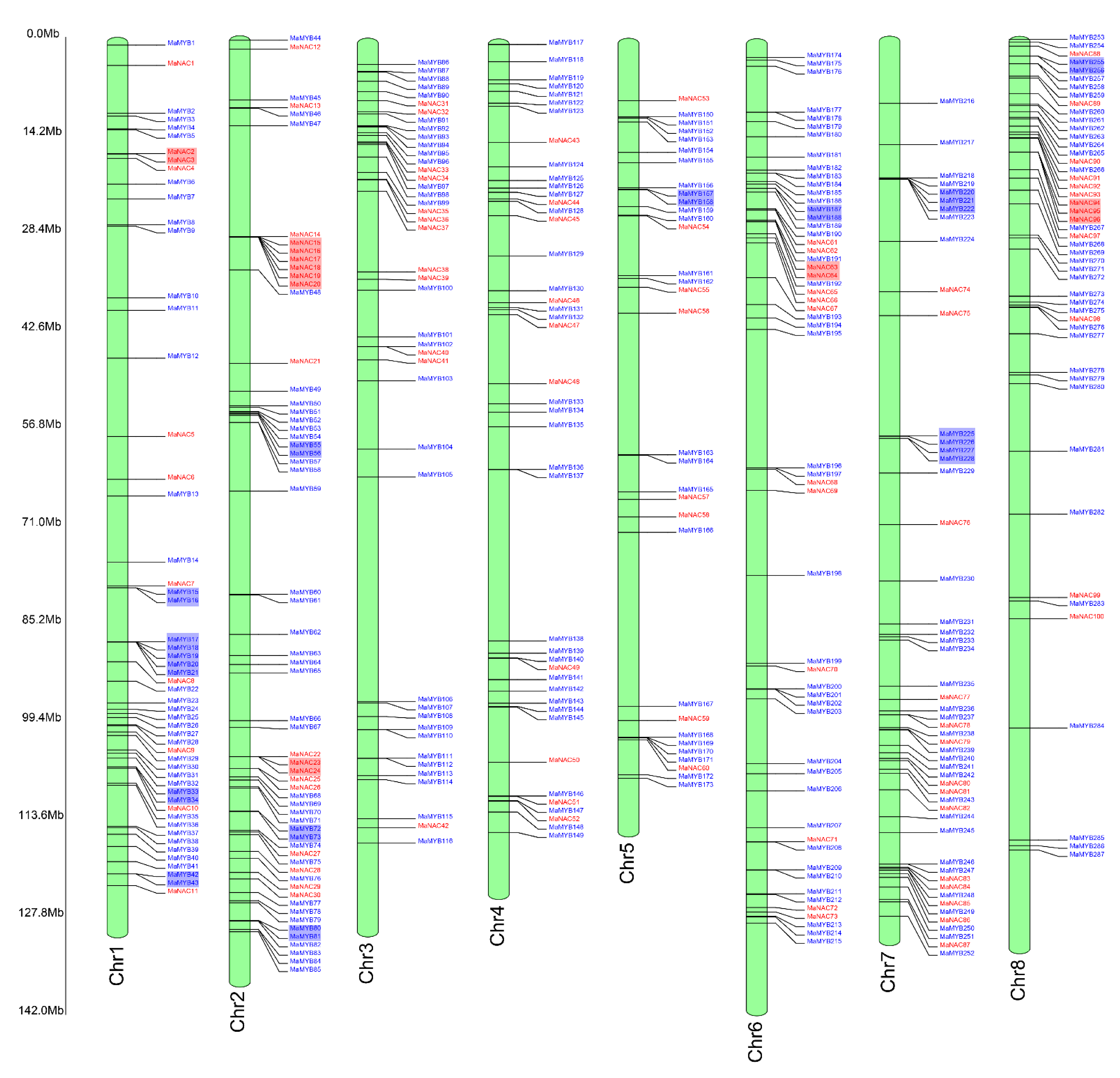
2.2. Genome Distribution and Gene Synteny Analysis
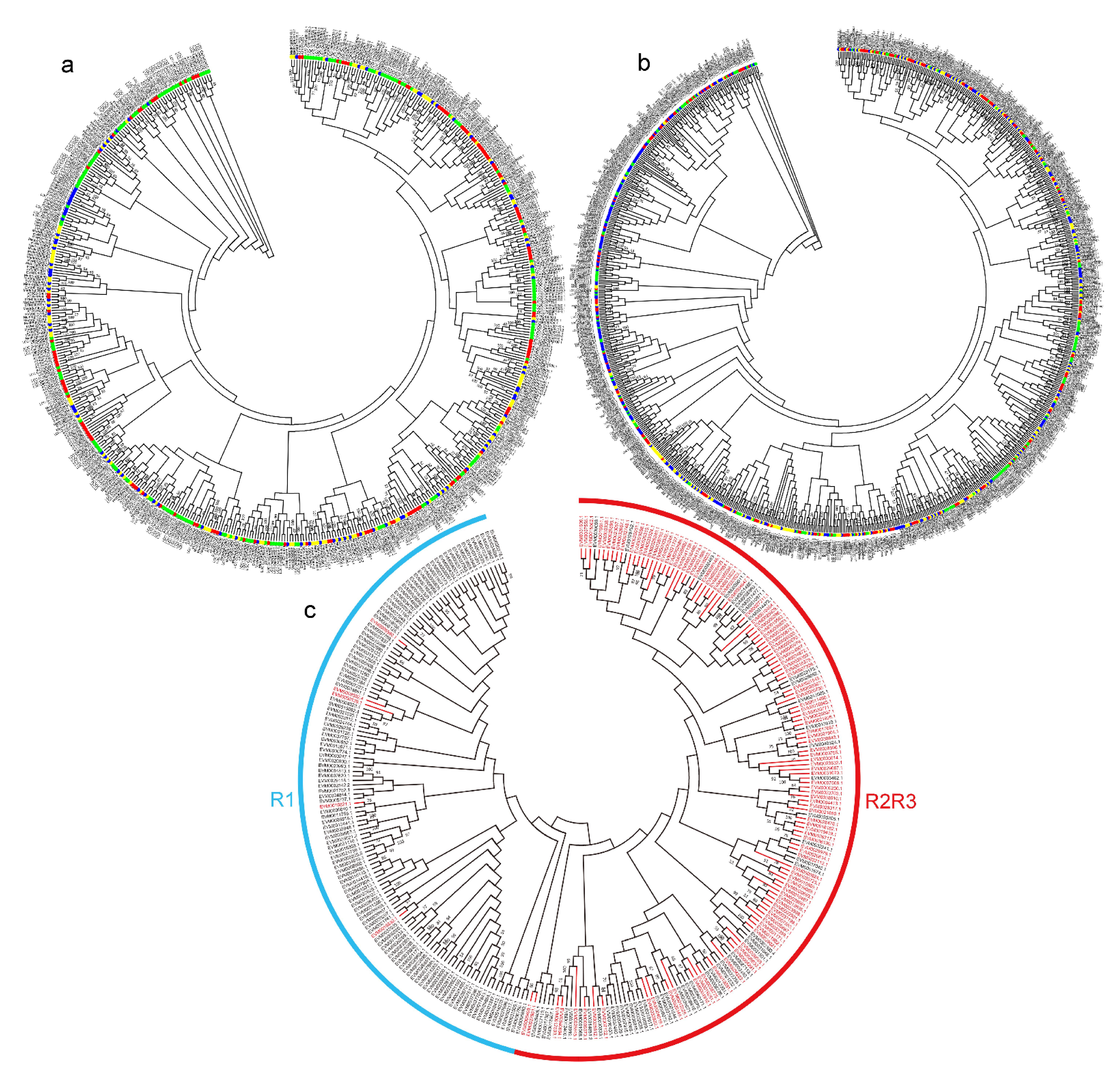
2.3. Phylogenetic Analysis of NAC and MYB TF Genes
2.4. Gene Structure and Protein Motif Analysis
2.5. Gene Promoter Region and Functional Analysis
2.6. Lignin Biosynthesis Genes Regulated by NAC Genes
2.7. Lignin Biosynthesis Genes Regulated by MYB Genes
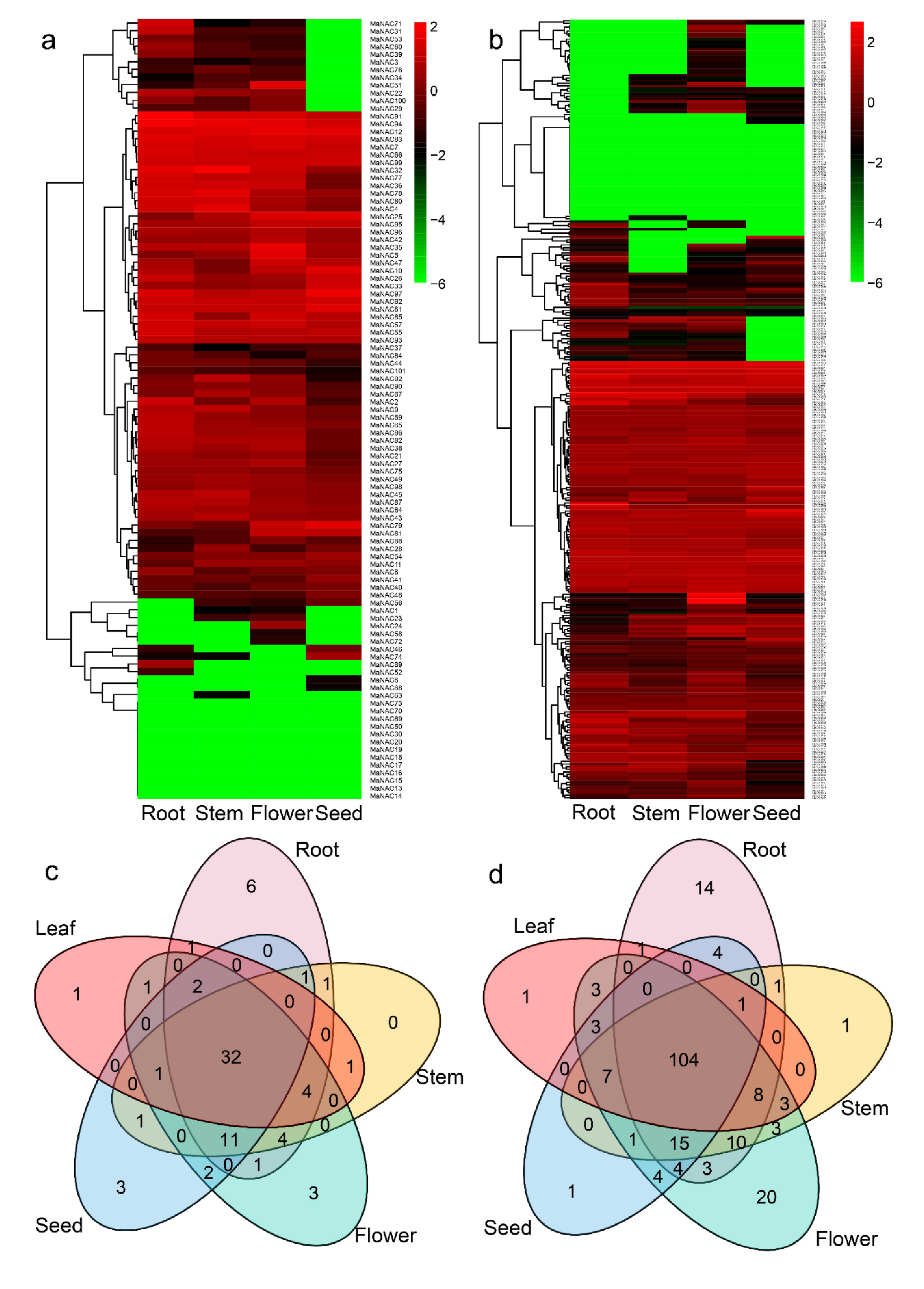
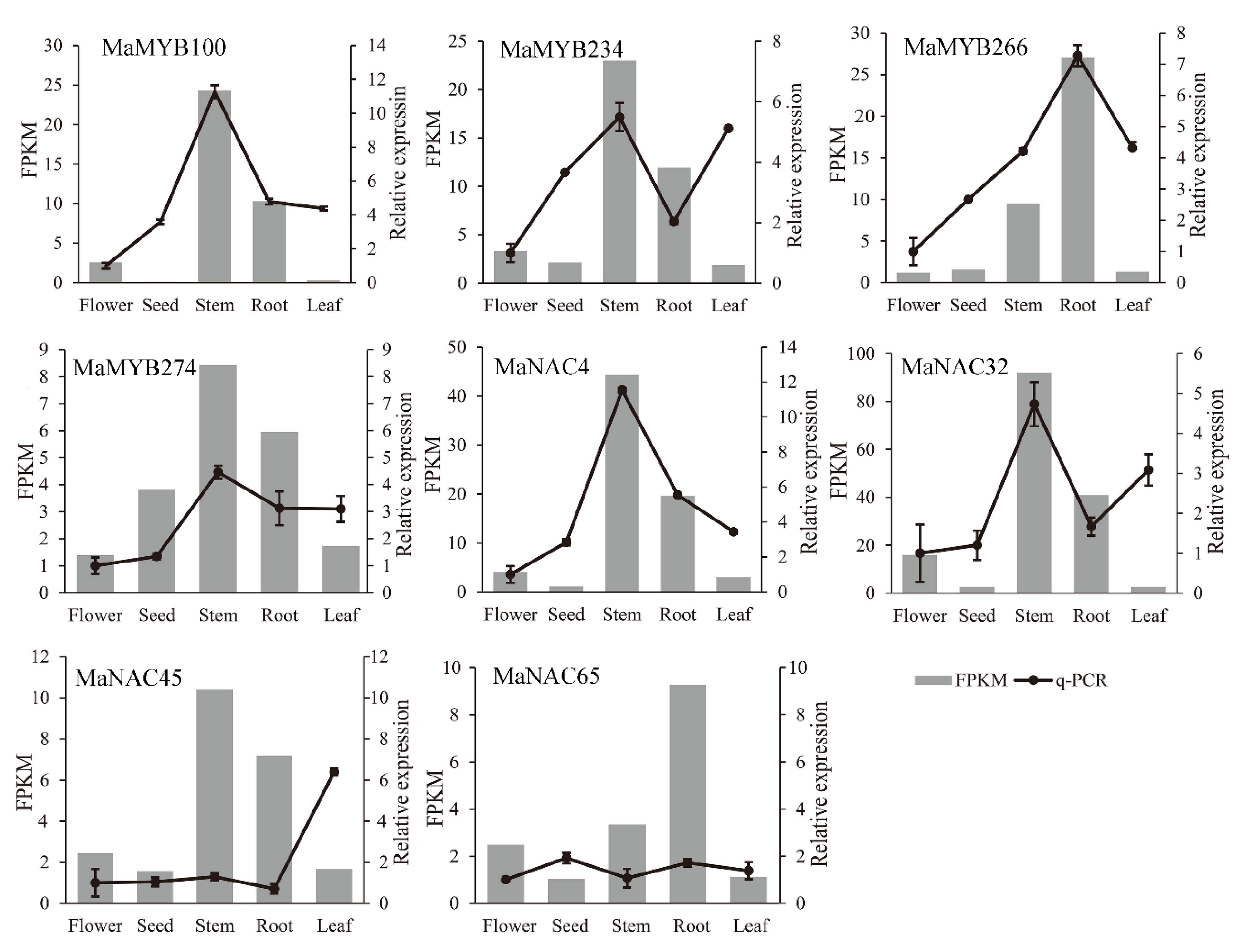
2.8. NAC and MYB Gene Expression and Coexpression Analysis
2.9. Network of Lignin Biosynthesis-Related MaNACs and MaMYBs
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of NAC and MYB Genes in M. albus
4.2. Chromosomal Location, Motif Analysis, and Gene Structure of the NAC and MYB Genes in M. albus
4.3. Phylogenetic Analysis
4.4. Analysis of the Promoter Regions of MaNAC and MaMYB Genes
4.5. Synteny Analysis of MYB and NAC Genes
4.6. Expression Analysis and Gene Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Tongkun, L.; Xiaoming, S.; Weike, D.; Zhinan, H.; Gaofeng, L.; Ying, L.; Xilin, H. Genome-Wide Analysis and Expression Patterns of NAC Transcription Factor Family Under Different Developmental Stages and Abiotic Stresses in Chinese Cabbage. Plant Mol. Biol. Rep. 2014, 32, 1041–1056. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC Family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kim, S.-G.; Kim, Y.-S.; Seo, P.J. Exploring membrane-associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res. 2007, 35, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Lee, S.; Seo, P.J.; Kim, S.K.; Kim, J.K.; Park, C.M. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 2010, 95, 56–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, M.K.; Hagedorn, P.H.; Torres-Zabala, M.D.; Grant, M.R.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Mol. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Kirik, V. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 2005, 132, 1477–1485. [Google Scholar] [CrossRef] [Green Version]
- Li, S.F.; Milliken, O.N.; Pham, H.; Seyit, R.; Napoli, R.; Preston, J.; Koltunow, A.M.; Parish, R.W. The Arabidopsis MYB5 Transcription Factor Regulates Mucilage Synthesis, Seed Coat Development, and Trichome Morphogenesis. Plant Cell 2009, 21, 72–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, A.A. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Park, C.M. The MYB96 Transcription Factor Mediates Abscisic Acid Signaling during Drought Stress Response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Z. Lignification: Flexibility, Biosynthesis and Regulation. Trends Plant Sci. 2016, 21, 713–721. [Google Scholar] [CrossRef]
- Weng, J.K.; Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 2010, 187, 273–285. [Google Scholar] [CrossRef]
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2018, 56, 82–87. [Google Scholar] [CrossRef]
- Minoru, K.; Makiko, U.; Nobuyuki, N.; Gorou, H.; Masatoshi, Y.; Jun, I.; Tetsuro, M.; Hiroo, F.; Taku, D. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.H.; Jeon, H.W.; Kim, W.C.; Kim, J.Y.; Han, K.H. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, G.; Su, Z.; Jinyue, L.; Cuihuan, Z.; Jie, W.; Yingping, C.; Chunxiang, F.; Xue, H.; Hang, H.; Qiao, Z. MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 2020, 182. [Google Scholar] [CrossRef] [Green Version]
- Smith, W.K.; Gorz, H.J. Sweetclover improvement. Adv. Agron. 1965, 17, 163–231. [Google Scholar] [CrossRef] [Green Version]
- Wunderlin, R. The Leguminosae: A source book of characteristics, uses, and nodulation. Econ. Bot. 1982, 36, 224. [Google Scholar] [CrossRef]
- Clark, A. Managing Cover Crops Profitably, 3rd ed.; Sustainable Agriculture Network: Beltsville, MD, USA, 2007. [Google Scholar]
- Jung, H.G.; Allen, M.S. Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J. Anim. Sci. 1995, 73, 2774–2790. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, D.; Ma, J.; Luo, K.; Di, H.; Liu, Z.; Zhang, J.; Wang, Y. Analysis of genetic diversity and population structure in accessions of the genus Melilotus. Ind. Crop. Prod. 2016, 85, 84–92. [Google Scholar] [CrossRef]
- Di, H.; Duan, Z.; Luo, K.; Zhang, D.; Wu, F.; Zhang, J.; Liu, W.; Wang, Y. Interspecific Phylogenic Relationships within Genus Melilotus Based on Nuclear and Chloroplast DNA. PLoS ONE 2015, 10, e0132596. [Google Scholar] [CrossRef]
- Zhang, J.; Hongyan, D.; Kai, L.; Zulfi, J.; Fan, W.; Zhen, D.; Alan, S.; Zhuanzhuan, Y.; Yanrong, W. Coumarin content, morphological variation, and molecular phylogenetics of Melilotus. Molecules 2018, 23, 810. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Ma, J.; Meng, Y.; Zhang, D.; Pascal Muvunyi, B.; Luo, K.; Di, H.; Guo, W.; Wang, Y.; Feng, B.; et al. Potential DNA barcodes for Melilotus species based on five single loci and their combinations. PLoS ONE 2017, 12, e0182693. [Google Scholar] [CrossRef]
- Luo, K.; Jahufer, M.Z.Z.; Zhao, H.; Zhang, R.; Wu, F.; Yan, Z.; Zhang, J.; Wang, Y. Genetic improvement of key agronomic traits in Melilotus albus. Crop Sci. 2017, 58, 1–10. [Google Scholar] [CrossRef]
- Wu, F.; Luo, K.; Yan, Z.; Zhang, D.; Yan, Q.; Zhang, Y.; Yi, X.; Zhang, J. Analysis of miRNAs and their target genes in five Melilotus albus NILs with different coumarin content. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Wu, F.; Zhang, D.; Dong, R.; Fan, Z.; Zhang, R.; Yan, Z.; Wang, Y.; Zhang, J. Transcriptomic profiling of Melilotus albus near-isogenic lines contrasting for coumarin content. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Assis, R. Rapid functional divergence after small-scale gene duplication in grasses. BMC Evol. Biol. 2019, 19. [Google Scholar] [CrossRef]
- Shariatipour, N.; Heidari, B. Meta-analysis of expression of the stress tolerance associated genes and uncover their cis-regulatory elements in rice (Oryza sativa L.). Open Bioinform. J. 2020, 13, 39–49. [Google Scholar] [CrossRef]
- Kim, W.; Kim, J.; Ko, J.; Kang, H.; Han, K. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 2014, 85, 589–599. [Google Scholar] [CrossRef]
- Kim, W.C.; Kim, J.; Ko, J.; Kim, J.; Han, K. Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J. Plant Physiol. 2013, 170, 1374–1378. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.C.; Ko, J.H.; Han, K.H. Identification of acis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 2012, 78, 489–501. [Google Scholar] [CrossRef]
- Kim, W.C.; Ko, J.H.; Kim, J.Y.; Kim, J.; Bae, H.J.; Han, K.H. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 2013, 73, 26–36. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Ye, Z.H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyoko, O.I.; Yoshihisa, O.; Hiroo, F. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Demura, T. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011, 5, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Ohman, D.; Demedts, B.; Kumar, M.; Gerber, L.; Sundberg, B. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. Cell Mol. Biol. 2013, 73, 63–76. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Zhou, J.; Mccarthy, R.L.; Ye, Z.H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, M.E.; Colmer, T.D.; Frost, K.; Henry, D.; Cornwall, D.; Hulm, E.; Deretic, J.; Hughes, S.R.; Craig, A.D. Diversity in the genus Melilotus for tolerance to salinity and waterlogging. Plant Soil 2008, 304, 89–101. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, S.G.; Park, J.E.; Park, H.Y.; Lim, M.H.; Chua, N.H.; Park, C.M. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.G.; Kim, S.Y.; Park, C.M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 2007, 226, 647–654. [Google Scholar] [CrossRef]
- Xie, Q. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goicoechea, M.; Lacombe, E.; Legay, S.; Mihaljevic, S.; Grima-Pettenati, J. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2005, 43, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Bedon, F.; Jacqueline, G.P.; John, M. Conifer R2R3-MYB transcription factors: Sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biol. 2007, 7. [Google Scholar] [CrossRef] [Green Version]
- Abe, H. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2002, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Yeon, K.; Xiaohua, L.; Sun Ju, K.; Haeng, K.; Jeongyeo, L.; Hyeran, K.; Sang, P. MYB transcription factors regulate glucosinolate biosynthesis in different rrgans of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 2013, 18, 8682–8695. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2007, 146, 623–635. [Google Scholar] [CrossRef] [Green Version]
- Maria, P.R.; Felix, J.; Eugenio, B.; Cathie, M. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 2005, 132, 359–370. [Google Scholar]
- Baumann, K.; Perez Rodriguez, M.; Bradley, D.; Venail, J.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Mihaela, P.; Pertea, G.M.; Antonescu, C.M.; Tsung-Cheng, C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Wu, F.; Zhang, J. NAC and MYB Families and Lignin Biosynthesis-Related Members Identification and Expression Analysis in Melilotus albus. Plants 2021, 10, 303. https://doi.org/10.3390/plants10020303
Chen L, Wu F, Zhang J. NAC and MYB Families and Lignin Biosynthesis-Related Members Identification and Expression Analysis in Melilotus albus. Plants. 2021; 10(2):303. https://doi.org/10.3390/plants10020303
Chicago/Turabian StyleChen, Lijun, Fan Wu, and Jiyu Zhang. 2021. "NAC and MYB Families and Lignin Biosynthesis-Related Members Identification and Expression Analysis in Melilotus albus" Plants 10, no. 2: 303. https://doi.org/10.3390/plants10020303





