Genetic and Epigenetic Changes during the Upward Expansion of Deyeuxia angustifolia Kom. in the Alpine Tundra of the Changbai Mountains, China
Abstract
1. Introduction
2. Results
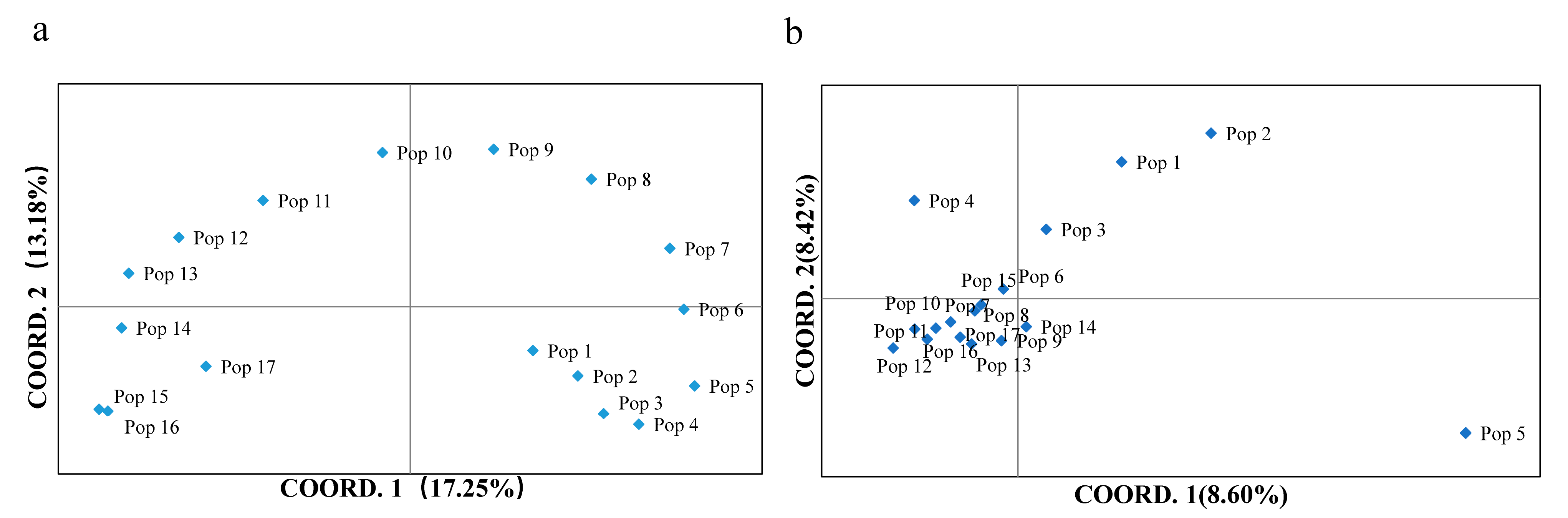
2.1. Genetic Diversity and Structure
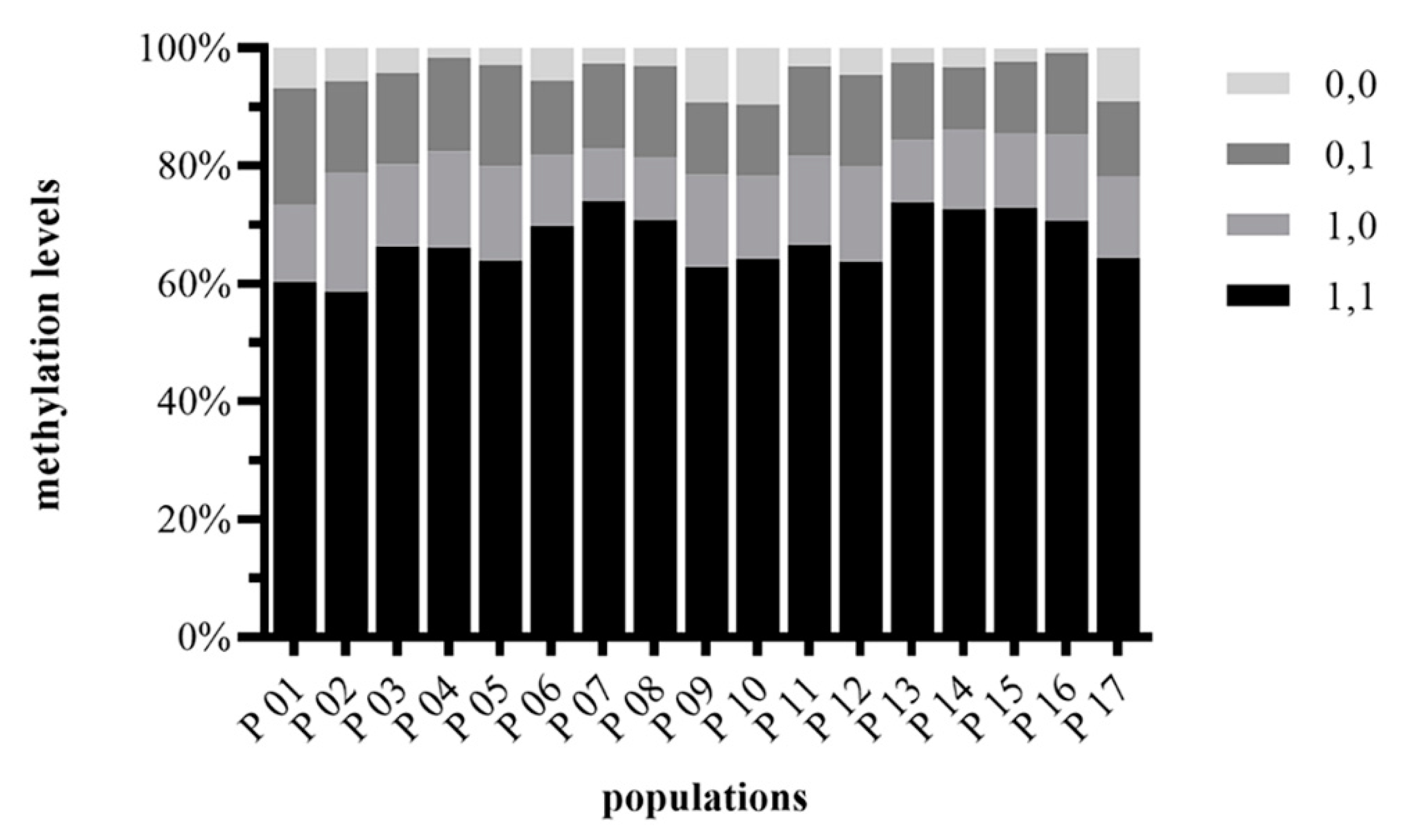
2.2. DNA Methylation Level
2.3. Epigenetic Diversity and Structure
2.4. Differences in Soil Properties
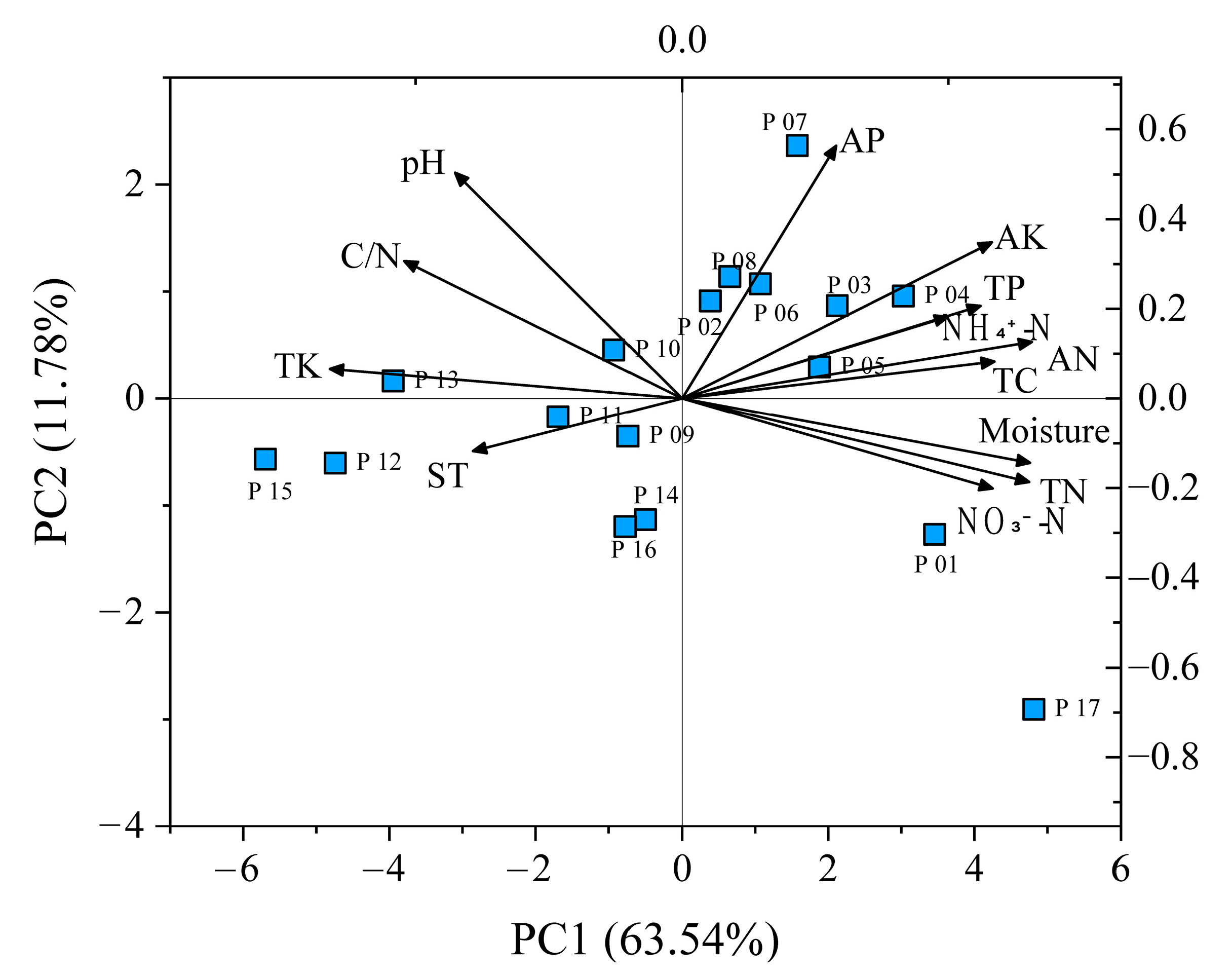
2.5. Environmental Drivers of Genetics and Epigenetics
3. Discussion
3.1. Genetic Diversity and Structure
3.2. Epigenetic Diversity and Structure
3.3. Environmental Drivers of Genetics and Epigenetics
4. Materials and Methods
4.1. Plant Materials
4.2. Soil Sampling
4.3. Plant Genomic DNA Extraction
4.4. AFLP Analysis
4.5. MSAP Analysis
4.6. Soil Properties
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Renwick, K.M.; Rocca, M.E. Temporal context affects the observed rate of climate-driven range shifts in tree species. Glob. Ecol. Biogeogr. 2015, 24, 44–51. [Google Scholar] [CrossRef]
- Du, H.; Liu, J.; Li, M.; Büntgen, U.; Yang, Y.; Wang, L.; Wu, Z.; He, H. Warming-induced upward migration of the alpine treeline in the Changbai Mountains, northeast China. Glob. Chang. Biol. 2018, 24, 1256–1266. [Google Scholar] [CrossRef]
- Shi, H.; Zhou, Q.; Xie, F.; He, N.; He, R.; Zhang, K.; Zhang, Q.; Dang, H. Disparity in elevational shifts of upper species limits in response to recent climate warming in the Qinling Mountains, North-central China. Sci. Total Environ. 2020, 706, 135718–135730. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wang, Z.; Ma, C.; He, H.; Xu, J.; Jin, Y.; Wang, H.; Zheng, X. Soil Mesofauna Respond to the Upward Expansion of Deyeuxia purpurea in the Alpine Tundra of the Changbai Mountains, China. Plants 2019, 8, 615. [Google Scholar] [CrossRef]
- Alvarez-Garrido, L.; Viñegla, B.; Hortal, S.; Powell, J.R.; Carreira, J.A. Distributional shifts in ectomycorrizhal fungal communities lag behind climate-driven tree upward migration in a conifer forest-high elevation shrubland ecotone. Soil Biol. Biochem. 2019, 137, 107545–107555. [Google Scholar] [CrossRef]
- Løkken, J.O.; Evju, M.; Söderström, L.; Hofgaard, A. Vegetation response to climate warming across the forest–tundra ecotone: Species-dependent upward movement. J. Veg. Sci. 2020, 31, 854–866. [Google Scholar] [CrossRef]
- Scheffers, B.R.; De Meester, L.; Bridge, T.C.L.; Hoffmann, A.A.; Pandolfi, J.M.; Corlett, R.T.; Butchart, S.H.M.; Pearce-Kelly, P.; Kovacs, K.M.; Dudgeon, D.; et al. The broad footprint of climate change from genes to biomes to people. Science 2016, 354, aaf7671-1–aaf7671-11. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.C.H. Foundations of invasion genetics: The Baker and Stebbins legacy. Mol. Ecol. 2015, 24, 1927–1941. [Google Scholar] [CrossRef]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L.; et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Li, L.F.; Li, Y.L.; Jia, Y.; Caicedo, A.L.; Olsen, K.M. Signatures of adaptation in the weedy rice genome. Nat. Genet. 2017, 49, 811–814. [Google Scholar] [CrossRef]
- Yuan, Y.; Jin, X.; Liu, J.; Zhao, X.; Zhou, J.; Wang, X.; Wang, D.; Lai, C.; Xu, W.; Huang, J.; et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Niu, X.M.; Xu, Y.C.; Li, Z.W.; Bian, Y.T.; Hou, X.H.; Chen, J.F.; Zou, Y.P.; Jiang, J.; Wu, Q.; Ge, S.; et al. Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl. Acad. Sci. USA 2019, 116, 6908–6913. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Segal, D.L.; Hoyle, G.L.; Schrey, A.W.; Verhoeven, K.J.F.; Richards, C.L. Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecol. Evol. 2015, 5, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Roda, F.; Ambrose, L.; Walter, G.M.; Liu, H.L.; Schaul, A.; Lowe, A.; Pelser, P.B.; Prentis, P.; Rieseberg, L.H.; Ortiz-Barrientos, D. Genomic evidence for the parallel evolution of coastal forms in the Senecio lautus complex. Mol. Ecol. 2013, 22, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013, 197, 314–322. [Google Scholar] [CrossRef]
- Kelager, A.; Pedersen, J.S.; Bruun, H.H. Multiple introductions and no loss of genetic diversity: Invasion history of Japanese Rose, Rosa rugosa, in Europe. Biol. Invasions 2012, 15, 1125–1141. [Google Scholar] [CrossRef]
- Mori, B.A.; Davis, C.S.; Evenden, M.L. Genetic diversity and population structure identify the potential source of the invasive red clover casebearer moth, Coleophora deauratella, in North America. Biol. Invasions 2016, 18, 3595–3609. [Google Scholar] [CrossRef]
- Xu, C.Y.; Julien, M.H.; Fatemi, M.; Girod, C.; Van Klinken, R.D.; Gross, C.L.; Novak, S.J. Phenotypic divergence during the invasion of Phyla canescensin Australia and France: Evidence for selection-driven evolution. Ecol. Lett. 2010, 13, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Medrano, M.; Herrera, C.M.; Bazaga, P. Epigenetic variation predicts regional and local intraspecific functional diversity in a perennial herb. Mol. Ecol. 2014, 23, 4926–4938. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Latzel, V.; Fischer, M.; Bossdorf, O. Understanding the evolutionary potential of epigenetic variation: A comparison of heritable phenotypic variation in epiRILs, RILs, and natural ecotypes of Arabidopsis thaliana. Hered 2018, 121, 257–265. [Google Scholar] [CrossRef]
- Bossdorf, O.; Richards, C.L.; Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 2007, 11, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Pei, C.; Liu, S.; Guo, X.; Du, N.; Guo, W. Genetic and epigenetic changes during the invasion of a cosmopolitan species (Phragmites australis). Ecol. Evol. 2018, 8, 6615–6624. [Google Scholar] [CrossRef]
- Lele, L.; Ning, D.; Cuiping, P.; Xiao, G.; Weihua, G. Genetic and epigenetic variations associated with adaptation to heterogeneous habitat conditions in a deciduous shrub. Ecol. Evol. 2018, 8, 2594–2606. [Google Scholar] [CrossRef]
- Crews, D.; Gore, A.C.; Hsu, T.S.; Dangleben, N.L.; Spinetta, M.; Schallert, T.; Anway, M.D.; Skinner, M.K. Transgenerational epigenetic imprints on mate preference. Proc. Natl. Acad. Sci. USA 2007, 104, 5942–5946. [Google Scholar] [CrossRef] [PubMed]
- Heard, E.; Martienssen Robert, A. Transgenerational Epigenetic Inheritance: Myths and Mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, D.L.; Huang, H.; Zhang, G.; He, L.; Pang, J.; Lozano-Duran, R.; Lang, Z.; Zhu, J.K. Epigenetic memory marks determine epiallele stability at loci targeted by de novo DNA methylation. Nat. Plants 2020, 6, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhou, S.; Gong, X.; Song, Y.; Van Nocker, S.; Ma, F.; Guan, Q. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol. J. 2018, 16, 672–687. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Mithani, A.; Belfield, E.J.; Mott, R.; Hurst, L.D.; Harberd, N.P. Environmentally responsive genome-wide accumulation of de novoArabidopsis thalianamutations and epimutations. Genome Res. 2014, 24, 1821–1829. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, Z.; Cui, X.; Ji, C.; Li, Y.; Zhang, P.; Liu, J.; Riaz, A.; Yao, P.; Liu, M.; et al. N6-Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses. Mol. Plant 2018, 11, 1492–1508. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M.; Torelli, A.; Marieschi, M.; Cozza, R. Role of DNA methylation in the chromium tolerance of Scenedesmus acutus (Chlorophyceae) and its impact on the sulfate pathway regulation. Plant Sci. 2020, 301, 110680. [Google Scholar] [CrossRef]
- Ni, P.; Li, S.; Lin, Y.; Xiong, W.; Huang, X.; Zhan, A. Methylation divergence of invasive Ciona ascidians: Significant population structure and local environmental influence. Ecol. Evol. 2018, 8, 10272–10287. [Google Scholar] [CrossRef]
- Guarino, F.; Cicatelli, A.; Brundu, G.; Improta, G.; Triassi, M.; Castiglione, S. The use of MSAP reveals epigenetic diversity of the invasive clonal populations of Arundo donax L. PLoS ONE 2019, 14, e0215096. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.; Xu, J.; Wu, Z. Investigation and mechanism analysis of Deyeuxia. angustifolia (Kom.) invading tundra on the western slope of Changbai Mountain. J. Mt. Sci. 2013, 31, 448–455. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Dong, W.; Fan, W.; Wu, Z. Comparison of the response of Rhododendron chrysanthum Pall. and Deyeuxia angustifolia Kom. to temperature rise on the forest line on the west slope of Changbai Mountain. Geogr. Res. 2015, 34, 1044–1052. [Google Scholar] [CrossRef]
- Xue, J.G.; Wu, Y.X. Investigation about vegetation restoration at typhoon ruined area in Jilin Changbai Mountain National Nature Reserve. Territ. Nat. Resour. Study 2009, 1, 95–96. [Google Scholar] [CrossRef]
- Zong, S.; Jin, Y.; Xu, J.; Wu, Z.; He, H.; Du, H.; Wang, L. Nitrogen deposition but not climate warming promotes Deyeuxia angustifolia encroachment in alpine tundra of the Changbai Mountains, Northeast China. Sci. Total Environ. 2016, 544, 85–93. [Google Scholar] [CrossRef]
- Li, L.; Xing, M.; Lv, J.; Wang, X.; Chen, X. Response of rhizosphere soil microbial to Deyeuxia angustifolia encroaching in two different vegetation communities in alpine tundra. Sci. Rep. 2017, 7, 43150. [Google Scholar] [CrossRef]
- Shi, W.; Chen, X.; Gao, L.; Xu, C.Y.; Ou, X.; Bossdorf, O.; Yang, J.; Geng, Y. Transient Stability of Epigenetic Population Differentiation in a Clonal Invader. Front. Plant Sci. 2019, 9, 1851. [Google Scholar] [CrossRef]
- Kanbe, T.; Akimoto, S. Allelic and genotypic diversity in long-term asexual populations of the pea aphid, Acyrthosiphon pisum in comparison with sexual populations. Mol. Ecol. 2009, 18, 801–816. [Google Scholar] [CrossRef]
- Lambertini, C.; Riis, T.; Olesen, B.; Clayton, J.S.; Sorrell, B.K.; Brix, H. Genetic diversity in three invasive clonal aquatic species in New Zealand. BMC Genet. 2010, 11, 52. [Google Scholar] [CrossRef]
- Loomis Eli, S.; Fishman, L. A Continent-Wide Clone: Population Genetic Variation of the Invasive Plant Hieracium aurantiacum (Orange Hawkweed; Asteraceae) in North America. Int. J. Plant Sci. 2009, 170, 759–765. [Google Scholar] [CrossRef][Green Version]
- DeWoody, J.; Rowe, C.A.; Hipkins, V.D.; Mock, K.E. “Pando” lives: Molecular genetic evidence of a giant aspen clone in central Utah. West. N. Am. Nat. 2008, 68, 493–497. [Google Scholar] [CrossRef]
- Foust, C.M.; Preite, V.; Schrey, A.W.; Alvarez, M.; Robertson, M.H.; Verhoeven, K.J.F.; Richards, C.L. Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol. Ecol. 2016, 25, 1639–1652. [Google Scholar] [CrossRef]
- Geng, Y.; van Klinken, R.D.; Sosa, A.; Li, B.; Chen, J.; Xu, C.-Y. The Relative Importance of Genetic Diversity and Phenotypic Plasticity in Determining Invasion Success of a Clonal Weed in the USA and China. Front. Plant Sci. 2016, 7, 213. [Google Scholar] [CrossRef]
- Malone, J.M.; Virtue, J.G.; Williams, C.; Preston, C. Genetic diversity of giant reed (Arundo donax) in Australia. Weed Biol. Manag. 2017, 17, 17–28. [Google Scholar] [CrossRef]
- Schulz, B.; Eckstein, R.L.; Durka, W. Epigenetic variation reflects dynamic habitat conditions in a rare floodplain herb. Mol. Ecol. 2014, 23, 3523–3537. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaf, A.; Wardenaar, R.; Neumann, D.A.; Taudt, A.; Shaw, R.G.; Jansen, R.C.; Schmitz, R.J.; Colomé-Tatché, M.; Johannes, F. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. USA 2015, 112, 6676–6681. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef]
- Heer, C.; Körner, C. High elevation pioneer plants are sensitive to mineral nutrient addition. Basic Appl. Ecol. 2002, 3, 39–47. [Google Scholar] [CrossRef]
- Jin, Y.; Xu, J.; Wang, Y.; Wang, S.; Chen, Z.; Huang, X.; Niu, L. Effects of nitrogen deposition on tundra vegetation undergoing invasion by Deyeuxia angustifolia in Changbai Mountains. Chin. Geogr. Sci. 2015, 26, 99–108. [Google Scholar] [CrossRef]
- Xu, W.; Liu, L.; He, T.; Cao, M.; Sha, L.; Hu, Y.; Li, Q.; Li, J. Soil properties drive a negative correlation between species diversity and genetic diversity in a tropical seasonal rainforest. Sci. Rep. 2016, 6, 20652. [Google Scholar] [CrossRef]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef]
- Vanden Broeck, A.; Cox, K.; Brys, R.; Castiglione, S.; Cicatelli, A.; Guarino, F.; Heinze, B.; Steenackers, M.; Vander Mijnsbrugge, K. Variability in DNA Methylation and Generational Plasticity in the Lombardy Poplar, a Single Genotype Worldwide Distributed Since the Eighteenth Century. Front. Plant Sci. 2018, 9, 1635. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.K.; Liu, G.X.; Zhang, B.G.; Zhang, G.S. Effects of environmental factors on population genetic diversity of Tamarix chinensis. Acta Pratacult. Sin. 2019, 28, 178–186. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, W.; Li, L.; You, J.; Ni, B.; Chen, X. Clonal plasticity and diversity facilitates the adaptation of Rhododendron aureum Georgi to alpine environment. PLoS ONE 2018, 13, e0197089. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Shi, X.; Li, J.; Guo, W.; Liu, C.; Chen, X. Genetic, Epigenetic, and HPLC Fingerprint Differentiation between Natural and Ex Situ Populations of Rhodiola sachalinensis from Changbai Mountain, China. PLoS ONE 2014, 9, e112869. [Google Scholar] [CrossRef]
- Chen, L.; Liu, L.; Mao, C.; Qin, S.; Wang, J.; Liu, F.; Blagodatskiy, S.; Yang, G.; Zhang, Q.; Zhang, D.; et al. Nitrogen availability regulates topsoil carbon dynamics after permafrost thaw by altering microbial metabolic efficiency. Nat. Commun. 2018, 9, 3951. [Google Scholar] [CrossRef]
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Resour. 2013, 13, 642–653. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]

| Elevation (m) | Latitude (°N) | Longitude (°E) | Nsp | ST (°C) | |
|---|---|---|---|---|---|
| P01 | 2216 | 41.99077 | 128.01663 | 3 | 14.3 |
| P02 | 2220 | 41.99142 | 128.01673 | 3 | 11.5 |
| P03 | 2242 | 41.99280 | 128.01647 | 3 | 16.0 |
| P04 | 2241 | 41.99272 | 128.01640 | 6 | 15.2 |
| P05 | 2240 | 41.99268 | 128.01627 | 4 | 15.9 |
| P06 | 2162 | 41.99052 | 128.01345 | 3 | 25.0 |
| P07 | 2163 | 41.99053 | 128.01325 | 3 | 15.3 |
| P08 | 2151 | 41.99018 | 128.01317 | 3 | 17.7 |
| P09 | 2148 | 41.99065 | 128.01085 | 5 | 16.5 |
| P10 | 2142 | 41.99053 | 128.01088 | 5 | 15.0 |
| P11 | 2140 | 41.99050 | 128.01078 | 5 | 17.6 |
| P12 | 2076 | 41.99030 | 128.00440 | 6 | 19.3 |
| P13 | 2077 | 41.99033 | 128.00435 | 6 | 20.4 |
| P14 | 2074 | 41.99020 | 128.00430 | 4 | 19.3 |
| P15 | 2047 | 41.98882 | 128.00275 | 6 | 25.7 |
| P16 | 2044 | 41.98880 | 128.00280 | 5 | 21.4 |
| P17 | 2041 | 41.98855 | 128.00267 | 5 | 16.0 |
| Sample Size | I-AFLP | PPL-AFLP | I-MSAP | PPL-MSAP | |
|---|---|---|---|---|---|
| P01 | 10 | 0.074 | 15.43% | 0.073 | 18.98% |
| P02 | 10 | 0.025 | 4.90% | 0.087 | 18.93% |
| P03 | 10 | 0.016 | 2.98% | 0.051 | 11.98% |
| P04 | 10 | 0.051 | 9.23% | 0.016 | 3.54% |
| P05 | 10 | 0.015 | 2.82% | 0.018 | 4.61% |
| P06 | 10 | 0.034 | 6.19% | 0.033 | 9.04% |
| P07 | 10 | 0.020 | 3.77% | 0.029 | 7.68% |
| P08 | 10 | 0.009 | 1.75% | 0.018 | 4.05% |
| P09 | 10 | 0.020 | 4.22% | 0.061 | 14.80% |
| P10 | 10 | 0.023 | 4.50% | 0.046 | 13.35% |
| P11 | 10 | 0.045 | 8.22% | 0.044 | 9.08% |
| P12 | 10 | 0.026 | 5.29% | 0.042 | 9.59% |
| P13 | 10 | 0.015 | 2.70% | 0.011 | 2.26% |
| P14 | 10 | 0.014 | 2.48% | 0.024 | 6.31% |
| P15 | 10 | 0.025 | 4.56% | 0.020 | 4.73% |
| P 16 | 10 | 0.028 | 5.29% | 0.006 | 1.36% |
| P17 | 10 | 0.164 | 31.14% | 0.075 | 16.20% |
| Mean | 10 | 0.035 | 6.79% | 0.038 | 9.21% |
| SE | 0.000 | 0.001 | 1.72% | 0.001 | 1.38% |
| Source | df | Est. Var. | % | p-Value | Phi-Statistics |
|---|---|---|---|---|---|
| AMOVA results for AFLP data | |||||
| Among Pops | 16 | 364.440 | 95% | 0.0001 | 0.946 |
| Within Pops | 153 | 20.851 | 5% | ||
| Total | 169 | 385.291 | 100% | ||
| AMOVA results for MSAP data | |||||
| Among Pops | 16 | 214.356 | 87% | 0.0001 | 0.866 |
| Within Pops | 153 | 33.065 | 13% | ||
| Total | 169 | 247.421 | 100% | ||
| Gen | Epi | |
|---|---|---|
| Epi | 0.045 | - |
| Geo | 0.648 *** | 0.137 |
| Env | 0.127 * | −0.099 |
| Controlling for Gen.dist | Controlling for Epi.dist | Controlling for Geo.dist | Controlling for Env.dist | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect of Env.dist | Effect of Geo.dist | Effect of Env.dist | Effect of Geo.dist | Effect of Gen.dist | Effect of Epi.dist | Effect of Env.dist | Effect of Gen.dist | Effect of Epi.dist | Effect of Geo.dist | |
| Gen.dist | - | - | 0.132 * | 0.649 *** | - | −0.058 | −0.120 | - | 0.059 | 0.647 *** |
| Epi.dist | −0.106 | 0.142 | - | - | −0.058 | - | −0.154 | 0.059 | - | 0.181 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, B.; You, J.; Li, J.; Du, Y.; Zhao, W.; Chen, X. Genetic and Epigenetic Changes during the Upward Expansion of Deyeuxia angustifolia Kom. in the Alpine Tundra of the Changbai Mountains, China. Plants 2021, 10, 291. https://doi.org/10.3390/plants10020291
Ni B, You J, Li J, Du Y, Zhao W, Chen X. Genetic and Epigenetic Changes during the Upward Expansion of Deyeuxia angustifolia Kom. in the Alpine Tundra of the Changbai Mountains, China. Plants. 2021; 10(2):291. https://doi.org/10.3390/plants10020291
Chicago/Turabian StyleNi, Biao, Jian You, Jiangnan Li, Yingda Du, Wei Zhao, and Xia Chen. 2021. "Genetic and Epigenetic Changes during the Upward Expansion of Deyeuxia angustifolia Kom. in the Alpine Tundra of the Changbai Mountains, China" Plants 10, no. 2: 291. https://doi.org/10.3390/plants10020291
APA StyleNi, B., You, J., Li, J., Du, Y., Zhao, W., & Chen, X. (2021). Genetic and Epigenetic Changes during the Upward Expansion of Deyeuxia angustifolia Kom. in the Alpine Tundra of the Changbai Mountains, China. Plants, 10(2), 291. https://doi.org/10.3390/plants10020291






