A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos
Abstract
1. Introduction
2. Results and Discussion
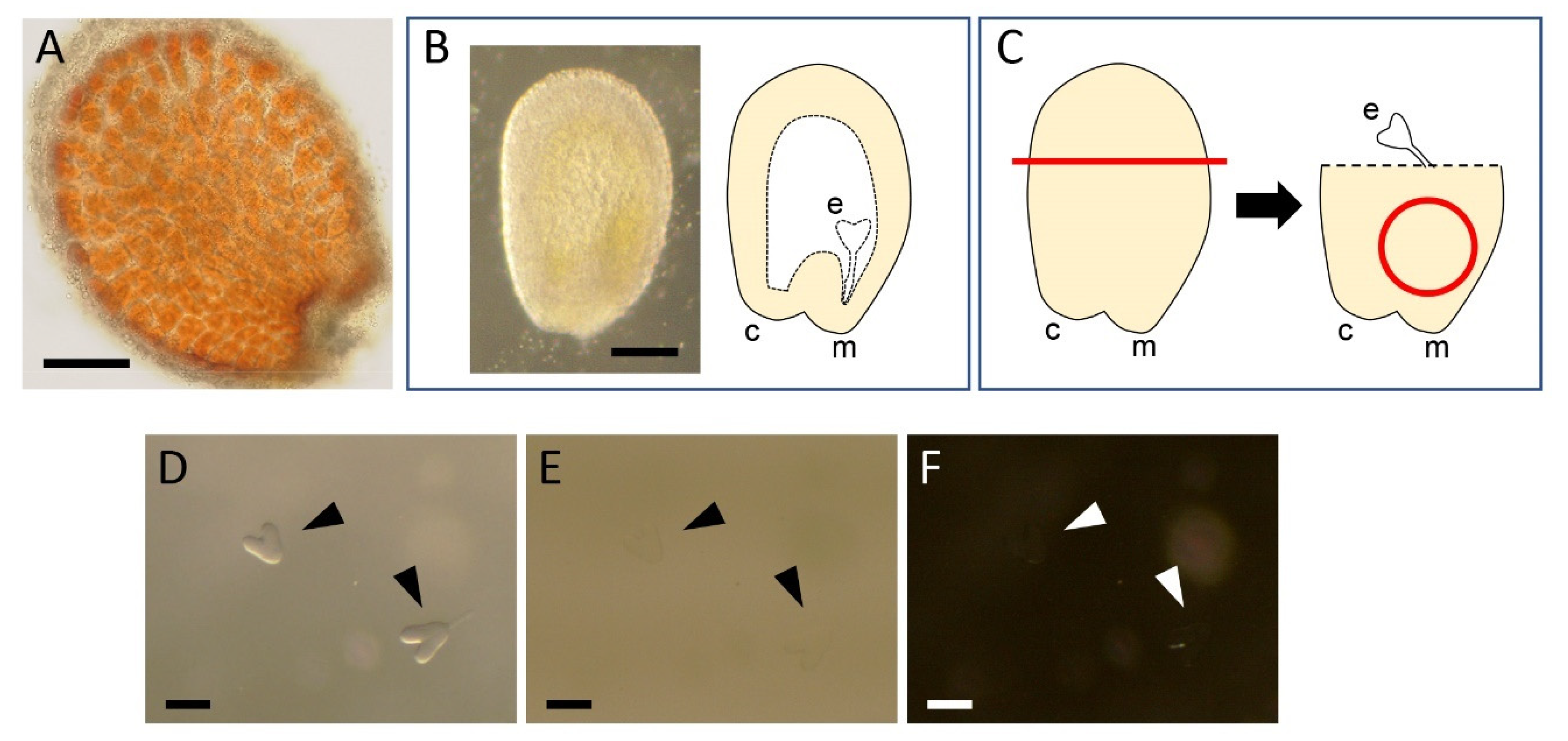
2.1. Dissection of Embryos
2.2. Fixation and Washing
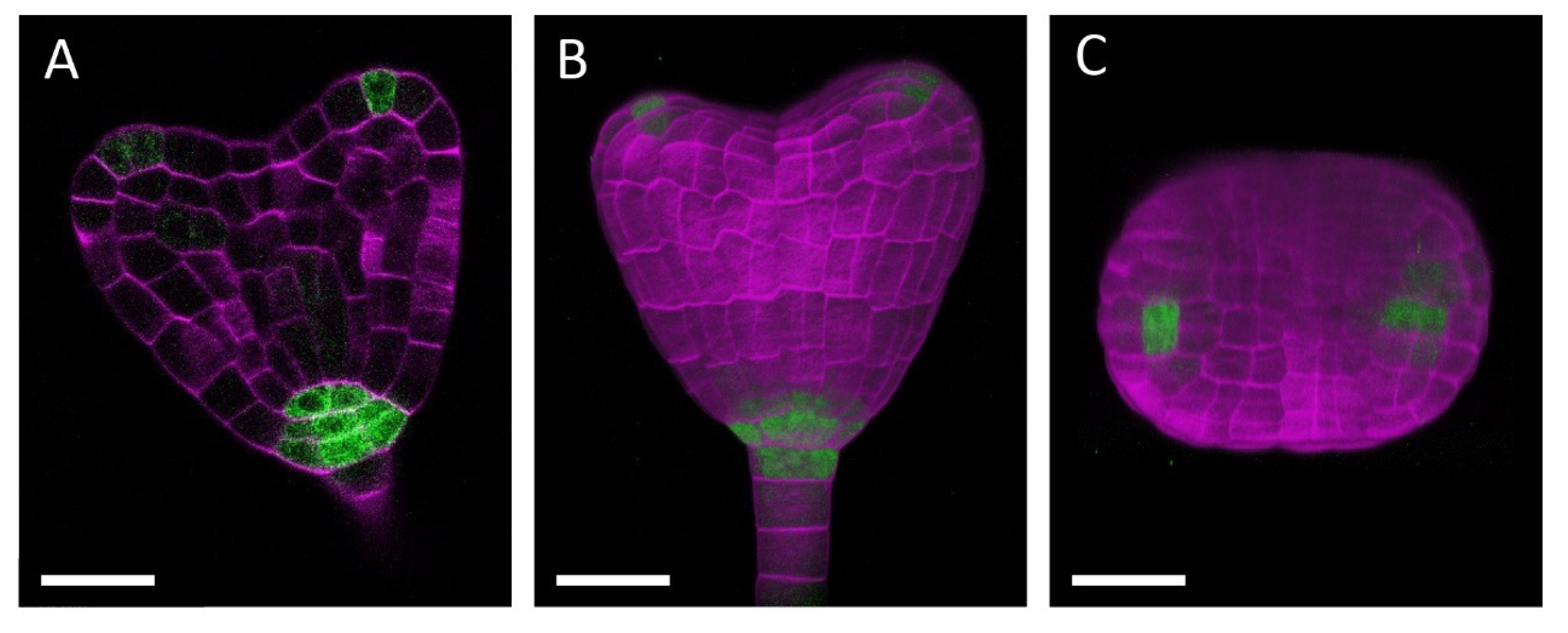
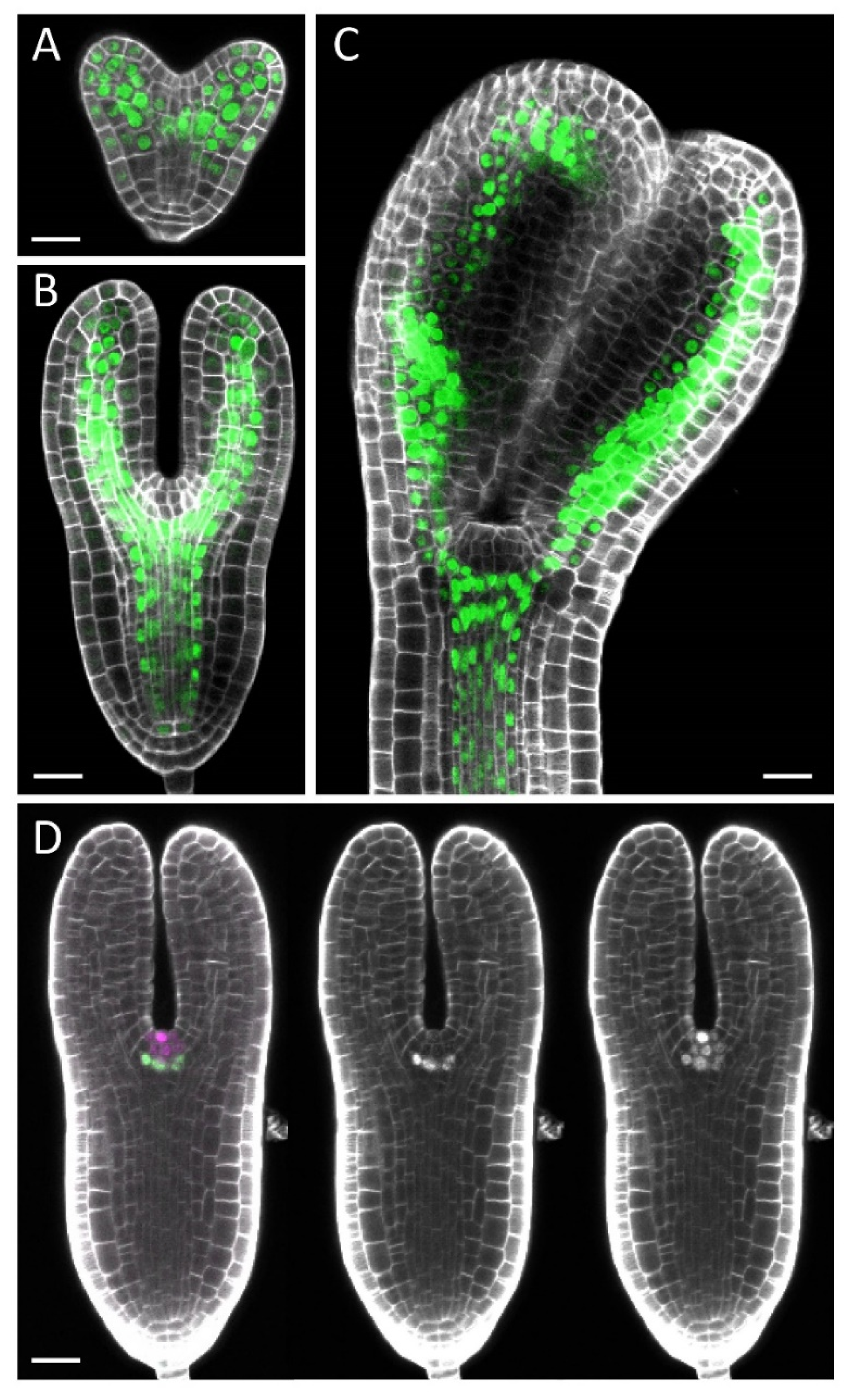
2.3. Clearing and Staining
2.4. Confocal Microscopy
3. Materials and Equipment
3.1. Plant Materials
3.2. Solutions
3.3. Equipment
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Smet, I.; Beeckman, T. Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Boscá, S.; Knauer, S.; Laux, T. Embryonic development in Arabidopsis thaliana: From the zygote division to the shoot meristem. Front. Plant Sci. 2011, 2, 93. [Google Scholar] [CrossRef] [PubMed]
- ten Hove, C.A.; Lu, K.J.; Weijers, D. Building a plant: Cell fate specification in the early Arabidopsis embryo. Development 2015, 142, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Scheres, B.; Wolkenfelt, H.; Willemsen, V.; Terlouw, M.; Lawson, E.; Dean, C.; Weisbeek, P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 1994, 120, 2475–2487. [Google Scholar]
- Barton, M.K.; Poethig, R.S. Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 1993, 119, 823–831. [Google Scholar]
- Mansfield, S.G.; Briarty, L.G. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 1991, 69, 461–476. [Google Scholar] [CrossRef]
- Chakrabortty, B.; Willemsen, V.; de Zeeuw, T.; Liao, C.Y.; Weijers, D.; Mulder, B.; Scheres, B. A plausible microtubule-based mechanism for cell division orientation in plant embryogenesis. Curr. Biol. 2018, 28, 3031–3043.e3032. [Google Scholar] [CrossRef]
- Yoshida, S.; Barbier de Reuille, P.; Lane, B.; Bassel, G.W.; Prusinkiewicz, P.; Smith, R.S.; Weijers, D. Genetic control of plant development by overriding a geometric division rule. Dev. Cell 2014, 29, 75–87. [Google Scholar] [CrossRef]
- van Dop, M.; Liao, C.Y.; Weijers, D. Control of oriented cell division in the Arabidopsis embryo. Curr. Opin. Plant Biol. 2015, 23, 25–30. [Google Scholar] [CrossRef]
- Bayer, M.; Slane, D.; Jürgens, G. Early plant embryogenesis—Dark ages or dark matter? Curr. Opin. Plant Biol. 2017, 35, 30–36. [Google Scholar] [CrossRef]
- Kurihara, D.; Mizuta, Y.; Sato, Y.; Higashiyama, T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 2015, 142, 4168–4179. [Google Scholar] [CrossRef] [PubMed]
- Bougourd, S.; Marrison, J.; Haseloff, J. An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J. 2000, 24, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, J.; Sakamoto, Y.; Nakagami, S.; Aida, M.; Sawa, S.; Matsunaga, S. Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol. 2016, 57, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Truernit, E.; Bauby, H.; Dubreucq, B.; Grandjean, O.; Runions, J.; Barthélémy, J.; Palauqui, J.C. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 2008, 20, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, A.; Tofanelli, R.; Strauss, S.; Cerrone, L.; Wolny, A.; Strohmeier, J.; Kreshuk, A.; Hamprecht, F.A.; Smith, R.S.; Schneitz, K. A digital 3D reference atlas reveals cellular growth patterns shaping the Arabidopsis ovule. eLife 2021, 10. [Google Scholar] [CrossRef]
- Ueda, M.; Kimata, Y.; Kurihara, D. Live-cell imaging of zygotic intracellular structures and early embryo pattern formation in Arabidopsis thaliana. In Plant Embryogenesis: Methods and Protocols; Bayer, M., Ed.; Springer: New York, NY, USA, 2020; pp. 37–47. [Google Scholar]
- Hughes, R. Determinants of Seed Size and Yield in Arabidopsis thaliana. Ph.D. Thesis, University of Bath, Bath, UK, 2009. [Google Scholar]
- Wilson, E.E.; Chambers, W.; Pelc, R.; Nothnagle, P.; Davidson, M.W. Stereomicroscopy in neuroanatomy. In Neurohistology and Imaging Techniques; Pelc, R., Walz, W., Doucette, J.R., Eds.; Springer: New York, NY, USA, 2020; pp. 245–274. [Google Scholar]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Benkova, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertova, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Barbier de Reuille, P.; Routier-Kierzkowska, A.L.; Kierzkowski, D.; Bassel, G.W.; Schüpbach, T.; Tauriello, G.; Bajpai, N.; Strauss, S.; Weber, A.; Kiss, A.; et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife 2015, 4, 05864. [Google Scholar] [CrossRef]
- Rademacher, E.H.; Möller, B.; Lokerse, A.S.; Llavata-Peris, C.I.; van den Berg, W.; Weijers, D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011, 68, 597–606. [Google Scholar] [CrossRef]
- Tucker, M.R.; Hinze, A.; Tucker, E.J.; Takada, S.; Jürgens, G.; Laux, T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development 2008, 135, 2839–2843. [Google Scholar] [CrossRef] [PubMed]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Brand, U.; Grünewald, M.; Hobe, M.; Simon, R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002, 129, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imoto, A.; Yamada, M.; Sakamoto, T.; Okuyama, A.; Ishida, T.; Sawa, S.; Aida, M. A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos. Plants 2021, 10, 190. https://doi.org/10.3390/plants10020190
Imoto A, Yamada M, Sakamoto T, Okuyama A, Ishida T, Sawa S, Aida M. A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos. Plants. 2021; 10(2):190. https://doi.org/10.3390/plants10020190
Chicago/Turabian StyleImoto, Ayame, Mizuki Yamada, Takumi Sakamoto, Airi Okuyama, Takashi Ishida, Shinichiro Sawa, and Mitsuhiro Aida. 2021. "A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos" Plants 10, no. 2: 190. https://doi.org/10.3390/plants10020190
APA StyleImoto, A., Yamada, M., Sakamoto, T., Okuyama, A., Ishida, T., Sawa, S., & Aida, M. (2021). A ClearSee-Based Clearing Protocol for 3D Visualization of Arabidopsis thaliana Embryos. Plants, 10(2), 190. https://doi.org/10.3390/plants10020190





