Abstract
MADS-box transcription factors are crucial regulators of inflorescence and flower development in plants. Therefore, the recent interest in this family has received much attention in plant breeding programs due to their impact on plant development and inflorescence architecture. The aim of this study was to investigate the role of HvMADS-box genes in lateral spikelet development in barley (Hordeum vulgare L.). A set of 30 spike-contrasting barley lines were phenotypically and genotypically investigated under controlled conditions. We detected clear variations in the spike and spikelet development during the developmental stages among the tested lines. The lateral florets in the deficiens and semi-deficiens lines were more reduced than in two-rowed cultivars except cv. Kristina. Interestingly, cv. Kristina, int-h.43 and int-i.39 exhibited the same behavior as def.5, def.6, semi-def.1, semi-def.8 regarding development and showed reduced lateral florets size. In HOR1555, HOR7191 and HOR7041, the lateral florets continued their development, eventually setting seeds. In contrast, lateral florets in two-rowed barley stopped differentiating after the awn primordia stage giving rise to lateral floret sterility. At harvest, the lines tested showed large variation for all central and lateral spikelet-related traits. Phylogenetic analysis showed that more than half of the 108 MADS-box genes identified are highly conserved and are expressed in different barley tissues. Re-sequence analysis of a subset of these genes showed clear polymorphism in either SNPs or in/del. Variation in HvMADS56 correlated with altered lateral spikelet morphology. This suggests that HvMADS56 plays an important role in lateral spikelet development in barley.
1. Introduction
Increasing the yield performance of cereal crops remains one of the major goals of plant breeding programs [1]. One of the strategies has been to increase the number of seed primordia per inflorescence. In rice [2], maize [3] and to a certain extent also in wheat [4,5], this has been achieved by enhancing the number of florets. In barley (Hordeum vulgare L.), however, grain number has been increased by suppressing floret sterility [6]. The inflorescence architecture of barley is unique among the Triticeae. The barley inflorescence is an unbranched spike [7], whose constituent units are called spikelets. Each spikelet contains a single floret and two glumes and is attached to the floral axis (the rachis). Floral patterning of the three spikelet primordia of immature barley inflorescence—one central and two laterals—is initiated at each rachis node. In wild barley (H. vulgare sp. spontaneum) and domesticated barley of the two-rowed type (H. vulgare sp. vulgare), lateral floret development is arrested, and only the central spikelets set grains [8].
Altering the fertility of lateral florets is one of the major strategies to increase the number of grains at each rachis node. It is likely that six-rowed barley (H. vulgare convar. hexastichon), with three fertile spikelets per rachis node, was derived from an ancestral two-rowed form (convar. distichon (L.) Alef.) [6]. Based on the central and lateral spikelet fertility, barley plants are categorized into four different groups [9]: (i) two-rowed barley having a fertile central spikelet flanked by two sterile lateral ones, (ii) deficiens barley, basically being a two-rowed barley in which the lateral spikelets are absent or extremely reduced; (iii) six-rowed barley having three fully fertile spikelets per node; (iv) labile-barley having the lateral spikelets fully developed or absent, fertile or sterile, even within one individual spike. Another row-type class named intermedium barley (H. vulgare convar. intermedium (Körn.) Mansf. (syn. H. intermedium Körn.)) displays various degrees of lateral spikelet fertility and seed development intermediate between two- and six-rowed types [6]. Another group of barley row-type, the so-called semi-deficiens-barley, has a two-rowed phenotype whose reduced lateral spikelets are longer than those in deficiens but smaller than those in true two-rowed barley.
The barley row-type is regulated by several loci of which some are identified; Six-rowed spike 1 (Vrs1 [syn = HvHox1], [7], Vrs2 [10], Vrs3 [11,12], Vrs4 [13], intermedium spike-c (int-c [syn = Vrs5], [14]. Out of these loci, four encode transcription factors; Vrs1 (homeodomain-leucine zipper class I (corresponds to HD-Zip I in Arabidopsis)), Vrs2 (SHORT INTERNODES), Vrs4 (RAMOSA2) and Vrs5 (TEOSINTE BRANCHED1), while Vrs3 encodes an enzyme (a histone H3K9 demethylase). Previous genetic analyses of the intermedium barley have revealed that other genes, independently of Vrs1, can increase the size of florets and even stimulate occasional grain setting in lateral spikelets [6]. The results showed that the six-rowed phenotype arises in various panels of intermedium barley carrying the two-rowed allele of Vrs1 in the presence of the six-rowed allele of Int-c, previously considered only as a modifier of lateral spikelet fertility. The six-rowed allele of Int-c probably arose before domestication and is associated with the enlargement of lateral florets in wild barley. Since this allele cannot overcome the lateral florets sterility in the genomic background of wild barleys, we infer the existence of other loci at which novel alleles or allelic combinations were selected for after domestication to increase the grain number of barley independently of Vrs1 [6]. Our understanding of the genetic basis of barley inflorescence development and growth has greatly increased over the recent decades. The spikelet arrangement patterning in two-rowed and wild barleys is regulated by the transcriptional regulator (Vrs2), which plays a role in controlling the levels of important developmental hormones along the spike [10,15,16].
Among the various transcription factors active in biological systems is the MADS-box family. The name is derived from MCM1 (in yeast), AG (in Arabidopsis), DEFICIENS (in Antirrhinum), and SRF (in mammals). These four protein families are considered as the first four discovered transcription factors [17,18]. Many of the MADS-box genes are crucial for floral initiation, development and growth and proposed to be the dynamic force of the floral diversity in many plants [19,20]. Therefore, a better understanding of MADS-box gene function can provide information on how different floral structures evolved and identify target genes for the improvement of crop breeding programs [21]. In Arabidopsis, most of the floral organs development genes encode MADS-box transcription factors. The MADS-box genes in rice, maize and barley showed crucial similarities to those in Arabidopsis, suggesting a similar function in floral development [22,23].
In the present work, we studied the relations between HvMADS-box genes and the phenotypic development of lateral spikelets using a set of spike-variation barley lines. We found that HvMADS56 could be considered as a novel gene that plays an important role in lateral spikelet development in barley.
2. Results
2.1. Phenotypic Status and Scanning Electron Microscopy (SEM) of the Tested Barley Lines
Thirty barley lines with a large variation in their spike morphology were selected to investigate the role of HvMADS-box genes in relation to their spike and spikelet development. Among these lines were two-rowed, deficiens, semi-deficiens and intermedium types. The 30 barley lines were grown under controlled greenhouse conditions. At harvest, plant height, number of tillers per plant, number of spikes per plant, main culm spike length and weight as well as central and lateral spikelet length, width, weight and area were measured (Table S1). The analysis of variance revealed highly significant differences for all studied traits. The coefficient of determination (R2) and heritability in broad-sense (Hb) ranged from 84.7 to 90.9% (for central seed width) and from 93.9 to 96.8% (for central seed area), respectively (Table S2). Plant height ranged between 59 cm in def.8 and 128 cm in HOR6407, with an average of 83 cm. Number of tillers per plant varied from 2.3 in int-l.81 to 22.0 in cv. Bonus with an average of 10.6. Number of spikes per plant ranged from 2.0 in int-l.81 to 21.3 in cv. Bonus with an average of 9.7. Spike length ranged from 4.4 cm in HOR10166 to 11.8 cm in cv. Bonus with an average of 9.2 cm. Spike weight varied from 0.34 g in def.5 to 2.84 g in HOR7191 with an average of 1.27 g. The seed weight of the central spikelet ranged between 0.12 g in def.5 and 1.45 g in cv. Bonus with an average of 0.87 g. Seed width of the central spikelet ranged between 3.17 mm in def.6 and 4.20 mm in cv. Bowman with an average of 3.68 mm. Seed length of the central spikelet ranged between 6.77 mm in HOR1555 and 16.30 mm in HOR6407 with an average of 10.45 mm. Seed area of the central spikelet varied between 18.13 mm2 in HOR1555 and 37.90 mm2 in int-l.81 with an average of 26.0 mm2. Seed numbers of the central spikelet ranged between 4.0 seeds in int-l.81 and 27.3 seeds in cv. Bonus with an average of 16.7 seeds. Thousand seed weight of the central spikelet ranged between 28.8 g in def.6 and 70.1 g in int-l.81 with an average of 52.2 g (Table S1).
Four lines showed fertile lateral spikelets and were setting seeds; hex-v.3, HOR1555, HOR7191 and HOR7041. All the other lines showed sterile lateral spikelets. Highly significant differences were observed for lateral spikelet weight (LSW), width (LSWi), length (LSL), area (LSA), and thousand seeds weight (in case of fertile lateral spikelet) or thousand spikelet weight (in case of sterile lateral spikelet) (TSW) (Table 1). Large estimates of Hb and R2 were obtained for all lateral spikelet-related traits (Table S2). Among the sterile lateral spikelets, the genotype def.2 showed the lowest values of lateral spikelet weight, spikelet width, spikelet length, spikelet area and TSW recording 0.00007 g, 0.54 mm, 3.46 mm, 0.97 mm2 and 0.07 g, respectively (Table 1). The genotype int-e.4 gave the highest values of spikelet weight, spikelet width, spikelet area and TSW recording 0.0026 g, 2.28 mm, 13.99 mm2 and 2.59 g, respectively. While the genotype int-f.19 gave the longest spikelet by length of 12.58 mm.

Table 1.
The statistical analysis of the lateral spikelet traits in the set of 30 barley lines used in this study. Mean squares (MS) of lateral spikelet weight (LSW), lateral spikelet width (LSWi), lateral spikelet length (LSL), lateral spikelet area (LSA) and thousand seed or spikelet weight (TSW) in the tested lines.
Among the four lines showing fertile lateral spikelets, hex-v.3, despite displaying the longest lateral spikelets (11.22 mm), exhibited the lowest values of lateral spikelet weight, width, area and TSW by 0.0224 g, 3.15 mm, 19.04 mm2 and 22.37 g, respectively (Table 1). The genotype HOR7191 showed the highest values for spikelet weight, width, area and TSW by 0.0477 g, 4.23 mm, 27.93 mm2 and 47.74 g, respectively. HOR1555 genotype gave the shortest spikelet by value of 9.77 mm.
For SEM analysis, for each individual line, five or more spikes were dissected from the developmental stages triple mound, glum primordia, stamen primordia, lemma primordia and awn primordia [24] from each of the lines. We detected clear variation among the tested lines (Figure 1). The lateral florets in the deficiens lines were more reduced than the semi-deficiens and two-rowed cultivars such as cv. Bonus. Interestingly, cv. Kristina, int-h.43 and int-i.39 showed the same appearance as def.5, def.6, semi-def.1, semi-def.8 in development and reduced lateral floret size. Up until the awn primordium stage, the lateral spikelets of the six-rowed lines HOR1555, HOR7191 and HOR7041 developed similar to those of two-rowed cultivars. After the awn primordia stage, however, lateral spikelets of these HOR lines continued their development, while those of two-rowed barley stopped differentiating at this stage, leading to lateral floret sterility.

Figure 1.
SEM of lateral spikelet development at awn primordia stage and spike form showing increase in the lateral spikelet size and development from left to right in the lines; deficiens phenotype (cv. Kristina, int-h.43, int-i39, def.5, def.6, semi-def.1, semi-def.8), semi-deficiens phenotype (semi-def.4, semi-def.5 and semi-def.7), two-rowed phenotype (cvs. Bonus, Bowman and Foma), intermedium phenotype (mutants int-a.1, int-c.5, int-e.20, int-e.4, int-f.19, int-l.81, int-m.85, HOR6211, HOR10166 and HOR6407) and Six-rowed phenotype (hex-v.3, HOR1555, HOR7191 and HOR7041).
2.2. HvMADS-box Genes Map
We identified the DNA and protein sequences of 108 different HvMADS-box genes located on all seven barley chromosomes; 13 on chromosome (chr) 1H, 14 on chr 2H, 17 on chr 3H, 9 on chr 4H, 13 on chr 5H, 19 on chr 6H and 23 on chr 7H (Figure 2). The gene annotation results showed that 30 of them are Agamous-like MADS-box transcription factors, 7 are K-box region and MADS-box transcription factor family proteins, and two are PISTILLATA-like MADS-box transcription factors. Fifty-one genes out of the 108 genes are orthologous to MADS-box genes in Arabidopsis or rice (or both). The other 57 genes could be unique to barley or yet unknown in other plants like Arabidopsis or rice (Table S3).
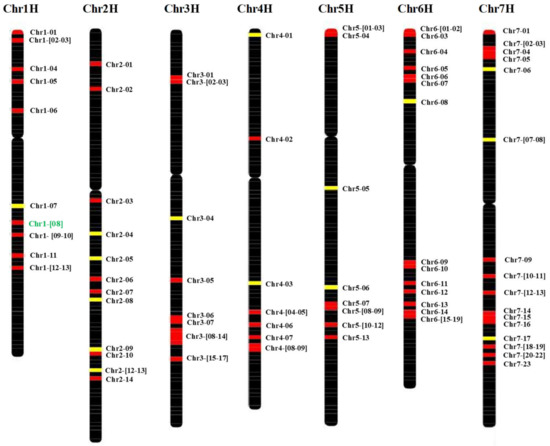
Figure 2.
Genetic map of the HvMADS-box genes. The map includes all seven barley chromosomes. Genes are indicated to the right of each interval position. High confidence (HC) genes are marked with red, while low confidence (LC) genes are colored with yellow. HvMADS56 is in green (Chr1-08). Full name of each gene is given in Table S4.
2.3. Phylogenetic Analysis of HvMADS-box Genes
We performed a phylogenetic analysis of the 108 HvMADS-box genes in barley based on their protein sequences. The results showed that these genes can be separated into three clades. The small clade includes 16 genes with 13 high confidence and 3 low confidence genes. The middle clade includes 27 genes with 16 high confidence and 11 low confidence genes. The remaining 65 genes make up the large clade, including 57 high confidence and 8 low confidence genes (Figure 3). The small clade (in purple color) includes HvMADS-box gene’ class AGL80 and one gene belonging to AGL65. The middle clade is divided into five sub-clades and includes many of the unknown yet MADS-box genes either in barley, rice or Arabidopsis. The last clade includes all HvMADS-box genes, which have been assigned a function through studies in barley or through studies with their orthologous proteins in rice and Arabidopsis (Figure 3 and Table S3).
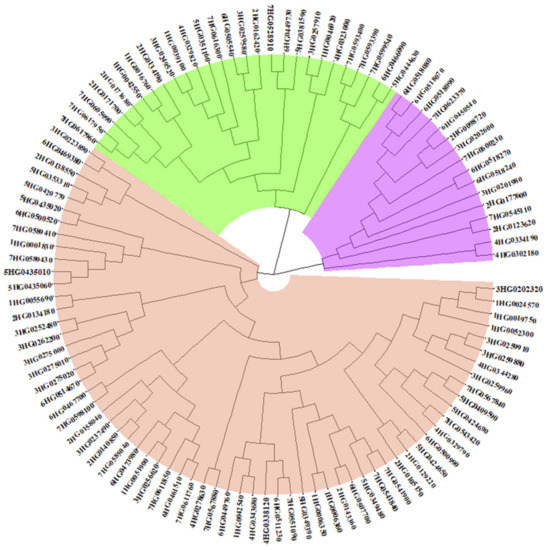
Figure 3.
Phylogenetic tree obtained for the 108 HvMADS-box genes. A total of 108 protein sequences were identified and annotated as MADS-box transcription factors. The genes names in the tree derived from their names in the barley reference genome, e.g., 5HG0409590 corresponds to HORVU.MOREX.r2.5HG0409590.
2.4. In Silico Analysis of Barley HvMADS-box Genes Expression
In order to understand how the different barley HvMADS-box transcription factors regulate floral initiation, development and growth, the Barlex database (https://apex.ipk-gatersleben.de/apex/f?p=284:10, accessed on 9 November 2021) was explored [25]. The Barlex database contains information on expression of genes in a wide range of barley tissues such as; 4-day embryos (EMB), roots from seedlings (ROO1), shoots from seedlings (LEA), young developing inflorescence (INF1), developing inflorescence (INF2), developing tillers (NOD), developing grains (CAR5), developing grain (CAR15), etiolated seedling (ETI), inflorescence lemma (LEM), inflorescence lodicule (LOD), dissected inflorescences palea (PAL), epidermal strips (EPI), inflorescences rachis (RAC), roots 28 days after planting (ROO2) and senescing leaves (SEN). It was found that 32 HvMADS-box genes were not expressed in any of the tested tissues, while 25 genes were expressed in all tissues. The remaining 51 genes showed different levels of expressions in one or more of the tested tissues (Table S3). Interestingly, none of the genes in the small clade showed any expression in the tested tissues except gene AGL65, which was expressed in all tissues. Of the genes in the second clade, eight showed no expression, six were expressed in all tissues, and the remaining 13 genes were expressed in one or more tissues. In the large clade, 12 genes showed no expression, 18 genes were expressed in all tissues, while the remaining 35 genes showed expression in one or more tissues (Table S3). In our study, we focused on the 14 HvMADS-box genes, which showed a high expression profile in the tissues or organs of the barley inflorescence (Figure 4). The results showed that HvMADS1, 3, 6, 7, 8, 13, 21, 29, 34 and 58 are specifically highly expressed in CAR5, while MADS2, 6, 7, 8, 22, 29 and 56 are found in LOD and MADS1, 5, 34 and 56 are highly presented in INF2.
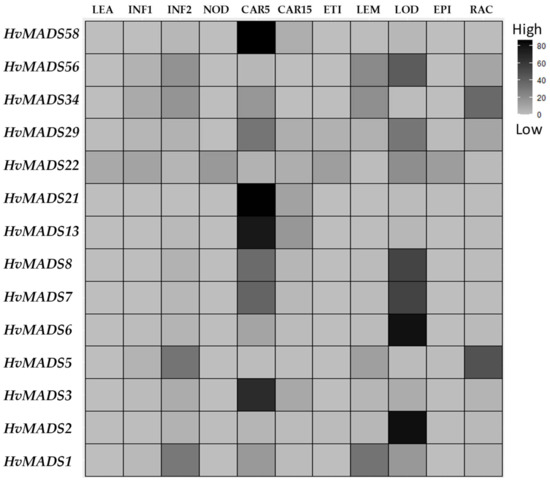
Figure 4.
Clustering analysis of 14 HvMADS-box transcription factor gene transcripts in different barley tissues. LEA, leaves 17 days after planting (DAP); developing inflorescence 5mm (INF1, about 30 DAP); developing inflorescence 10–15mm (INF2, about 50 DAP); developing tillers (NOD), developing grain 5 days after anthesis (CAR5), developing grain 15 days after anthesis (CAR15), etiolated seedling (ETI), inflorescence lemma (LEM), inflorescence lodicule (LOD), epidermal strips (EPI), inflorescences rachis (RAC). Raw data from the https://apex.ipk-gatersleben.de/apex/f?p=284:10 website (accessed on 9 November 2021).
2.5. MADS-box Genes Polymorphism and Lateral Spikelet Development
MADS-box genes, which were expressed mainly in tissues related to spikelet development, identified by our in silico analysis, were re-sequenced in all 30 lines in the present study to investigate the DNA sequence polymorphism that may affect spikelet development among these lines. The sequence variations in these genes showed polymorphism was largely absent among the tested lines except for HvMADS3, 6, 8, 34, 21 and 56 (Table 2).

Table 2.
Sequence variations and polymorphism at Hv-MADS3, Hv-MADS6, HvMADS8, HvMADS34, HvMADS21 and HvMADS56, including the nucleotide (nt) position of each among the tested lines.
In HvMADS3 (HORVU.MOREX.r2.3HG0202320), the results showed nucleotide substitutions from G to A in def.4 at nucleotide position 84. This single nucleotide polymorphism (SNP) did not change the protein. In contrast, a G to C substitution at nucleotide position 104 in HOR1555 and HOR7041 was a nonsynonymous SNP causing a change from serine to threonine (Table 2). The sequence polymorphism observed in HvMADS6 (HORVU.MOREX.r2.6HG0500990) was an 18 bp deletion from nucleotide 627 to 644 in HOR6211 and HOR6407 and a 21 bp deletion from nucleotide 627 to 647 in HOR10166. From the sequence data of HvMADS8 (HORVU.MOREX.r2.5HG0409590), we found a GGCGGCGGCGGC insertion in cv. Bonus in the promotor region before the ATG start codon. In addition, a synonymous SNP from C to T in HOR7041 was observed.
The re-sequence data of HvMADS34 (HORVU.MOREX.r2.5HG0424690) identified an SNP from G to A in the promotor region and a synonymous SNP from A to C at nucleotide position nine in HOR1555 and HOR7191. Re-sequencing of HvMADS21 (HORVU.MOREX.r2.1HG0052300) identified three synonymous SNPs from G to C at nucleotide position 123 in cv. Kristina, def.4, def.5 and def.6, semi-def.8, int-h.43 and int-l.81, and from A to G at nucleotide position 411 in cv. Kristina, def.4, semi-def.5, int-h.43, int-i.39 and HOR10166 and from G to T at nucleotide position 573 in HOR1555, HOR7191 and HOR7041. In addition, two insertions were observed: GGC at nucleotide positions 702-704 in HOR1555, HOR7191 and HOR7041 and C at nucleotide position 702, causing a frameshift and immature stop codon in cv. Foma, semi-def.5, semi-def.6, int-c.5, int-e.20, int-f.19 and hex-v.3. The results of re-sequence HvMADS56 (HORVU.MOREX.r2.1HG0042540) revealed a nonsynonymous SNP from T to A at nucleotide position 83, causing a change from leucine to glutamine in semi-def.4, semi-def.5 and semi-def.6, a synonymous SNP from C to A at nucleotide position 105 in def.4, def.5 and def.6, semi-def.8, int-h.43, int-i.39, HOR10166, HOR1555, HOR6211, HOR6407, HOR7191 and HOR7041. In addition, the results showed a G deletion at nucleotide position 321, causing a frameshift and immature stop codon in cv. Kristina, def.5, def.6, semi-def.8, int-h.43, int-i.39, HOR10166, HOR1555, HOR6211, HOR6407, HOR7191 and HOR7041, as well as a 67 bp deletion in semi-def.1 (Table 2). The SNP analysis revealed a correlation between lines with contrasting lateral spikelet phenotype and the SNPs and in/del polymorphisms detected in the HvMADS6 and HvMADS56. Therefore, the G deletion at nucleotide position 321 in HvMADS56 may be responsible for the significant lateral spikelet reduction in cv. Kristina, def.5, def.6, Semi-def.8, int-h.43, int-i.39, while the 67 bp deletion may account for the effect seen in semi-def.1. The lateral spikelet reduction in the three lines, HOR10166, HOR6211 and HOR6407, might be due to the 18 bp deletion in HOR6211 and HOR6407 and the 21 bp deletion in HOR10166 in HvMADS6 as well as the G deletion at nucleotide position 321 in all of the three lines in HvMADS56.
3. Discussion
The aim of the present study was to further elucidate the genetic basis of barley spike and spikelet development to increase seed yield. We used 30 different lines with variations in lateral spikelet formation. Genetic and phenotypic variation remains a cornerstone in plant breeding. Understanding the mechanisms of the relative fertility of central and lateral spikelets will provide novel solutions to enhanced grain yield in barley [11]. This could be achieved by increasing the number of seeds per spike which is a major goal of modern plant breeding programs [26]. The MADS-box genes family is considered to represent the first discovered transcription factor proteins [17,18]. In plants, MADS-box proteins are involved in various developmental processes during flowering [27,28,29] and are proposed to be the driving force behind floral diversity [19,20]. Therefore, a better understanding of their role and function in barley could help improve cereal breeding programs.
In the current study, we found a large variation among the tested lines for all central and lateral spikelet-related traits. All these traits had a broad-sense heritability of over 90% (Table 1 and Table S2), which confirmed that genetic effects are the major determinant of the phenotypic variance of these traits in barley. Our results are consistent with those obtained by [30,31], describing different fertility degrees for lateral spikelets in the intermedium-spike barley collection ranging from completely infertile (two-rowed like) to full fertile (six-rowed like). Our scanning electron micrographs of the tested lines revealed clear variation in the lateral spikelet development, especially at the awn primordia stage.
Kuijer et al. [23] analyzed 34 MADS-box genes and noticed that they regulate floret, spikelet and spike development in barley. Our phylogenetic analysis further complemented these results by revealing the highly conserved nature of 108 HvMADS-box genes in barley, more than half of which (76 genes) were expressed in different barley tissues. Expression data of many of these genes (Figure 4) from individual floral organs such as INF1, INF2, LEM, LOD and PAL suggested a role in floret and/or spikelet and spike development in barley. These results are consistent with those obtained by Schilling et al. [32] in wheat and Ciaffi et al. [33] in rice, maize and wheat.
We re-sequenced 14 of the HvMADS-box genes to investigate their role in lateral spikelet development. Sequence analysis of the 30 barley lines identified HvMADS6 and HvMDS56 as likely regulators of lateral spikelet development (Table 1 and Table 2). The G deletion and the 67 bp deletion in HvMADS56 and 18 bp and 21 bp deletions in HvMADS6 were likely candidates for inducing the reduced stage of lateral spikelets in cv. Kristina, def.5, def.6, semi-def.1, semi-def.8, int-h.43, int-i.39, HOR10166, HOR6211 and HOR6407.
HvMADS56, which is located on barley chromosome 1H, is very similar to the putative ortholog in rice (OsMADS56, on chromosome 10) and putative ortholog in Brachypodium (Bradi3g32090, on chromosome 3) [34,35,36] composed seven exons and six introns [32]. The annotation result of these genes showed that they contain the characteristic K-domain and belong to the MADS-box (type II) family [37]. Blasting the DNA and protein sequences of HvMADS56 to the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 9 November 2021) revealed that, HvMADS56 is similar to Arabidopsis SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) gene. Our in silico analysis showed that a maximum expression of HvMADS56 (HvSOC1) was in inflorescence development organs INF2, LEM, LOD, PAL and RAC. These expression results were in agreement with what was found in Arabidopsis [38]. Papaefthimiou et al. [39] studied HvSOC1-like1 and HvSOC1-like2 and found that both are expressed at different stages during the reproductive phase portentous their possible insinuation in seed development. This clearly suggests an important role of HvMADS56 (HvSOC1) in floret and spikelet development prior to the formation and setting of seeds. From the previous studies, SOC1 is known as a MADS-box transcription factor that is conserved and multifunctional protein in monocotyledons and dicotyledons [40,41,42,43,44,45], regulating flowering time, floral meristem patterning and determinacy [46,47,48]. It is also well known that hormones regulate floral organ patterning and spike and spikelet development in barley [10,16]. We suggest that the HvMADS56 (HvSOC1) gene is functionally acting upstream of floral meristem identity genes in barley. In Arabidopsis, Agamous-like genes such as AGL6, 17 and 24 and SOC1 interact and upregulate each other. The SOC1 gene regulates AP1, LFY, SEP3 and other flowering time genes (Figure 5). The effects of SOC1 have also been reported in other species [49,50]. We hypothesize that the activation of the floral meristem identity genes and subsequent plant flowering in barley are affected by the gibberellic acid pathway either directly or indirectly through HvSOC1 (Figure 5). This is in line with what is known about the role of SOC1 and GA in regulating flowering and inflorescence development in Arabidopsis and Orchid [49,50].
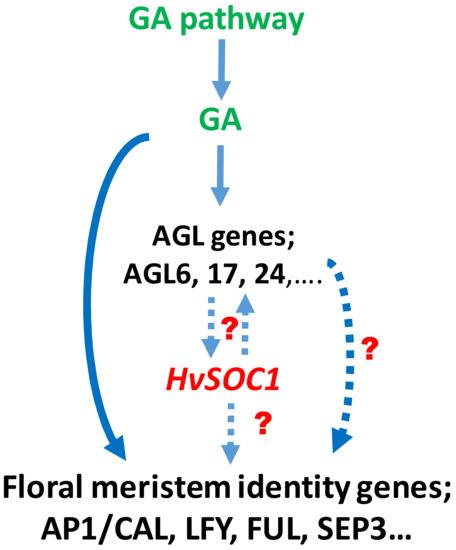
Figure 5.
Suggested role of HvMADS56 (HvSOC1) in regulating spikelet development in barley through its direct effect on the floral meristem identity genes or through stimulation of the AGAMOUS-like (AGL) genes activity. Hypothetical interactions that need further investigations in barley are indicated by dotted lines.
4. Conclusions
In order to address the recent focus of crop breeding programs on flower development and its effect on yield, we studied the role of MADS-box proteins in inflorescence development in barley. Our results highlight the importance of HvMADS56 in lateral spikelet development as well as floret and spikelet development prior to the formation and setting of seeds. The loss-of-function of HvMADS56 due to the two deletions (a G and a 67 bp deletion) might cause the reduction and late spikelet development in some of the tested lines in this study. We also found that some other lines showed a reduction in lateral spikelet size, but they did not carry a polymorphism at the tested MADS-box genes. Thus, we conclude that HvMADS56 (HvSOC1) not only plays an important role in floret and spikelet development but also other novel genes might be involved as well. Further investigations will be needed to solidify this conclusion and identify the relation between HvMADS56 and the other genes underlying the spike and spikelet development in barley.
5. Materials and Methods
5.1. Plant Materials
Seeds of 30 spring barley lines (mutants, accessions and cultivars) with variation in lateral spikelet size were studied; two-rowed (4 lines; Foma, Bowman, Bonus, Kristina), deficiens (5 lines; def.2, def.4 to def.6 and def.8), semi-deficiens (5 lines; semi-def.1, semi-def.4, semi-def.5, semi-def.7 and semi-def.8), intermedium (int) mutants (9 lines; int-a.1, int-c.5, int-e.4, int-e.20, int-f.19, int-h.43, int-i.39, int-l.81, and int-m.85), hexachiton mutants (hex-v.3) and 6 accessions from the natural barley collection classified as intermedium barley [9], barley accessions (Hordeum vulgare L. convar. intermedium (Körn.) Mansf.). All the lines were obtained from NordGen (the Nordic Genetic Resource Center, Alnarp, Sweden), except the 6 lines of the natural collection that were obtained from the German Federal ex situ Genebank hosted at IPK Gatersleben, Germany (Table S5).
5.2. Growth Conditions and Spike Phenotyping
Three replicates from each accession of the 30 lines were germinated and grown in the greenhouse facilities at the Department of Biology, Lund University, Lund, Sweden, between March and August 2019. The plants were grown in potting soil from SW Horto, Sweden (swhorto.se, article number 744704) in 1.5-L pots (a single plant/pot, 14 cm diameter and 14 cm height) under long-day conditions, 16 h light/8 h dark and temperature 20 ± 2 °C during the day and 16 ± 2 °C during the night. After harvest, the main culm spike of each plant was scored for central and lateral seed size; seeds weight, seed length, seed width, seed area, thousand seed weight, as well as plant seed number and plant seed weight, plant height (from the soil surface to the base of the spike), spike length (from the base to the tip of the spike without awns) and number per plant and number of tillers per plant. In addition to the lines which did not show fertility for the lateral spikelets, spikelet weight, spikelet length, spikelet width and spikelet area of 20 lateral florets from ten rachis nodes from the middle of the spike were measured after harvesting. The traits were measured with a MARVIN Seed Analyzer (GTA Sensorik, Neubrandenburg, Germany).
5.3. Scanning Electron Microscopy
Immature spike tissues (from five or more plants) at different spike developmental stages; triple mound, glum primordia, stamen primordia, lemma primordia and awn primordia were used for scanning electron microscopy (SEM) (Hisco Europe, Ratingen, Germany), which was conducted as previously described by Lolas et al. [51].
5.4. Genomic DNA Isolation, Amplification and Sequencing
Leaf samples were collected in 96-well plates from seedlings with 2–3 leaves. Genomic DNA extraction and polymerase chain reaction (PCR) amplifications were performed as described in Matyszczak et al. [52]. For each of the tested MADS-box genes in this study, primer pairs were designed to obtain sequence data for the whole gene by Sanger sequencing. Primer sequences, annealing temperatures and fragment lengths for all genes are provided in Table S6. PCR amplification profile with initial denaturation step: 3 min at 96 °C followed by the main program for 40 cycles at 96 °C for 40 s, 60 °C for 40 s and extension at 72 °C for 2 min, followed by a final extension for 12 min at 72 °C. The PCR products were tested on 1% agarose gel. For Sanger-sequencing, 2 µL of Exoprostar was added to 5 µL of the PCR products. The mixture was incubated at 37 °C for 15 min followed by 80 °C for 15 min. After that, 8 µL H2O and 2 µL of 10 μM primer were added to the Mixture and sent to the sequencing service offered by Eurofins Genomics, Germany.
5.5. Sequence Analysis and Sequence Homology Searches
Sequencher 5.2.3 DNA sequence assembly software (Gene Codes Corporation) was used for DNA sequencing analysis, quality score assignments and construction of contigs. Multiple sequence alignments were carried out using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 9 November 2021). Barley DNA sequences of the MADS-box genes with respective genetic and physical locations were extracted from [53,54] using the barley genome explore (Barlex) (https://apex.ipk-gatersleben.de/apex/f?p=284:10::::::, accessed on 9 November 2021). The positions of the MADS-box genes on all barley chromosomes were plotted using the R package ChromoMap [55].
5.6. Phylogenetic Analysis
MADS-box protein sequences were isolated using the barley genome explorer (Barlex) (https://apex.ipk-gatersleben.de/apex/f?p=284:10::::::, accessed on 9 November 2021). Protein sequences (108) were isolated and annotated as the MADS-box transcription factor. A multiple sequence alignment was performed using the R-package DECIPHER [56], and then a circular phylogenetic tree was generated using the R-package ggtree [57].
5.7. Expression Data Analysis
Transcript data of the MADS-box genes for the different barley plant tissues were isolated from the https://apex.ipk-gatersleben.de/apex/f?p=284:10:::::: website as FPKM—fragments per kilobase of exon model per million reads mapped (accessed on 9 November 2021). Source: [25].
5.8. Statistical Analysis
The analysis of variance (ANOVA) of a randomized complete block design (RCBD) experiment with three replications of the agronomic and central spikelet-related traits and with four replications of the lateral spikelet-related traits was performed using SAS software v.9.2 with PROC GLM procedure [58]. Means were compared by a Fisher’s least significant difference (LSD) procedure at 0.05 level of significance. Broad-sense heritability (Hb) estimates were calculated under control according to [59].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10122825/s1. Table S1: Summary statistics of the agronomic, central spikelets and lateral spikelets related traits in the 30 tested lines; Table S2: The statistical analysis of variance, coefficient of determination (R2), broad-sense heritability (Hb) and coefficient of variation (CV%) of the agronomic traits, central spikelet traits and lateral spikelet traits in the set of 30 barley lines used in this study; Table S3: List of MADS-box genes including gene ID, gene sequence, protein sequence, tissue of expression, chromosomal position and start and end of each gene on the barley chromosomes; Table S4: Genes ID for the map in Figure 2; Table S5: List of tested lines used in the study; Table S6: Primer sequences, annealing temperatures and fragment lengths for the tested genes.
Author Contributions
H.M.Y. conceived the study; H.M.Y., M.A.S., M.A., Q.K.H., I.U., T.R., D.S. and S.Z. performed experiments; H.M.Y., M.A.S. and M.A. analyzed data; K.P., A.B. and M.H. helped in the interpretation of results; H.M.Y., M.A.S. and I.U. wrote the paper with input from all coauthors. All authors have read and agreed to the published version of the manuscript.
Funding
German Research Foundation (DFG YO 304/1-1) to H.M.Y., SusCrop-ERA-NET project BARISTA (BMBF: 031B0811B) to K.P. and H.M.Y., the Swedish Research Council (VR 2018-05117 to M.H.), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS 2018-01026) to M.H., the Erik Philip-Sörensen Foundation to M.H. and the Royal Physiographic Society in Lund to D.S., H.Y. and S.Z.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Acknowledgments
This work was supported by the German Research Foundation (DFG YO 304/1-1 to H.Y.), the Swedish Research Council (VR 2018-05117 to M.H.), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS 2018-01026 to M.H.), the Erik Philip-Sörensen Foundation (M.H.), the Royal Physiographic Society in Lund (D.S., H.Y., S.Z.). We are also thankful to Udda Lundqvist for providing germplasm.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Donald, C.M. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Plant science: Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Bommert, P.; Nagasawa, N.S.; Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013, 45, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaya, O.; Pont, C.; Sibout, R.; Martinek, P.; Badaeva, E.; Murat, F.; Chosson, A.; Watanabe, N.; Prat, E.; Gautier, N.; et al. Frizzy panicle drives supernumerary spikelets in bread wheat. Plant Physiol. 2015, 167, 189–199. [Google Scholar] [CrossRef] [Green Version]
- Poursarebani, N.; Seidensticker, T.; Koppolu, R.; Trautewig, C.; Gawroński, P.; Bini, F.; Govind, G.; Rutten, T.; Sakuma, S.; Tagiri, A.; et al. The genetic basis of composite spike form in barley and ‘miracle-wheat’. Genetics 2015, 201, 155–165. [Google Scholar] [CrossRef]
- Youssef, H.M.; Mascher, M.; Ayoub, M.A.; Stein, N.; Kilian, B.; Schnurbusch, T. Natural diversity of inflorescence architecture traces cryptic domestication genes in barley (Hordeum vulgare L.). Genet. Resour. Crop Evol. 2017, 64, 843–853. [Google Scholar] [CrossRef]
- Forster, B.P.; Franckowiak, J.D.; Lundqvist, U.; Lyon, J.; Pitkethly, I.; Thomas, W.T.B. The barley phytomer. Ann. Bot. 2007, 100, 725–733. [Google Scholar] [CrossRef] [Green Version]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, K.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 2007, 104, 1424–1429. [Google Scholar] [CrossRef] [Green Version]
- Mansfeld, R. Das morphologische System der Saatgerste, Hordeum vulgare L. sl. Züchter 1950, 64, 843–853. [Google Scholar] [CrossRef]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef]
- Bull, H.; Casao, M.C.; Zwirek, M.; Flavell, A.J.; Thomas, W.T.B.; Guo, W.; Zhang, R.; Rapazote-Flores, P.; Kyriakidis, S.; Russell, J.; et al. Barley SIX-ROWED SPIKE3 encodes a putative Jumonji C-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat. Commun. 2017, 8, 936. [Google Scholar] [CrossRef] [Green Version]
- Van Esse, G.W.; Walla, A.; Finke, A.; Koornneef, M.; Pecinka, A.; von Korff, M. Six-rowed spike3 (VRS3) is a histone demethylase that controls lateral spikelet development in Barley. Plant Physiol. 2017, 174, 2397–2408. [Google Scholar] [CrossRef] [Green Version]
- Koppolu, R.; Anwar, N.; Sakuma, S.; Tagiri, A.; Lundqvist, U.; Pourkheirandish, M.; Rutten, T.; Seiler, C.; Himmelbach, A.; Ariyadasa, R.; et al. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. USA 2013, 110, 13198–13203. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, L.; Comadran, J.; Druka, A.; Marshall, D.F.; Thomas, W.T.B.; MacAulay, M.; MacKenzie, K.; Simpson, C.; Fuller, J.; Bonar, N.; et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 2011, 43, 169–172. [Google Scholar] [CrossRef]
- Boden, S.A. How hormones regulate floral architecture in barley. Nat. Genet. 2017, 49, 8–9. [Google Scholar] [CrossRef]
- Youssef, H.M.; Hansson, M. Crosstalk among hormones in barley spike contributes to the yield. Plant Cell Rep. 2019, 38, 1013–1016. [Google Scholar] [CrossRef] [Green Version]
- Shore, P.; Sharrocks, A.D. The MADS-Box Family of Transcription Factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef]
- Lawton-Rauh, A.L.; Alvarez-Buylla, E.R.; Purugganan, M.D. Molecular evolution of flower development. Trends Ecol. Evol. 2000, 15, 144–149. [Google Scholar] [CrossRef]
- Theißen, G.; Saedler, H. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Hirano, H.Y. Function and diversification of MADS-box genes in rice. Sci. World J. 2006, 6, 1923–1932. [Google Scholar] [CrossRef]
- Callens, C.; Tucker, M.R.; Zhang, D.; Wilson, Z.A. Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 2018, 69, 2435–2459. [Google Scholar] [CrossRef]
- Yoshida, H.; Nagato, Y. Flower development in rice. J. Exp. Bot. 2011, 62, 4719–4730. [Google Scholar] [CrossRef] [Green Version]
- Kuijer, H.N.J.; Shirley, N.J.; Khor, S.F.; Shi, J.; Schwerdt, J.; Zhang, D.; Li, G.; Burton, R.A. Transcript Profiling of MIKCc MADS-Box Genes Reveals Conserved and Novel Roles in Barley Inflorescence Development. Front. Plant Sci. 2021, 12, 1834. [Google Scholar] [CrossRef]
- Kirby, E.J.M.; Appleyard, M. Cereal Development Guide, 2nd ed.; NAC Cereal Unit: Stoneleigh, UK, 1987; 85p. [Google Scholar]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX—The barley draft genome explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, S.; Lundqvist, U.; Kakei, Y.; Thirulogachandar, V.; Suzuki, T.; Hori, K.; Wu, J.; Tagiri, A.; Rutten, T.; Koppolu, R.; et al. Extreme suppression of lateral floret development by a single amino acid change in the VRS1 transcription factor. Plant Physiol. 2017, 175, 1720–1731. [Google Scholar] [CrossRef] [Green Version]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [Green Version]
- Pelucchi, N.; Fornara, F.; Favalli, C.; Masiero, S.; Lago, C.; Pè, E.M.; Colombo, L.; Kater, M.M. Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 2002, 15, 113–122. [Google Scholar] [CrossRef]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, G.; Ren, X.; Du, B.; Cheng, Y.; Wang, Y.; Li, C.; Sun, D. Dissecting the genetic basis of grain size and weight in barley (Hordeum vulgare L.) by QTL and comparative genetic analyses. Front. Plant Sci. 2019, 10, 469. [Google Scholar] [CrossRef]
- Youssef, H.M.; Allam, M.; Boussora, F.; Himmelbach, A.; Milner, S.G.; Mascher, M.; Schnurbusch, T. Dissecting the genetic basis of lateral and central spikelet development and grain traits in intermedium-spike barley (Hordeum vulgare convar. intermedium). Plants 2020, 9, 1655. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciaffi, M.; Paolacci, A.R.; Tanzarella, O.A.; Porceddu, E. Molecular aspects of flower development in grasses. Sex. Plant Reprod. 2011, 24, 247–282. [Google Scholar] [CrossRef] [PubMed]
- Bolot, S.; Abrouk, M.; Masood-Quraishi, U.; Stein, N.; Messing, J.; Feuillet, C.; Salse, J. The ‘inner circle’ of the cereal genomes. Curr. Opin. Plant Biol. 2009, 12, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Martis, M.; Hedley, P.E.; Šimková, H.; Liu, H.; Morris, J.A.; Steuernagel, B.; Taudien, S.; Roessner, S.; Gundlach, H.; et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 2011, 23, 1249–1263. [Google Scholar] [CrossRef] [Green Version]
- Wicker, T.; Mayer, K.F.X.; Gundlach, H.; Martis, M.; Steuernagel, B.; Scholz, U.; Šimková, H.; Kubaláková, M.; Choulet, F.; Taudien, S.; et al. Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 2011, 23, 1706–1718. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of constans target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [Green Version]
- Papaefthimiou, D.; Kapazoglou, A.; Tsaftaris, A.S. Cloning and characterization of SOC1 homologs in barley (Hordeum vulgare) and their expression during seed development and in response to vernalization. Physiol. Plant. 2012, 146, 71–85. [Google Scholar] [CrossRef]
- Lee, H.; Suh, S.S.; Park, E.; Cho, E.; Ahn, J.H.; Kim, S.G.; Lee, J.S.; Kwon, Y.M.; Lee, I. The AGAMOUS-lIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kim, J.; Han, J.J.; Han, M.J.; An, G. Functional analyses of the flowering time gene OsMADS50, the putative Suppressor of Overexpression of CO1/Agamous-Like 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004, 38, 754–764. [Google Scholar] [CrossRef]
- Lee, J.; Oh, M.; Park, H.; Lee, I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 2008, 55, 832–843. [Google Scholar] [CrossRef]
- Cseke, L.J.; Zheng, J.; Podila, G.K. Characterization of PTM5 in aspen trees: A MADS-box gene expressed during woody vascular development. Gene 2003, 318, 55–67. [Google Scholar] [CrossRef]
- Ferrario, S.; Busscher, J.; Franken, J.; Gerats, T.; Vandenbussche, M.; Angenent, G.C.; Immink, R.G.H. Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 2004, 16, 1490–1505. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Song, I.J.; Fukuda, T.; Yokoyama, J.; Maki, M.; Ochiai, T.; Kameya, T.; Kanno, A. Characterization of TrcMADS1 gene of Trillium camtschatcense (Trilliaceae) reveals functional evolution of the SOC1/TM3-like gene family. J. Plant Res. 2005, 118, 229–234. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, J.; Bracha-Drori, K.; Yalovsky, S.; Ito, T.; Yu, H. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 2007, 134, 1901–1910. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of Floral Patterning by Flowering Time Genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [Green Version]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; García-Ponce, B.; Sánchez, M.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-Box Genes Are Key Components of Genetic Regulatory Networks Involved in Abiotic Stress and Plastic Developmental Responses in Plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef] [Green Version]
- Teo, Z.W.N.; Zhou, W.; Shen, L. Dissecting the Function of MADS-Box Transcription Factors in Orchid Reproductive Development. Front. Plant Sci. 2019, 10, 1474. [Google Scholar] [CrossRef]
- Lolas, I.B.; Himanen, K.; Grønlund, J.T.; Lynggaard, C.; Houben, A.; Melzer, M.; Lijsebettens, M.V.; Grasser, K.D. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010, 61, 686–697. [Google Scholar] [CrossRef]
- Matyszczak, I.; Tominska, M.; Zakhrabekova, S.; Dockter, C.; Hansson, M. Analysis of early-flowering genes at barley chromosome 2H expands the repertoire of mutant alleles at the Mat-c locus. Plant Cell Rep. 2020, 39, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Mascher, M.; Richmond, T.A.; Gerhardt, D.J.; Himmelbach, A.; Clissold, L.; Sampath, D.; Ayling, S.; Steuernagel, B.; Pfeifer, M.; D’Ascenzo, M.; et al. Barley whole exome capture: A tool for genomic research in the genus Hordeum and beyond. Plant J. 2013, 76, 494–505. [Google Scholar] [CrossRef] [Green Version]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Anand, L.; Rodriguez Lopez, C.M. ChromoMap: An R package for Interactive Visualization and Annotation of Chromosomes. bioRxiv 2019, 605600. [Google Scholar] [CrossRef] [Green Version]
- Wright, E.S. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- SAS Institute Inc. The SAS System for Windows, Release 9.0; SAS Institute: Cary, NC, USA, 2002. [Google Scholar]
- Padi, F.K. Genotype × environment interaction and yield stability in a cowpea-based cropping system. Euphytica 2007, 158, 11–25. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).