Chromosome and Genome Diversity in the Genus Trifolium (Fabaceae)
Abstract
1. Introduction
2. Trifolium Genomes, Chromosomes, and Chromosome Number Variation
3. Current State of Knowledge on Trifolium Genomes
4. Hybridization in the Clover Genus
5. Chromosome Identification in Trifolium
6. Chromosomal Distribution of Ribosomal DNA Genes
7. Other Repetitive and Single-Copy Markers
8. Comparative Mapping in Legumes
9. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Zohary, M.; Heller, D. The Genus Trifolium, 1st ed.; Israel Academy of Sciences and Humanities: Jerusalem, Israel, 1984; pp. 1–610. [Google Scholar]
- Gillet, J.M.; Taylor, N.L. The World of Clovers, 1st ed.; Iowa State University Press: Ames, IA, USA, 2001; pp. 1–457. [Google Scholar]
- Ellison, N.W.; Liston, A.; Steiner, J.J.; Williams, W.M.; Taylor, N.L. Molecular phylogenetics of the clover genus (Trifolium-Leguminosae). Mol. Phylogenet. Evol. 2006, 39, 688–705. [Google Scholar] [CrossRef] [PubMed]
- Zohary, M. Origin and evolution in the genus Trifolium. Bot. Notiser. 1972, 125, 501–511. [Google Scholar]
- Taylor, N.L. Clovers Around the World. In Agronomy Monographs; Taylor, N.L., Ed.; Soil Science Society of America: Madison, WI, USA, 1985; Volume 25, pp. 1–6. [Google Scholar]
- Panitsa, M.; Trigas, P.; Iatrou, G.; Sfenthourakis, S. Factors affecting plant species richness and endemism on land-bridge islands—An example from the East Aegean Archipelago. Acta Oceol. 2010, 36, 431–437. [Google Scholar] [CrossRef]
- Scoppola, A.; Tirado, J.L.; Gutiérrez, F.M.; Magrini, S. The genus Trifolium (Fabaceae) in South Europe: A critical review on species richness and distribution. Nord. J. Bot. 2018, 36, e01723. [Google Scholar] [CrossRef]
- Boissier, E. Trifolium. In Flora Orientalis; H. Georg: Basileae, Switzerland, 1872; pp. 110–156. [Google Scholar]
- Hossain, M. A revision of Trifolium in the nearer East. Notes R. Bot. Gard. Edinb. 1961, 23, 387–481. [Google Scholar]
- Lavin, M.; Doyle, J.J.; Palmer, J.D. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 1990, 44, 390–402. [Google Scholar]
- Liston, A. Use of the polymerase chain reaction to survey for the loss of the inverted repeat in the legume chloroplast genome. In Advances in Legume Systematics; Crisp, M.D., Doyle, J.J., Eds.; Royal Botanic Gardens: Kew, UK, 1995; Volume 7, pp. 31–40. [Google Scholar]
- Watson, L.E.; Sayed-Ahmed, H.; Badr, A. Molecular phylogeny of Old World Trifolium (Fabaceae). Plant Syst. Evol. 2000, 224, 153–171. [Google Scholar] [CrossRef]
- Steele, K.; Wojciechowski, M. Phylogenetic analyses of tribes Trifolieae and Vicieae, based on sequences of the plastid gene matK (Papilionoideae: Leguminosae). Adv. Legume Syst. 2003, 1, 355–370. [Google Scholar]
- Sveinsson, S.; Cronk, Q. Evolutionary origin of highly repetitive plastid genomes within the clover genus (Trifolium). BMC Evol. Biol. 2014, 14, 228. [Google Scholar] [CrossRef]
- Kintl, A.; Elbl, J.; Lošák, T.; Vaverková, M.; Nedělník, J. Mixed intercropping of wheat and white clover to enhance the sustainability of the conventional cropping system: Effects on biomass production and leaching of mineral nitrogen. Sustainability 2018, 10, 3367. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.-K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Kim, J.-S.; Islam-Faridi, M.N.; Klein, P.E.; Stelly, D.M.; Price, H.J.; Klein, R.R.; Mullet, J.E. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: Distribution of euchromatin, heterochromatin, genes and recombination in comparison to cice. Genetics 2005, 171, 1963–1976. [Google Scholar] [CrossRef]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef]
- Schubert, I. Chromosome Evolution. Curr. Opin. Plant Biol. 2007, 10, 109–115. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.P.; Schwarzacher, T. Organisation of the plant genome in chromosomes: Organisation of the plant genome in chromosomes. Plant J. 2011, 66, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Liu, B.; Segal, G.; Abbo, S.; Levy, A.A.; Vega, J.M. Rapid elimination of low-copy dna sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 1997, 147, 1381–1387. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Hiscock, S.J. Hybrid speciation in plants: New insights from molecular studies. New Phytol. 2005, 165, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F.; Schnabel, A.; Seelanan, T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 1995, 92, 280–284. [Google Scholar] [CrossRef]
- Plant DNA C-Value Database. Available online: https://cvalues.science.kew.org/ (accessed on 20 August 2021).
- Taylor, N.L.; Quesenberry, K.H.; Anderson, M.K. Genetic system relationships in Trifolium. Econ. Bot. 1979, 33, 431–441. [Google Scholar] [CrossRef]
- Index to Plant Chromosome Numbers. Available online: http://legacy.tropicos.org/Project/IPCN (accessed on 3 June 2021).
- Salimpour, F.; Sharifnia, F.; Mostafavi, G.; Hajrasoliha, S.; Ukhneh, E. chromosome counts and determination of ploid levels in iranian species of Trifolium. Chromosome Bot. 2008, 3, 53–63. [Google Scholar] [CrossRef][Green Version]
- Uslu, E. Karyology of Nine Trifolium L. Taxa from Turkey. Caryologia 2012, 65, 304–310. [Google Scholar] [CrossRef]
- Chromosome Counts Database. Available online: http://ccdb.tau.ac.il/ (accessed on 20 August 2021).
- Vižintin, L.; Javornik, B.; Bohanec, B. Genetic characterization of selected Trifolium species as revealed by nuclear DNA content and ITS rDNA region analysis. Plant Sci. 2006, 170, 859–866. [Google Scholar] [CrossRef]
- Vozárová, R.; Macková, E.; Vlk, D.; Řepková, J. Variation in ribosomal DNA in the genus Trifolium (Fabaceae). Plants 2021, 10, 1771. [Google Scholar] [CrossRef]
- Lawrence, M. Population Genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Ann. Bot.-Lond. 2000, 85, 221–226. [Google Scholar] [CrossRef]
- Abberton, M.T. Interspecific hybridization in the genus Trifolium. Plant Breed. 2007, 126, 337–342. [Google Scholar] [CrossRef]
- Williams, W.M.; Ellison, N.W.; Ansari, H.A.; Verry, I.M.; Hussain, S. Experimental evidence for the ancestry of allotetraploid Trifolium repens and creation of synthetic forms with value for plant breeding. BMC Plant Biol. 2012, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Egan, L.M.; Hofmann, R.W.; Ghamkhar, K.; Hoyos-Villegas, V. Prospects for Trifolium improvement through germplasm characterisation and pre-breeding in New Zealand and beyond. Front. Plant Sci. 2021, 12, 653191. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.M.F. Inbreeding and fertility in egyptian clover, Trifolium alexandrinum. J. Pharmacogn. Phytother. 2012, 4, 16–25. [Google Scholar]
- Schifino, M.T.; Moreas-Fernandes, M.I.B. Cytological comparision of diploid and autotetraploid Trifolium riograndense Burkart (Leguminosae). Rev. Bras. Genet. IX 1986, 4, 637–643. [Google Scholar]
- Schifino-Wittmann, M.T.; Moraes-Fernandes, B. Chromosome numbers, karyotypes and meiotic behavior of populations of some Trifolium (Leguminosae) species. Rev. Brazil Genet. 1988, 11, 379–390. [Google Scholar]
- Sheidai, M.; Hamta, A.; Mofidabadi, A.J.; NooriDaloii, M.R. Karyotypic study of Trifolium species and cultivars in Iran. J. Sci. Islam. Repub. Iran 1998, 9, 215–222. [Google Scholar]
- Khatoon, S.; Ali, S.I. Chromosome numbers and polyploidy in the legumes of Pakistan. Pak. J. Bot. 2006, 38, 935–945. [Google Scholar]
- Conterato, I.F.; Schifino-Wittmann, M.T.; Dall′Agnol, M. Seed dimorphism, chromosome number and karyotype of the amphicarpic species Trifolium argentinense Speg. Genet. Resour. Crop. Evol. 2010, 57, 727–731. [Google Scholar] [CrossRef]
- Isobe, S.; Klimenko, I.; Ivashuta, S.; Gau, M.; Kozlov, N.N. First RFLP linkage map of red clover (Trifolium pratense L.) based on cDNA probes and its transferability to other red clover germplasm. Theor. Appl. Genet. 2003, 108, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Isobe, S.; Asamizu, E.; Ohmido, N.; Kataoka, R.; Nakamura, Y.; Kaneko, T.; Sakurai, N.; Okumura, K.; Klimenko, I.; et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res. 2005, 12, 301–364. [Google Scholar] [CrossRef]
- Isobe, S.; Kolliker, R.; Hisano, H.; Sasamoto, S.; Wada, T.; Klimenko, I.; Okumura, K.; Tabata, S. Construction of a consensus linkage map for red clover (Trifolium pratense L.). BMC Plant Biol. 2009, 9, 57. [Google Scholar] [CrossRef]
- Zhang, Y.; Sledge, M.K.; Bouton, J.H. Genome mapping of white clover (Trifolium repens L.) and comparative analysis within the Trifolieae using cross-species SSR markers. Theor. Appl. Genet. 2007, 114, 1367–1378. [Google Scholar] [CrossRef]
- Ghamkhar, K.; Isobe, S.; Nichols, P.G.H.; Faithfull, T.; Ryan, M.H.; Snowball, R.; Sato, S.; Appels, R. The first genetic maps for subterranean clover (Trifolium subterraneum L.) and comparative genomics with T. pratense L. and Medicago Truncatula Gaertn. to identify new molecular markers for breeding. Mol. Breed. 2012, 30, 213–226. [Google Scholar] [CrossRef]
- Isobe, S.N.; Hisano, H.; Sato, S.; Hirakawa, H.; Okumura, K.; Shirasawa, K.; Sasamoto, S.; Watanabe, A.; Wada, T.; Kishida, Y.; et al. Comparative genetic mapping and discovery of linkage disequilibrium across linkage groups in white clover (Trifolium repens L.). G3-Genes Genomes Genet. 2012, 2, 607–617. [Google Scholar] [CrossRef]
- Jones, E.S.; Hughes, L.J.; Drayton, M.C.; Abberton, M.T.; Michaelson-Yeates, T.P.T.; Bowen, C.; Forster, J.W. An SSR and AFLP molecular marker-based genetic map of white clover (Trifolium repens L.). Plant Sci. 2003, 165, 531–539. [Google Scholar] [CrossRef]
- Barrett, B.; Griffiths, A.; Schreiber, M.; Ellison, N.; Mercer, C.; Bouton, J.; Ong, B.; Forster, J.; Sawbridge, T.; Spangenberg, G.; et al. a microsatellite map of white clover. Theor. Appl. Genet. 2004, 109, 596–608. [Google Scholar] [CrossRef]
- Griffiths, A.G.; Barrett, B.A.; Simon, D.; Khan, A.K.; Bickerstaff, P.; Anderson, C.B.; Franzmayr, B.K.; Hancock, K.R.; Jones, C.S. An integrated genetic linkage map for white clover (Trifolium repens L.) with alignment to Medicago. BMC Genom. 2013, 14, 388. [Google Scholar] [CrossRef]
- Ištvánek, J.; Jaroš, M.; Křenek, A.; Řepková, J. Genome assembly and annotation for red clover (Trifolium pratense; Fabaceae). Am. J. Bot. 2014, 101, 327–337. [Google Scholar] [CrossRef]
- De Vega, J.J.; Ayling, S.; Hegarty, M.; Kudrna, D.; Goicoechea, J.L.; Ergon, Å.; Rognli, O.A.; Jones, C.; Swain, M.; Geurts, R.; et al. Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 2015, 5, 17394. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Dluhošová, J.; Ištvánek, J.; Nedělník, J.; Řepková, J. Red clover (Trifolium pratense) and zigzag clover (T. medium)—A picture of genomic similarities and differences. Front. Plant. Sci. 2018, 9, 724. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Kaur, P.; Shirasawa, K.; Nichols, P.; Nagano, S.; Appels, R.; Erskine, W.; Isobe, S.N. Draft genome sequence of subterranean clover, a reference for genus Trifolium. Sci. Rep. 2016, 6, 30358. [Google Scholar] [CrossRef]
- Griffiths, A.G.; Moraga, R.; Tausen, M.; Gupta, V.; Bilton, T.P.; Campbell, M.A.; Ashby, R.; Nagy, I.; Khan, A.; Larking, A.; et al. Breaking Free: The genomics of allopolyploidy-facilitated niche expansion in white clover. Plant Cell 2019, 31, 1466–1487. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M. Clovers. In Evolution of Crop Plants, 1st ed.; Simmonds, N.W., Ed.; Longman: London, UK, 1976; Volume 4, pp. 79–98. [Google Scholar]
- Ansari, H.A.; Ellison, N.W.; Williams, W.M. Molecular and cytogenetic evidence for an allotetraploid origin of Trifolium dubium (Leguminosae). Chromosoma 2008, 117, 159–167. [Google Scholar] [CrossRef]
- Neumann, P.; Navrátilová, A.; Schroeder-Reiter, E.; Koblížková, A.; Steinbauerová, V.; Chocholová, E.; Novák, P.; Wanner, G.; Macas, J. Stretching the rules: Monocentric chromosomes with multiple centromere domains. PLoS Genet. 2012, 8, e1002777. [Google Scholar] [CrossRef]
- Macas, J.; Novák, P.; Pellicer, J.; Čížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 2015, 10, e0143424. [Google Scholar] [CrossRef]
- Ávila Robledillo, L.; Koblížková, A.; Novák, P.; Böttinger, K.; Vrbová, I.; Neumann, P.; Schubert, I.; Macas, J. Satellite DNA in Vicia faba is characterized by remarkable diversity in its sequence composition, association with centromeres, and replication timing. Sci. Rep. 2018, 8, 5838. [Google Scholar] [CrossRef]
- Taylor, N.L.; Quesenberry, K.H. Red Clover Science; Kluwer Academic: Dordrecht, The Netherlands, 1996; Volume 28. [Google Scholar]
- Williams, W.M. Trifolium interspecific hybridisation: Widening the white clover gene pool. Crop Pasture Sci. 2014, 65, 1091. [Google Scholar] [CrossRef]
- Řepková, J.; Nedbálková, B.; Holub, J. Regeneration of plants from zygotic embryos after interspecific hybridization within the genus Trifolium and electrophoretic evaluation of hybrids. Sci. Stud. Res. Inst. Fodd. Plants 1991, 12, 7–14. [Google Scholar]
- Řepkova, J.; Jungmannová, B.; Jakešová, H. Identification of barriers to interspecific crosses in the genus Trifolium. Euphytica 2006, 151, 39–48. [Google Scholar] [CrossRef]
- Fuchs, J.; Strehl, S.; Brandes, A.; Schweizer, D.; Schubert, I. Molecular-cytogenetic characterization of the Vicia faba genome—Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res. 1998, 6, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Kühne, M.; Schubert, I. Assignment of linkage groups to pea chromosomes after karyotyping and gene mapping by fluorescent in situ hybridization. Chromosoma 1998, 107, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, R.; Hara, M.; Kato, S.; Isobe, S.; Sato, S.; Tabata, S.; Ohmido, N. Integration of linkage and chromosome maps of red clover (Trifolium pratense L.). Cytogenet. Genome Res. 2012, 137, 60–69. [Google Scholar] [CrossRef]
- De Oliveira Bustamante, F.; do Nascimento, T.H.; Montenegro, C.; Dias, S.; do Vale Martins, L.; Braz, G.T.; Benko-Iseppon, A.M.; Jiang, J.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Oligo-FISH barcode in beans: A new chromosome identification system. Theor. Appl. Genet. 2021, 134, 3675–3686. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, C.; do Vale Martins, L.; de Oliveira Bustamante, F.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Comparative cytogenomics reveals genome reshuffling and centromere repositioning in the legume tribe Phaseoleae. bioRxiv 2021. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef]
- Flavell, R.B. The structure and control of expression of ribosomal RNA genes. Oxf. Surv. Plant Mol. Cell Biol. 1986, 3, 251–274. [Google Scholar]
- Dvořák, J.; Zhang, H.-B.; Kota, R.S.; Lassner, M. Organization and evolution of the 5S ribosomal rna gene family in wheat and related species. Genome 1989, 32, 1003–1016. [Google Scholar] [CrossRef]
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148. [Google Scholar] [CrossRef]
- Raina, S.N.; Mukai, Y. Detection of a variable number of 18S-5.8S-26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome 1999, 42, 52–59. [Google Scholar] [CrossRef]
- Pedrosa-Harand, A.; de Almeida, C.C.S.; Mosiolek, M.; Blair, M.W.; Schweizer, D.; Guerra, M. Extensive ribosomal DNA amplification during andean common bean (Phaseolus vulgaris L.) evolution. Theor. Appl. Genet. 2006, 112, 924–933. [Google Scholar] [CrossRef]
- Chung, M.-C.; Lee, Y.-I.; Cheng, Y.-Y.; Chou, Y.-J.; Lu, C.-F. Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theor. Appl. Genet. 2008, 116, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Roa, F.; Guerra, M. Non-Random Distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet. Genome Res. 2015, 146, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Dluhošová, J.; Řepková, J.; Jakešová, H.; Nedělník, J. Impact of interspecific hybridization of T. pratense × T. medium and backcrossing on genetic variability of progeny. Czech J. Genet. Plant 2016, 52, 125–131. [Google Scholar] [CrossRef]
- Falistocco, E.; Marconi, G.; Falcinelli, M. Comparative cytogenetic study on Trifolium subterraneum (2n = 16) and Trifolium israeliticum (2n = 12). Genome 2013, 56, 307–313. [Google Scholar] [CrossRef]
- Ansari, H. Molecular cytogenetic organization of 5S and 18S-26S rDNA loci in white clover (Trifolium repens L.) and related species. Ann. Bot. Lond. 1999, 83, 199–206. [Google Scholar] [CrossRef]
- Vicient, C.M.; Suoniemi, A.; Anamthawat-Jonsson, K.; Tanskanen, J.; Beharav, A.; Nevo, E.; Schulman, A.H. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. The Plant Cell. 1999, 11, 1769. [Google Scholar] [CrossRef]
- Macas, J.; Neumann, P.; Navrátilová, A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom. 2007, 8, 427. [Google Scholar] [CrossRef]
- Macas, J.; Kejnovský, E.; Neumann, P.; Novák, P.; Koblížková, A.; Vyskot, B. Next generation sequencing-based analysis of repetitive DNA in the model dioecious [corrected] plant Silene latifolia. PLoS ONE 2011, 6, e27335. [Google Scholar] [CrossRef]
- Tenaillon, M.I.; Hufford, M.B.; Gaut, B.S.; Ross-Ibarra, J. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol. Evol. 2011, 3, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Piednoël, M.; Aberer, A.J.; Schneeweiss, G.M.; Macas, J.; Novak, P.; Gundlach, H.; Temsch, E.M.; Renner, S.S. Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol. Biol. Evol. 2012, 29, 3601–3611. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Hřibová, E.; Neumann, P.; Koblížková, A.; Doležel, J.; Macas, J. Genome-wide analysis of repeat diversity across the family Musaceae. PLoS ONE 2014, 9, e98918. [Google Scholar] [CrossRef]
- Neumann, P.; Koblížková, A.; Navrátilová, A.; Macas, J. Significant expansion of Vicia Pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 2006, 173, 1047–1056. [Google Scholar] [CrossRef]
- Kulikova, O.; Gualtieri, G.; Geurts, R.; Kim, D.-J.; Cook, D.; Huguet, T.; De Jong, J.H.; Fransz, P.F.; Bisseling, T. Integration of the FISH pachytene and genetic maps of Medicago truncatula: FISH pachytene and genetic maps of M. truncatula. Plant J. 2001, 27, 49–58. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Jacobus, B.H.; SanMiguel, P.; Walling, J.G.; Yuan, Y.; Shoemaker, R.C.; Young, N.D.; Jackson, S.A. Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 2005, 170, 1221–1230. [Google Scholar] [CrossRef]
- Jiang, J.; Birchler, J.A.; Parrott, W.A.; Kelly Dawe, R. A molecular view of plant centromeres. Trends Plant Sci. 2003, 8, 570–575. [Google Scholar] [CrossRef]
- Zhu, J.; Ellison, N.; Rowland, R. Chromosomal localization of a tandemly repeated DNA sequence in Trifolium repens L. Cell Res. 1996, 6, 39–46. [Google Scholar] [CrossRef]
- Bucknell, T.T. Comparative cytogenetics in the genus Trifolium section Trifolium (clover). Master’s Thesis, Massey University, Palmerston North, New Zealand, 1999. [Google Scholar]
- Ansari, H.A.; Ellison, N.W.; Griffiths, A.G.; Williams, W.M. A lineage-specific centromeric satellite sequence in the genus Trifolium. Chromosome Res. 2004, 12, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of Similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Galasso, I.; Schmidt, T.; Pignone, D.; Heslop-Harrison, J.S. The molecular cytogenetics of Vigna unguiculata (L.) Walp: The physical organization and characterization of 18S-5.8S-25S rRNA genes, 5S rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theoret. Appl. Genet. 1995, 91, 928–935. [Google Scholar] [CrossRef]
- Gill, N.; Findley, S.; Walling, J.G.; Hans, C.; Ma, J.; Doyle, J.; Stacey, G.; Jackson, S.A. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 2009, 151, 1167–1174. [Google Scholar] [CrossRef]
- Findley, S.D.; Cannon, S.; Varala, K.; Du, J.; Ma, J.; Hudson, M.E.; Birchler, J.A.; Stacey, G. A fluorescence in situ hybridization system for karyotyping soybean. Genetics 2010, 185, 727–744. [Google Scholar] [CrossRef]
- Tek, A.L.; Kashihara, K.; Murata, M.; Nagaki, K. Functional Centromeres in Soybean Include Two Distinct Tandem Repeats and a Retrotransposon. Chromosome Res. 2010, 18, 337–347. [Google Scholar] [CrossRef]
- Zatloukalová, P.; Hřibová, E.; Kubaláková, M.; Suchánková, P.; Šimková, H.; Adoración, C.; Kahl, G.; Millán, T.; Doležel, J. Integration of genetic and physical maps of the chickpea (Cicer arietinum L.) genome using flow-sorted chromosomes. Chromosome Res. 2011, 19, 729–739. [Google Scholar] [CrossRef]
- Iwata, A.; Tek, A.L.; Richard, M.M.S.; Abernathy, B.; Fonsêca, A.; Schmutz, J.; Chen, N.W.G.; Thareau, V.; Magdelenat, G.; Li, Y.; et al. Identification and characterization of functional centromeres of the common bean. Plant J. 2013, 76, 47–60. [Google Scholar] [CrossRef]
- Yu, F.; Dou, Q.; Liu, R.; Wang, H. A conserved repetitive DNA element located in the centromeres of chromosomes in Medicago genus. Genes Genom. 2017, 39, 903–911. [Google Scholar] [CrossRef]
- Karafiátová, M.; Bartoš, J.; Kopecký, D.; Ma, L.; Sato, K.; Houben, A.; Stein, N.; Doležel, J. Mapping nonrecombining regions in barley using multicolor FISH. Chromosome Res. 2013, 21, 739–751. [Google Scholar] [CrossRef]
- Danilova, T.V.; Friebe, B.; Gill, B.S. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor. Appl. Genet. 2014, 127, 715–730. [Google Scholar] [CrossRef]
- Ebeed, H.T. Omics approaches for developing abiotic stress tolerance in wheat. In Wheat Production in Changing Environments; Springer: Singapore, 2019. [Google Scholar]
- Simpson, P.R.; Newman, M.-A.; Davies, D.R. Detection of legumin gene DNA sequences in pea by in situ hybridization. Chromosoma 1988, 96, 454–458. [Google Scholar] [CrossRef]
- Schaff, D.A.; Koehler, S.M.; Matthews, B.F.; Bauchan, G.R. In situ hybridization of β-tubulin to alfalfa chromosomes. J. Hered. 1990, 81, 480–483. [Google Scholar] [CrossRef]
- Danilova, T.V.; Birchler, J.A. Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: Resolution, sensitivity, and banding paint development. Chromosoma 2008, 117, 345–356. [Google Scholar] [CrossRef]
- Jiang, J.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 4487–4491. [Google Scholar] [CrossRef] [PubMed]
- Lapitan, N.L.V.; Brown, S.E.; Kennard, W.; Stephens, J.L.; Knudson, D.L. FISH physical mapping with barley BAC clones. Plant J. 1997, 11, 149–156. [Google Scholar] [CrossRef]
- Peterson, D.G.; Lapitan, N.L.; Stack, S.M. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 1999, 152, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.-B.; Bodeau, J.; Fransz, P.F.; Williamson, V.M.; van Kammen, A.; de Jong, J.H.; Zabel, P. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively: Theor. Appl. Genet. 1999, 98, 365–370. [Google Scholar] [CrossRef]
- Islam-Faridi, M.N.; Childs, K.L.; Klein, P.E.; Hodnett, G.; Menz, M.A.; Klein, R.R.; Rooney, W.L.; Mullet, J.E.; Stelly, D.M.; Price, H.J. A molecular cytogenetic map of sorghum Chromosome 1. Fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 2002, 161, 345–353. [Google Scholar] [CrossRef]
- Lee, H.-R.; Eom, E.-M.; Lim, Y.-P.; Bang, J.-W.; Lee, D.-H. Construction of a garlic BAC library and chromosomal assignment of BAC clones using the FISH technique. Genome 2003, 46, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.; Kulikova, O.; Penmetsa, R.V.; Bisseling, T.; Cook, D.R.; Frugoli, J. An integrated physical, genetic and cytogenetic map around the sunn locus of Medicago truncatula. Genome 2003, 46, 665–672. [Google Scholar] [CrossRef]
- Pedrosa, A.; Sandal, N.; Stougaard, J.; Schweizer, D.; Bachmair, A. Chromosomal map of the model legume Lotus japonicus. Genetics 2002, 161, 1661–1672. [Google Scholar] [CrossRef]
- Pedrosa-Harand, A.; Kami, J.; Gepts, P.; Geffroy, V.; Schweizer, D. Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosome Res. 2009, 17, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Fonsêca, A.; Ferreira, J.; dos Santos, T.R.B.; Mosiolek, M.; Bellucci, E.; Kami, J.; Gepts, P.; Geffroy, V.; Schweizer, D.; dos Santos, K.G.B.; et al. Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosome Res. 2010, 18, 487–502. [Google Scholar] [CrossRef]
- Schubert, I.; Fransz, P.F.; Fuchs, J.; de Jong, J.H. Chromosome painting in plants. Method Cell Sci. 2001, 23, 57–69. [Google Scholar] [CrossRef]
- Lysak, M.A.; Fransz, P.F.; Ali, H.B.; Schubert, I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001, 28, 689–697. [Google Scholar] [CrossRef]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Betekhtin, A.; Jenkins, G.; Hasterok, R. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS ONE 2014, 9, e115108. [Google Scholar] [CrossRef]
- Iovene, M.; Wielgus, S.M.; Simon, P.W.; Buell, C.R.; Jiang, J. Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 2008, 180, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Amarillo, F.I.E.; Bass, H.W. A transgenomic cytogenetic sorghum (Sorghum propinquum) bacterial artificial chromosome fluorescence in situ hybridization map of maize (Zea mays L.) Pachytene chromosome 9, evidence for regions of genome hyperexpansion. Genetics 2007, 177, 1509–1526. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Braz, G.T.; He, L.; Zhao, H.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Torres, G.A.; Jiang, J. Comparative oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2018, 208, 513–523. [Google Scholar] [CrossRef]
- Qu, M.; Li, K.; Han, Y.; Chen, L.; Li, Z.; Han, Y. Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet. Genome Res. 2017, 153, 158–164. [Google Scholar] [CrossRef]
- Albert, P.S.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Kao, Y.-H.; Wang, C.-J.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sun, S.; Wu, Y.; Zhou, Y.; Gu, S.; Yu, H.; Yi, C.; Gu, M.; Jiang, J.; Liu, B.; et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 2020, 101, 112–121. [Google Scholar] [CrossRef]
- Šimoníková, D.; Němečková, A.; Karafiátová, M.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa spp.). Front. Plant Sci. 2019, 10, 1503. [Google Scholar] [CrossRef]
- Li, G.; Zhang, T.; Yu, Z.; Wang, H.; Yang, E.; Yang, Z. An efficient oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 2021, 105, 978–993. [Google Scholar] [CrossRef]
- Do Vale Martins, L.; de Oliveira Bustamante, F.; da Silva Oliveira, A.R.; da Costa, A.F.; de Lima Feitoza, L.; Liang, Q.; Zhao, H.; Benko-Iseppon, A.M.; Muñoz-Amatriaín, M.; Pedrosa-Harand, A.; et al. BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 2021, 130, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Cui, C.; Liu, H.; Fu, L.; Li, L.; Dai, X.; Qin, L.; Wang, S.; Han, S.; Xu, J.; et al. Development of an oligonucleotide dye solution facilitates high throughput and cost-efficient chromosome identification in peanut. Plant Methods 2019, 15, 69. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, E.M.; Fonsêca, A.; Almeida, C.; Dos Santos, K.G.B.; Pedrosa-Harand, A. Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor. Appl. Genet. 2012, 124, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
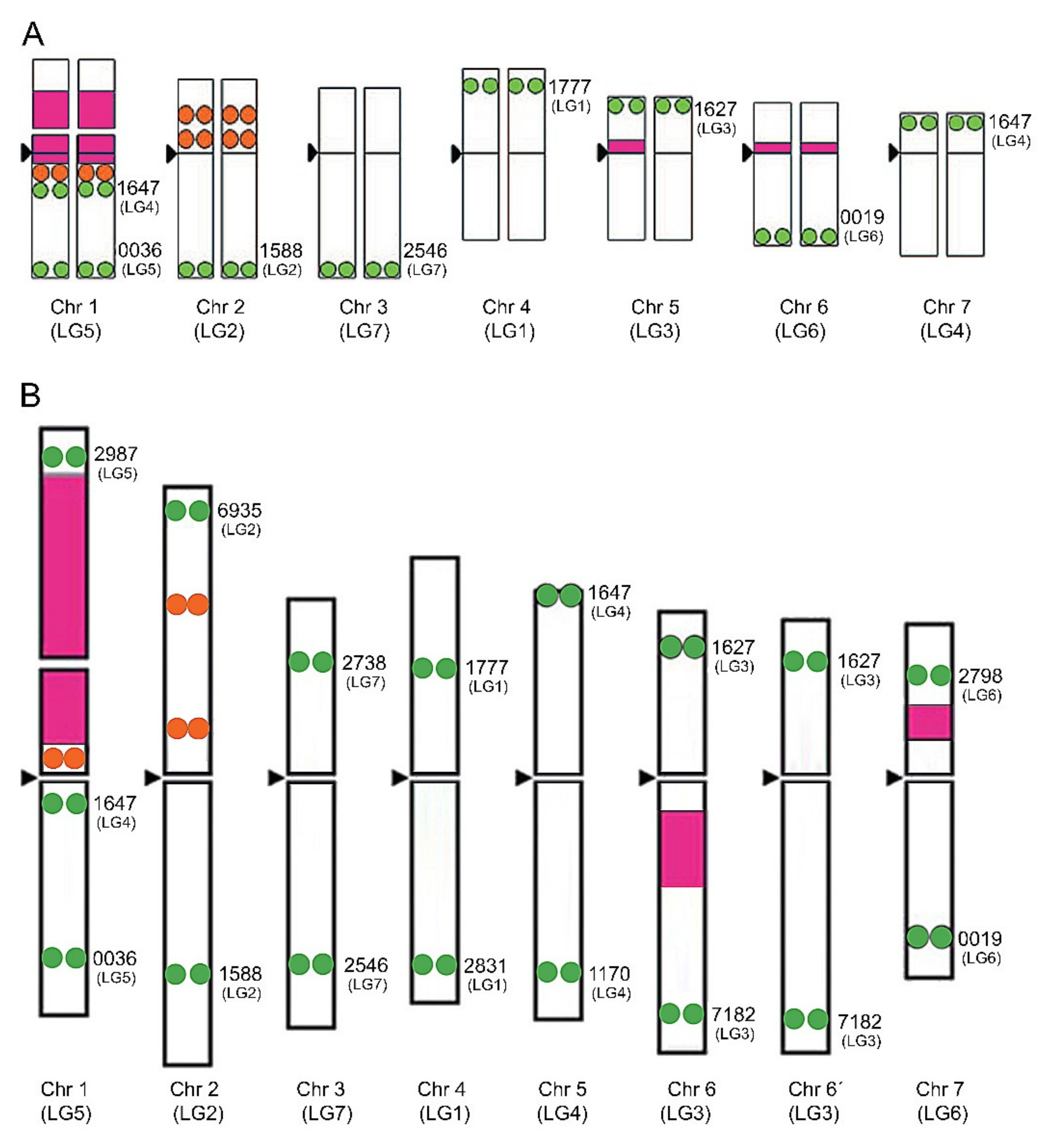
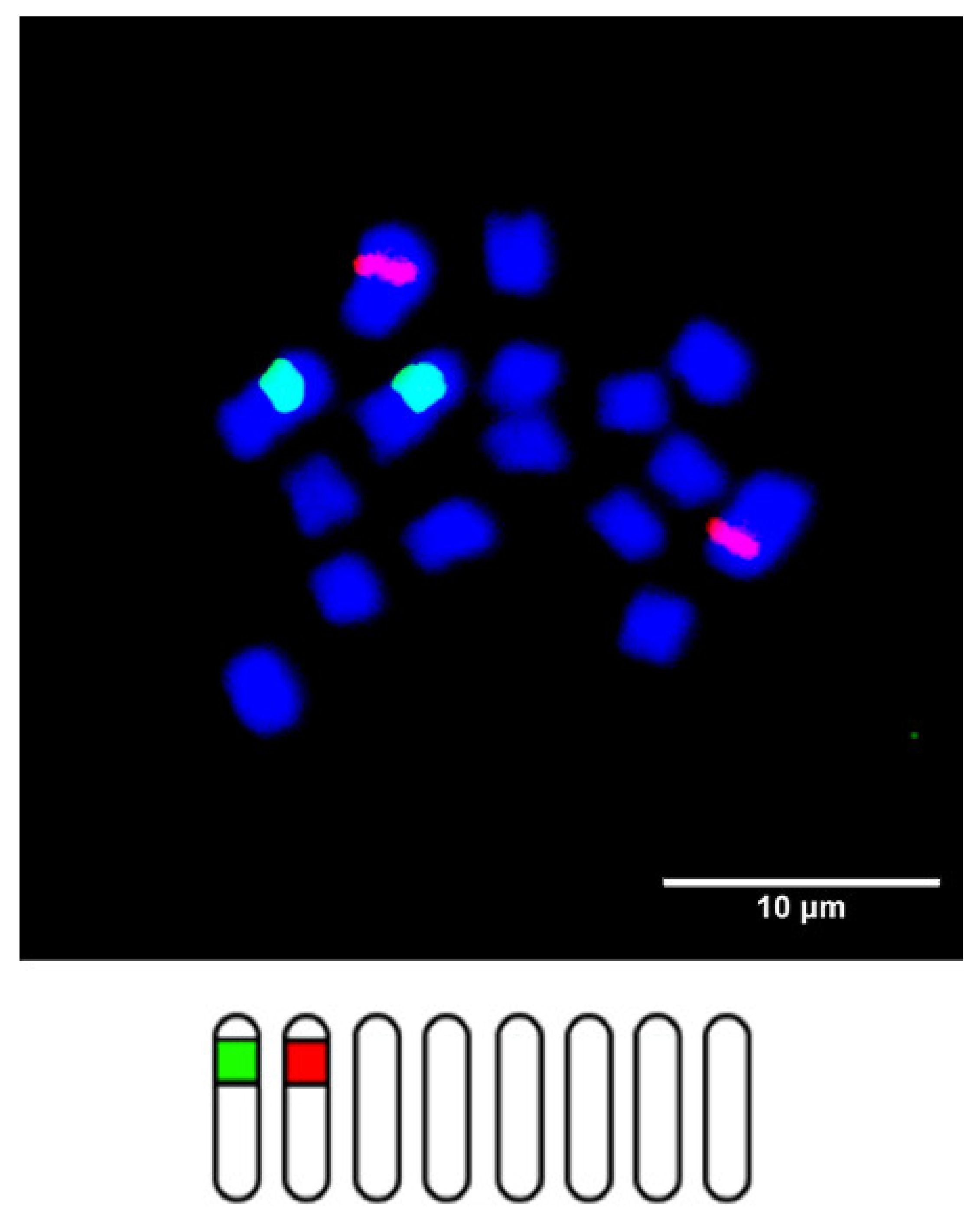
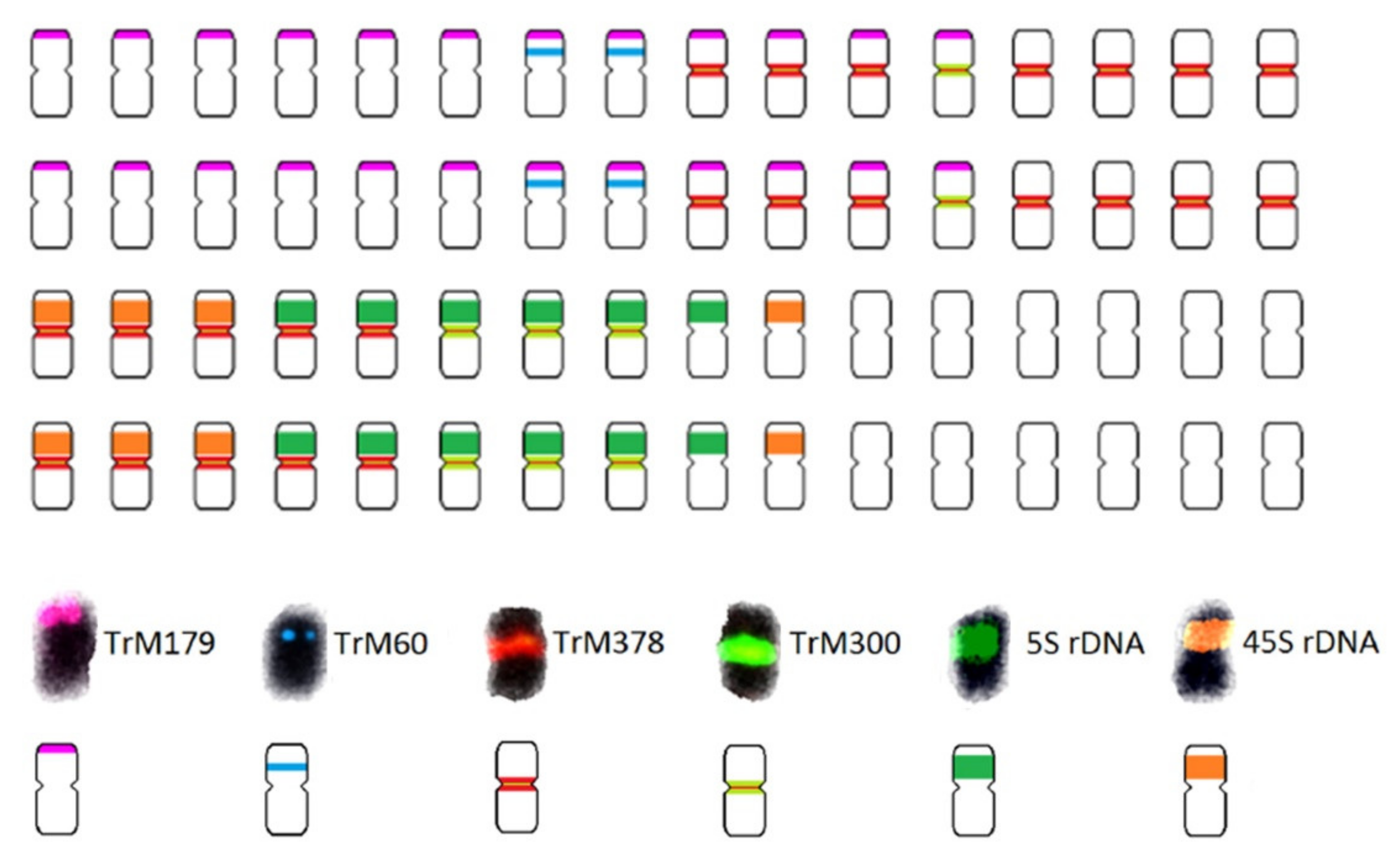
| Scheme 2. | Common Name | Section | Origin | 2n 1 | 1C Content (Mb) 2 |
|---|---|---|---|---|---|
| Rifolium resupinatum L. | Persian clover | Vesicastrum | Eurasian | 2x = 14, 16, 4x = 32 | 507.6 |
| Trifolium incarnatum L. | Crimson clover | Trifolium | Eurasian | 2x = 14 | 652.3 |
| Trifolium repens L. | White clover | Trifoliastrum | Eurasian | 4x = 32 | 546.7 |
| Trifolium pratense L. | Red clover | Trifolium | Eurasian | 2x = 14, 4x = 28 | 417.6 |
| Trifolium alexandrinum L. | Egyptian clover | Trifolium | Eurasian | 2x = 16 | 541.8 |
| Trifolium subterraneum L. | Subclover | Trichocephalum | Eurasian | 2x = 16 | 542.8 |
| Trifolium tembense Fres. | Tembien clover | Vesicastrum | African | 2x = 16 | 818.6 |
| Subgenus/Section | Trifolium Species | 2n | Loci Number per 2n | Reported in | |
|---|---|---|---|---|---|
| 5S | 25S | ||||
| CHRONOSEMIUM | |||||
| T. aureum | 2x = 16 | 4 | 2 | Vozárová et al. [31] | |
| 2x = 14 | 4 | 2 | |||
| T. badium | 2x = 14 | 2 | 4 | ||
| 2 | 2 | ||||
| T. campestre | 2x = 14 | 2 | 2 | Ansari et al. [58] | |
| T. micranthum | 2x = 16 | 2 | 2 | ||
| T. dubium | 4x = 30 | 4 | 4 | ||
| TRIFOLIUM | |||||
| TRIFOLIUM | T. alpestre | 2x = 16 | 10 | 2 | Vozárová et al. [31] |
| 11 | 2 | ||||
| T. arvense | 2x = 14 | 2 | 2 | ||
| T. bocconei | 2x = 12 | 2 | 2 | ||
| T. cherleri | 2x = 10 | 4 | 10 | ||
| T. diffusum | 2x = 16 | 2 | 2 | ||
| T. hirtum | 2x = 10 | 6 | 2 | ||
| T. ligusticum | 2x = 12 | 2 | 2 | ||
| 2x = 14 | 2 | 2 | |||
| T. pallidum | 2x = 16 | 4 | 2 | ||
| T. purpureum | 2x = 14 | 2 | 2 | ||
| T. rubens | 2x = 16 | 4 | 2 | ||
| T. squamosum | 2x = 16 | 4 | 2 | ||
| T. stellatum | 2x = 12 | 4 | 2 | ||
| 2x = 14 | 4 (2w) | 2 | |||
| T. pannonicum | 16x = 128 | 16 | 16 | ||
| T. pratense | 4x = 28 | 8 | 8 | Dluhošová et al. [80] | |
| 2x = 14 | 4 | 5 | Sato et al. [43] | ||
| T. medium | 8x = 64 | 12 | 8 | Dluhošová et al. [80] | |
| TRICHOCEPHALUM | T. subterraneum subsp. subterraneum | 2x = 16 | 2 | 4 (2w) | Vozárová et al. [31] |
| T. subterraneumsubsp. subterraneum | 2x = 16 | 2 | 2 | Falistocco et al. [81] | |
| T. subterraneum subsp. brachycalycinum | 2x = 16 | 2 | 4 (2w) | ||
| T. israeliticum | 2x = 12 | 10 | 4 | ||
| VESICASTRUM | T. fragiferum | 2x = 16 | 2 | 2 | Vozárová et al. [31] |
| T. resupinatum | 2x = 16 | 2 | 2 | ||
| 2x = 14 | 2 | 2 | |||
| T. spumosum | 2x = 16 | 2 | 2 | ||
| 4 | 2 | ||||
| TRIFOLIASTRUM | T. glomeratum | 2x = 16 | 2 | 2 | Vozárová et al. [31] |
| T. montanum | 2x = 16 | 2 | 2 | ||
| T. occidentale | 2x = 16 | 4 | 2 | Ansari [82] | |
| T. pallescens | 2x = 16 | 2 | 2 | Vozárová et al. [31] | |
| T. thalii | 2x = 16 | 2 | 2 | ||
| T. repens | 4x = 32 | 4 | 2 | Ansari [82] | |
| T. uniflorum | 4x = 32 | 4 | 4 | ||
| T. nigrescens subsp. nigrescens | 2x = 16 | 2 | 2 | ||
| T. nigrescens subsp. petrisavii | 2x = 16 | 2 | 2 | ||
| T. ambiguum | 2x = 16 | 2 | 2 | ||
| T. hybridum | 2x = 16 | 2 | 2 | ||
| T. isthmocarpum | 2x = 16 | 2 | 6 | ||
| INVOLUCRARIUM | T. chilense | 2x = 16 | 4 | 2 | Vozárová et al. [31] |
| T. microdon | 2x = 16 | 2 | 2 | ||
| T. microcephalum | 2x = 16 | 16 | 16 | ||
| 16 | 2 | ||||
| PARAMESUS | T. glanduliferum | 2x = 16 | 4 | 2 | Vozárová et al. [31] |
| 5 (1w) | 2 | ||||
| 4 | 2 | ||||
| T. strictum | 2x= 14 | 2 | 2 | ||
| LUPINASTER | T. lupinaster | 4x = 28 | 8 | 4 | Vozárová et al. [31] |
| 4x = 32 | 8 | 4 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukjanová, E.; Řepková, J. Chromosome and Genome Diversity in the Genus Trifolium (Fabaceae). Plants 2021, 10, 2518. https://doi.org/10.3390/plants10112518
Lukjanová E, Řepková J. Chromosome and Genome Diversity in the Genus Trifolium (Fabaceae). Plants. 2021; 10(11):2518. https://doi.org/10.3390/plants10112518
Chicago/Turabian StyleLukjanová, Eliška, and Jana Řepková. 2021. "Chromosome and Genome Diversity in the Genus Trifolium (Fabaceae)" Plants 10, no. 11: 2518. https://doi.org/10.3390/plants10112518
APA StyleLukjanová, E., & Řepková, J. (2021). Chromosome and Genome Diversity in the Genus Trifolium (Fabaceae). Plants, 10(11), 2518. https://doi.org/10.3390/plants10112518






