Abstract
Trifolium L. is an economically important genus that is characterized by variable karyotypes relating to its ploidy level and basic chromosome numbers. The advent of genomic resources combined with molecular cytogenetics provides an opportunity to develop our understanding of plant genomes in general. Here, we summarize the current state of knowledge on Trifolium genomes and chromosomes and review methodologies using molecular markers that have contributed to Trifolium research. We discuss possible future applications of cytogenetic methods in research on the Trifolium genome and chromosomes.
1. Introduction
The Fabaceae family (Leguminosae, legume or bean family) is the third-largest flowering plant family, after the Asteraceae and Orchidaceae families [1,2]. It is agronomically important, as it can form a symbiotic association with nitrogen-fixing bacteria. Several species from this family serve as genetic model organisms (e.g., Medicago truncatula Gaertn., Pisum sativum L., and Lotus japonicus L.). With more than 250 species, the clover genus, Trifolium, is one of the largest genera in this family [1,2,3]. This herbaceous genus acquired its name in reference to the characteristic form of the leaf, usually consisting of three leaflets (trifoliolate), and includes both annual and perennial species occurring natively across a large range of biotopes from meadows and open woodlands to semi-deserts and mountain ridges in temperate and, to a lesser extent, subtropical regions. The genus’s origin has been estimated to have occurred in the Early Miocene, 16–23 million years ago, and its center of origin was first assumed to be in California with its subsequent spread into Asia and hence to Europe and Africa [4]. Later, a new hypothesis was proposed of clovers originating in the Mediterranean region due to their species diversity, including the diversity in their chromosome numbers, and because the greatest occurrence of their endemic species is found in this area, with a secondary center of distribution in North America and East Africa [3,5,6,7]. By contrast, native clovers are absent from Australia and Southeast Asia.
Attempts have been made to divide this genus into natural groups. In the 19th century, Bossier [8] divided the genus into seven sections. A century later, eight subgenera were recognized and revised [1,9]. Better insight into phylogeny and the origin of the genus was facilitated by molecular analyses. These showed that Trifolium is a member of a large clade of legumes that lack one copy of the chloroplast inverted repeat [10,11], and a further molecular phylogenetic analysis of the internal transcribed spacer (ITS) and chloroplast genes provided evidence that most of these proposed sections are not monophyletic [12,13]. The most recent subgeneric classification, based on phylogenetic analyses of 218 species’ ribosomal ITS and chloroplast trnL intron sequences, was proposed by Ellison et al. [3], who divided the genus into two subgenera, Chronosemium and Trifolium, with the further subdivision of Trifolium into eight sections—Glycyrrhizum (2 species), Paramesus (2 species), Lupinaster (3 species), Trifolium (73 species), Trichocephalum (9 species), Vesicastrum (54 species), Trifoliastrum (20 species), and Involucrarium (72 species). In 2014, Trifolium phylogenetic analyses were conducted, based on highly unusual Trifolium plastomes [14].
The economic importance of this genus lies in its agricultural utilization. Historically, clovers, and especially red clovers, have been cultivated in rotation with other crops to maintain soil fertility due to their ability to establish a mutualistic relationship with root-nodulating and nitrogen-fixing bacteria. Their value was later diminished by the advent of nitrogen fertilizers, but the global need for sustainable and conservation agriculture is bringing this historical approach back into focus. Nowadays, many Trifolium species are extensively cultivated as fodder plants (Trifolium pratense L., Trifolium repens L., Trifolium hybridum L., and Trifolium resupinatum L.), and also as green manure crops to enhance soil fertility and sustainability [15]. Further knowledge about the genomes of both wild and cultivated clovers and an understanding of their evolution will prove to be of great benefit in the future of clover breeding.
2. Trifolium Genomes, Chromosomes, and Chromosome Number Variation
Genetic information within the cells of plants and animals alike experience changes during evolution. Chromosome structure changes through rearrangements involving DNA fissions, fusions, duplications, deletions, insertions, inversions, translocations, and the expansion of repetitive sequences [16,17,18,19,20]. These rearrangements are further heightened by polyploidization events that multiply a single genome (autopolyploidy) or combine two or more divergent genomes (allopolyploidy). These events are accompanied and followed not only by an increase in chromosome sets, but also by dynamic and stochastic changes in genome organization, including altered gene expression or transposon activation as well as structural rearrangements [21,22,23]. Therefore, plant chromosomes and karyotypes show significant variability and even closely related species can differ greatly, and Trifolium is no exception.
The Trifolium genus has small to medium genome sizes, ranging from 337.1 Mb in Trifolium ligusticum Balb. ex Loisel. to 5669.3 Mb in Trifolium pannonicum Jacq. per 1C value [24] and includes species with different chromosome numbers and ploidy levels. Chromosome numbers have been published for more than 184 Trifolium species [1,25,26,27,28] and the Chromosome Counts Database compiles 318 species for the Trifolium genus entry [29]. Agriculturally important Trifolium spp., their chromosome numbers, and their estimated genome sizes are summarized in Table 1. The ancestral chromosome constitution within the Trifolium genus is considered to be diploid, with 2n = 16 [3]. While 80% of the already analysed species have a basic chromosome number of x = 8, corresponding with the ancestral state, species forming aneuploid series (x = 7, 6, 5) have been identified in 31 species, 11 of which have both aneuploid and diploid or polyploid counts [3,30]. Ellison et al. [3] found an uneven distribution of aneuploidy across the phylogeny to be present in subgenus Chronosemium, sections of Trichocephalum, Trifoliastrum, and Vesicastrum, although most commonly in sections of Trifolium [3]. Vozárová et al. [31] found aneuploidy in sections of Lupinaster [31]. In sharp contrast to aneuploidy, polyploidy has been observed to be evenly distributed across the phylogeny, with all of the large clades having at least one inferred origin of polyploidy, the largest number (10) occurring in section Involucrarium [3]. In total, polyploidy has been observed in 24 species including tetraploidy, hexaploidy, octoploidy, dodecaploidy, and hexadecaploidy, and some species have been found to have both diploid and polyploid or have had different polyploid counts [3].

Table 1.
Selected important Trifolium crops, their origins, chromosome numbers, and genome sizes.
Perennial species in the Trifolium genus are known to be mostly self-incompatible. For instance, the agronomically important perennials T. repens, T. pratense, T. hybridum, and Trifolium ambiguum M. Bieb. have a gametophytic system of incompatibility determined by one locus, with both T. pratense and T. repens possessing large numbers of alleles [32]. The mating system of these self-incompatible crops leads to the development of highly heterozygous populations [33,34]. On the other hand, annual species, in general, are mostly self-compatible (e.g., Trifolium subterraneum L. and Trifolium alexandrinum L.), even though some may require a stimulating agent, such as insects or tripping, to produce high quantities of seeds [35,36].
The Trifolium chromosomes are small in size, usually metacentric or submetacentric [1,27,37,38,39,40]. To date, acrocentric chromosomes have been observed only in Trifolium argentinense Speg. [41]. Telocentric chromosomes have not yet been confirmed in the genus. These are generally rare in plants and eukaryotes, probably due to instability, which is attributed to the reduced centromere size and incompleteness of kinetochores that occur through the mis-division of centromeres in normal chromosomes.
3. Current State of Knowledge on Trifolium Genomes
Further research on clover genomes at the sequence level, through the localization of genes, was first enabled by genetic mapping. Since the beginning of the millennium, linkage maps have been published for three agronomically important cultivated clover species. A restriction fragment length polymorphism (RFLP) markers linkage map, high-density linkage maps with gene-associated microsatellite markers, and a consensus linkage map have been made available for red clover [42,43,44]. The latter has also been applied in white clover T. repens and T. subterraneum for comparative mapping [45,46,47].
In white clover, four independent genetic linkage maps were published [45,47,48,49], followed by an integrated linkage map published in 2013 [50]. The integrated map of T. repens includes 1109 loci, 18 candidate genes, and 1 morphological marker and covers 97% of the genome at a moderate density of one locus per 1.2 cM. The alignment of the integrated map with the M. truncatula genome assembly revealed substantial collinearity, and identified an interchromosomal rearrangement, whereby the M. truncatula chromosomes Mt-2 and Mt-6 were split across the T. repens chromosomes Tr-2 and Tr-6. In 2012, the first genetic linkage map for T. subterraneum was constructed showing great synteny between T. subterraneum and T. pratense, and proving transferability of T. pratense and T. repens molecular markers to T. subterraneum [46]. The advancement of next-generation sequencing assisted in acquiring sequences of numerous legume species, including clovers. To date, whole genome sequences of six clover species have been made available, and include both cultivated (T. pratense, T. repens) and wild (Trifolium medium L., T. subterraneum, Trifolium occidentale Coombe, Trifolium pallescens Schreb.) species.
The first clover draft genome sequence was made available for cultivated red clover (T. pratense, tetraploid variety Tatra) and was published by Ištvánek et al. [51]. The authors presented 314.6 Mbp of an estimated genome size 418 Mbp and annotated and predicted functions for 47,398 protein-coding genes, within a total of 64,761 predicted genes [30]. A year later, the genome sequence of another T. pratense, this time the diploid variety Milvus, was published [52]. Its assembly consists of 309 Mbp, with 164.2 Mb in seven chromosome-length sequences or pseudo-molecules. Among the annotated genes, 22,042 of a total 40,868 were anchored onto the seven chromosomes, making red clover the second forage legume with a genome assembly at a pseudo-molecule level just behind the model species M. truncatula. The authors also anchored the acquired data to the M. truncatula reference sequence [53] for the physical map construction and synteny analysis. Macrosynteny was observed in 248 synteny blocks carrying 12,278 gene pairs, with chromosomes 1 and 6 being almost entirely syntenic with M. truncatula chromosomes 1 and 7 and the remaining five chromosomes having large synteny blocks with two or three M. truncatula chromosomes [52]. The synteny between the red clover and M. truncatula is significantly less conserved than between the white clover and M. truncatula. The reason for this variability is most probably the difference in their basic chromosome numbers (white clover and M. truncatula x = 8, red clover x = 7) [47,52].
In 2014, a genome sequence of the wild zigzag clover T. medium, with an estimated genome size of 3154 Mbp [30], was published. To date, it remains the largest Trifolium genus sequencing project [54]. The authors assembled a partial genomic sequence of 492.7 Mb. The final assembly is very fragmented, due to, among other reasons, the large haploid size of the zigzag clover genome, its polyploid state, cross-pollination, and a high proportion of repetitive sequences equal to nearly half of the (46.67%) the genome size. Although the assembly was not sufficient for comprehensive annotation, the authors did provide a characterization and a comparative analysis of repeat content between T. medium and T. pratense.
Genome sequences of T. subterraneum were published in 2016 [55]. Those authors assembled genomic sequence of 471.8 Mb, covering 85.4% of the T. subterraneum genome and the annotated 30,543 of 42,706 predicted genes. A comparative analysis of T. subterraneum and of related legume species from the subfamily Papilionoideae revealed that the whole chromosome 1 in T. subterraneum is highly conserved across all analyzed species [55]. Moreover, the authors observed a clear syntenic relationship with M. truncatula chromosomes and revealed single synteny blocks for three pseudomolecules (Tsub-1 and Mt-1, Tsub-3 and Mt-3, and Tsub-5 and Mt-5). On the other hand, the authors observed the occurrence of duplication in T. subterraneum chromosome 2, compared to M. truncatula and T. pratense chromosome 2, suggesting that duplication occurs after the divergence of T. pratense and T. subterraneum. Alongside acquiring the genome sequences of T. pratense and T. subterraneum, the authors constructed high-density SNP linkage maps [52,55]
Genome sequences of an allotetraploid white clover were published in 2019 along with genome sequences of its extant relatives of the parental progenitors, namely, the western clover T. occidentale and pale clover T. pallescens [56]. Their assemblies spanned 437 Mb for western clover, 383 Mb for pale clover, and 841 Mb for white clover, accounting for 82%, 72%, and 72% of the estimated genome sizes (530, 534 and 1174 Mb), respectively. Based on the data, the authors estimate that T. repens originated ca 15,000 to 28,000 years ago during the last glaciation in Europe through multiple hybridization events in the contact zones of the progenitors in glacial refugia. Multiple allopolyploidization events indicate a high compatibility of the parental genomes and resulted in a noticeable diversity of newly developed T. repens populations. However, the authors show that subgenomes of the progenitors have maintained integrity and independence as well as gene expression activity. Overall, the allopolyploidization event of two highly specialized species enabled the newly developed species T. repens to colonize different niches and expand worldwide.
4. Hybridization in the Clover Genus
Ellison et al. [3] carried out a comprehensive DNA-based phylogenetic analysis, and only five or six instances of apparent hybrid speciation were found [3]. Overall, interspecific hybridization does not appear to play a significant role in the evolution of clovers, and this may well be associated with their predominant adaptation to insect pollination [57].
Despite the strong genetic barriers to interspecific hybridization, origin through hybridization was suggested in the cases of Trifolium dubium Sibth., T. repens, T. pannonicum, and T. medium [3,54]. To date, parental origin was identified in tetraploids white clover (T. repens), derived from hybridization of T. occidentale and T. pallescens [3,34], and suckling clover T. dubium, combining the genomes of Trifolium campestre Schreb. and Trifolium micranthum Viv. [58]. The ancestry of T. pannonicum seems to be complex, given that Ellison et al. [3] suggested its parental species to be distantly related to section Trifolium—one of which is probably related to Trifolium patulum Tausch. and Trifolium squamosum L., with the other unknown or perhaps extinct. For T. medium, its allopolyploid origin was proposed based on the existence of different types of centromeric repeats [54]. On the other hand, legumes reveals an exceptional diversity and promiscuity of satellite repeats in general [59,60,61].
Economically important clovers, such as white clover (T. repens) and red clover (T. pratense) are subject to breeding programs to enhance traits such as yield, flowering, nutrient use efficiency, stress tolerance, and resistance to specific diseases. The introduction of any specific trait via artificial interspecific hybridization is difficult to accomplish, however, and has generally only been achieved between closely related taxa, often with the aid of embryo-rescue techniques or the use of bridge species [62,63], as previously reviewed by Abberton [33]. Despite the great efforts directed to the introgression of desirable traits into cultivated clovers by crossing, only a few specific attempts have been successful.
To date, 11 related clover species were discovered to be capable of being integrated into the wider gene pool of white clover by interspecific hybridization (Trifolium nigrescens Viv., T. occidentale, Trifolium isthmocarpum Brot., Trifolium uniflorum L., T. ambiguum, T. hybridum, Trifolium thalii Vill., T. pallescens, Trifolium montanum L., Trifolium argutum Banks & Sol., and Trifolium semipilosum Fresen.) [63]. In the case of red clover, considerably less work has been carried out in its artificial hybridization with other taxa. Red clover has been successfully hybridized with five species so far (Trifolium sarosiense Hazsl., Trifolium alpestre L., T. ambiguum, Trifolium diffusum Ehrh., and T. medium; reviewed by Abberton [33]), but the only viable progeny has resulted from hybrids between tetraploid T. pratense cv. Tatra and octoploid T. medium [64,65]. As the ability to produce interspecific hybrids reflects the compatibility of parental genomes, further knowledge about genomes and of their similarities and differences can strongly support breeding efforts in Trifolium.
5. Chromosome Identification in Trifolium
In legumes with large chromosomes, such as Pisum sativum L. or Vicia faba L., individual chromosomes can be distinguished by ordinary karyotyping or banding methods [66,67], although the process is rather complicated in species with small chromosomes, such as Trifolium. Both repetitive and low- or single-copy sequences are important tools for chromosome identification in cytogenetic studies. Usually, a mix of different probes is used, which can include localizing ribosomal DNA (rDNA) sites, telomeric probes, large plasmid, bacteriophage, or bacterial artificial chromosomes (BACs) containing specific single-copy or repetitive inserts. Based on 5S rDNA, 25S rDNA, and seven bacterial artificial chromosome probes containing microsatellite markers with a known position, a cytogenetic map has been constructed for red clover (Figure 1).
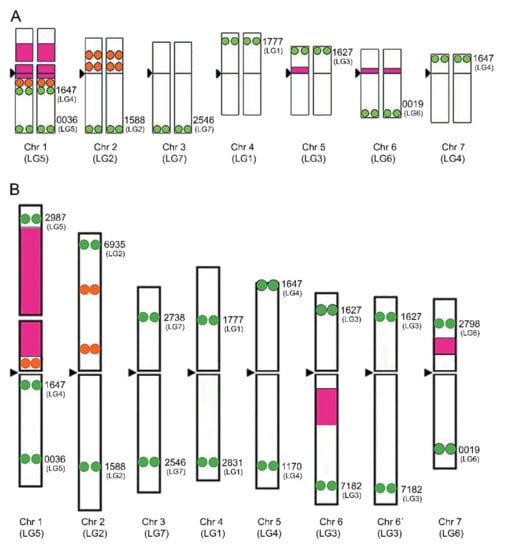
Figure 1.
Cytogenetic map of T. pratense based on the hybridization pattern of probes derived from 5S rDNA (orange circles) and 26S rDNA (pink boxes) and localization of 7 (A) or 14 (B) BAC clones corresponding to chromosome-specific microsatellite markers (green circles) (adapted from Sato et al. [43] and Kataoka et al. [68].
The easy design and production of oligonucleotide libraries has presented new opportunities to plant cytogenetics. Recently, an oligonucleotide barcode system was developed to identify all cowpea and common bean chromosomes [69,70]. Despite the availability of genome sequences for selected Trifolium spp., however, oligonucleotide libraries have not yet been exploited for Trifolium research.
6. Chromosomal Distribution of Ribosomal DNA Genes
Because ribosomal genes are among the best-researched regions of eukaryotic genomes, fluorescence in situ hybridization (FISH) analyses, using rDNA genes as probes, have been conducted in numerous plant species, including Trifolium. The 35S and 5S ribosomal genes are located independently in one or several loci as tandem repeats, ranging from hundreds to thousands of copies in higher vascular plant genomes [71]. While polycistronic gene 35S consists of 18S-5.8S-25S rDNA and occurs on chromosomal regions known as nucleolus organizer regions (NORs), the 5S rDNA gene is usually independent from NORs [72,73]. Ribosomal genes have undergone rapid evolution in their means of altering the number of copies and their localization on chromosomes [74,75,76,77]. Therefore, rDNA genes have been proven to act as excellent cytogenetic markers for karyotype analysis, and they have been widely used to examine and understand phylogenetic relationships, chromosomal organization, and evolution in many plant species.
Roa and Guerra [78,79] found that, in angiosperms rDNA sites, most often number one or two 45S and one 5S per haploid genome. The localization of 45S rDNA sites was observed preferentially on the short arm and in the terminal region of chromosomes in general, but genera with predominant proximal localization were found in some families, including Fabaceae (Arachis, Lens). On the other hand, 5S rDNA localization varies in different angiosperm families. In Fabaceae, 5S rDNA is preferentially found in the proximal region.
To date, the numbers and positions of rDNA loci on chromosomes have been reported for 42 Trifolium species (Table 2; adapted from Vozárová et al. [31]). Based on ancestral state reconstruction, Vozárová et al. [31] suggested the occurrence of one 5S and one 26S locus per haploid genome separately as an ancestral condition for the whole genus. The ancestral karyotype referencing the basic chromosome number and rDNA loci constitution may resemble the karyotype of T. diffusum (Figure 2).

Table 2.
Reported chromosome numbers 5S and 25S rRNA loci numbers in Trifolium species (adapted from Vozárová et al. [31]).

Figure 2.
Fluorescence in situ hybridization image and schematic karyotype of T. diffusum with 5S (red) and 26S (green) rDNA probes suggested to represent the ancestral state in the Trifolium genus (adapted from Vozárová et al. [31]).
7. Other Repetitive and Single-Copy Markers
The repetitive elements of both a satellite and dispersed character have been used as cytogenetic markers to define karyotypes in plants because these sequences constitute large proportions of most plant genomes. Even though in some species the satellite repeats can represent as much as 10–20% of the genome, the bulk of the repeat fraction usually consists of mobile elements, specifically long terminal repeats (LTR) retrotransposons [16,83,84,85,86,87,88]. The accumulation of LTR retrotransposons along with multiple rounds of polyploidization events are crucial in genome size expansion [18,89]. Repetitive elements are often clustered in heterochromatic regions, such as pericentromeres, and are crucial for maintaining genome stability. Pericentromeric regions and their repetitive content have been researched in M. truncatula and Glycine max (L.) Merill. model legumes related to Trifolium [90,91].
Most satellite repeats occupy centromeric, pericentromeric, subtelomeric, or telomeric regions. Typical plant centromeres are composed of centromere-specific satellite repeats, and often act as the dominant component, as well as retrotransposons [92]. In the 1990s, the C-banding technique was used to examine highly repetitive sequences in plant chromosomes. C-banding discriminates the constitutive heterochromatin, which is generally found in the centromeric, although sometimes in the telomeric or interstitial, region and consists largely of highly repetitive DNA. Zhu et al. [93] reconstructed a T. repens karyotype using C-banding for constitutive heterochromatin identification and by analyzing a 350 bp tandemly repeated DNA sequence. The analyses revealed 13 pairs of metacentric and 3 pairs of submetacentric chromosomes in T. repens karyotype. The C-bands were identified around the centromeric regions of 8 pairs of chromosomes, and no terminal or interstitial C-bands were observed. A probe derived from 350 bp tandemly repeated DNA sequence, hybridized to a centromeric region of 12 chromosome pairs, representing some of the chromosomes that correspond with C-bands. Similarly, karyotypes with C-bands were constructed by Bucknell [94] in Trifolium hirtum All. and T. incarnatum. The author observed C-bands in the centromeric and pericentromeric regions of T. hirtum and in the centromeric region and proximal-end of the satellite body in T. incarnatum.
Ansari et al. [95] analyzed the structure of a 350 bp centromeric satellite repeat identified by Zhu et al. [93], and its presence in the genome of 17 Trifolium species and subspecies, revealing a lineage-specific centromeric satellite repeat unique to species from section Lotoidea with Mediterranean origins. An in situ hybridization probe, derived from the analyzed centromeric satellite repeat, hybridized to all chromosomes of T. uniflorum, T. occidentale, and Trifolium nigrescens ssp. petrisavii (Clem.) Holmboe and ssp. nigrescens, to four chromosome pairs in T. pallescens (note that together with T. occidentale it corresponds to the hybridization pattern in T. repens [93]), to two chromosome pairs in T. isthmocarpum, and to one chromosome pair in Trifolium michelianum Savi, T. montanum, and T. ambiguum. However, C-banding conducted by those authors revealed the presence of C-bands on all chromosomes of T. ambiguum, thus indicating the coexistence of more than one family of the centromeric satellite repeats in this species. Similarly, the coexistence of two or more centromeric satellite repeats can be expected in other species whereby a probe derived from the analyzed centromeric satellite repeat hybridizes to only some chromosome pairs. According to the so-called “library hypothesis” proposed by Fry and Salser [96], related species share a collection of satellite sequences that may expand and contract in evolution. Ansari et al. [95] suggested that at least a partial homogenization and the amplification of this repeat family occurred before the radiation of this lineage and, during its evolution, diverse expansion and the reduction of specific satellite repeats resulted in differentiation and the presumable coexistence of more than one family of centromeric satellite repeats in some species.
The availability of genome sequences enables the further characterization of repetitive elements in plant genomes. Dluhošová et al. [54] conducted a thorough comparison between red clover and zigzag clover sequencing reads [54]. Repeat content characterization revealed a striking difference between the overall repeat content in both species and in the prevalence of individual DNA transposon lineages. The proportion of fully annotated repetitive elements was 33% in T. pratense and almost 47% in T. medium. However, due to a possible underestimation of the overall repeat content caused by the low number of reads included in the analysis, the repetitive elements may comprise as much as 70% of the T. medium genome. The major differences in the prevalence of individual repetitive elements showed a reduction of Ty1/Copia (12.22% in T. pratense and 7.80% in T. medium) and DNA transposons (6.07% in T. pratense and 2.89% in T. medium) and a remarkable rate of expansion (6.65% in T. pratense and 26.29% in T. medium) of Ty3/Gypsy retrotransposons in the genome of T. medium, compared to T. pratense. The authors also identified 7 and 45 species-specific clusters for T. pratense and T. medium, respectively, of which 6 and 11 were successfully validated. The newly discovered Trifolium-specific tandem repeats contained one centromeric and three pericentromeric repeats in T. pratense and two centromeric, one pericentromeric, and one subtelomeric repeat in T. medium. Based on the hybridization pattern of probes derived from 5S and 26S rDNA and on all the newly discovered T. medium-specific tandem repeats, the authors divided 64 T. medium chromosomes into 11 categories (Figure 3). Moreover, based on the presence of the two identified centromeric repeats on only half of the chromosomes, an allopolyploid origin of T. medium was suggested. A considerable variability and promiscuity in the satellite repeats has been observed in many Fabeae species generally [59,60,61], however, and the coexistence of more than one family of centromeric satellite repeats, emerging from the diverse expansion and reduction of specific satellite repeats in evolution, has already been proposed for some Trifolium species (e.g., T. ambiguum and T. pallescens) [95]. In addition to T. pratense, T. medium, and T. repens, centromere-specific satellite repeats have been found in many related legumes, such as cowpea, soybean, chickpea, common bean, or alfalfa and their related species [97,98,99,100,101,102,103].

Figure 3.
Schematic karyotype of T. medium based on the hybridization pattern of probes derived from 5S (dark green) and 26S (orange) rDNA, subtelomeric repeat TrM179 (pink), pericentromeric repeat TrM60 (blue), and two centromeric repeats TrM378 (red) and TrM300 (light green). Centromeric repeats Trm378 and TrM300 are colocalized on the same chromosomes, 24 chromosomes have a higher proportion of TrM378 while 8 chromosomes have a higher proportion of TrM300 (adapted from Dluhošová et al. [54]).
In large and complex plant genomes, a remarkable proportion of which can consist of repetitive sequences, unique motifs suitable as cytogenetic markers are restricted to regions that are only a few kb long, and that usually coincide with genes [104,105,106]. Successful reports of detecting single-copy gene sequences in legumes related to Trifolium include a legumin gene that us 13.5 kb long in pea [107], and a β-tubulin gene sequence that is 10 kb long in alfalfa [108]. The localization of single-copy DNA sequences has been hampered, however, by technical difficulties, mostly relating to the low sensitivity of short-length probes [109]. Since the turn of the century, this challenge has been overcome by advances in single-gene FISH protocols, modern microscopy resolution and by using large-insert DNA clones, usually BACs, anchored by the targeted small DNA probes [110,111,112,113,114,115,116].
Since then, BAC clones have been used for the physical mapping of single-copy sequences in T. pratense, including in constructing T. pratense cytogenetic maps [43,68] and in related legumes, including the model species M. truncatula [90] and L. japonicus [117] as well as the agronomically important species Phaseolus vulgaris L. [118,119] and Glycine spp. [99].
8. Comparative Mapping in Legumes
The hybridization of species-specific elements to their related species might provide insight into a species’ evolution and reveal rearrangements. The hybridization of probes to large chromosome regions is convenient for these analyses. Despite the common utilization of large chromosome segments or whole chromosomes acquired by flow sorting or microdissection in animal and human cytogenetics [120], these techniques are not applicable in plants due to their high complexity and the large number of repeated structures of various types. Instead, cytogenetic analyses in plants have been developed to exploit synthetic oligonucleotide libraries (oligo-FISH, oligo-painting) and high-capacity DNA vectors, such as BACs, carrying large inserts of DNA (BAC-FISH, BAC-painting).
As mentioned above, BAC-FISH is a powerful tool for chromosome identification and has been successfully used to mark specific chromosomes in many plant families, including legumes. The application of BACs has been extended to identify large chromosome regions, such as chromosome arms or whole chromosomes (BAC-painting). However, the availability of whole genome sequences, obtained from BAC to BAC sequencing and a sufficient pool of low- or single-copy BAC clones, is required for the successful development of chromosome BAC probes. Thus, BAC-painting is suitable for species with small genomes and low proportions of repetitive fraction, and to date it remains established only in the family Brassicaceae [121,122] and for Brachypodium in the family Poaceae [123], even though attempts have bene made to extend this method to other families [124,125].
Significant progress has recently been made in karyotype research in plants for the application of artificially synthetized oligonucleotides (oligo-FISH, further extended to oligo-painting) [126]. Short and unique oligonucleotides (usually 45–50 bp) can be computationally identified and designed specifically for a chromosomal region or even an entire chromosome. The hybridization of specific oligo pools to their related species has proven to be a powerful tool to reveal karyotypic and chromosomal evolution [126,127,128,129,130,131,132,133]. To date, this method has been used in a single legume species, Arachis [134], but the availability of the genome sequences of selected Trifolium spp. Creates opportunities for utilizing oligonucleotide libraries in Trifolium karyotype and evolutionary research.
9. Conclusions and Future Prospects
Improving clover productivity as a means of boosting yields and nitrogen fixation efficiency is therefore a central focus of plant breeders today. Information is mainly limited to the related legume model species, however, and unraveling the genome organization and understanding the evolution of clover are essential for greater breeding efficiency.
Advances in clover research within the genomics era have assisted in the development of an impressive array of genomic resources, including complete genome sequences of some clovers and related legumes. As high-throughput sequencing has revolutionized genome sequencing with its ultralow cost and overwhelmingly large data output, more and more new plant species sequences, as well as species’ resequences, supported by a large range of bioinformatic tools, provide us with more data applicable for more efficient breeding strategies. The combination of genomic and bioinformatic data with molecular cytogenetics may provide a more developed understanding of plant genomes in general.
Ribosomal DNA and other repetitive sequences have been widely used as plant cytogenetic markers, and recently, the development of large-DNA clones carrying target sequences, such as BACs, has facilitated the easier localization of low- or single-copy DNA sequences [110,111,112,113,114,115,116]. BAC-FISH has been applied successfully in clover karyotype characterization, and cross-species BAC-FISH has helped to identify chromosome structure and rearrangements in clover relatives such as the common and lima bean [135]. However, the extension of BAC-FISH to the BAC-painting of large chromosome regions is suitable only for species with small genomes and low proportions of repetitive fractions, and it has not been successfully established beyond crucifers [121,122], with the singular exception of Brachypodium [123].
The remarkable progress in plant genome research relating to reference sequences production and artificial DNA synthesis has provided an alternative chromosome painting technique. In silico designed and artificially synthesized oligonucleotide pools have already been applied successfully in various plant species to characterize chromosomal rearrangements [126,127,128,129,130,131,132,133]. The availability of the Trifolium genome and reference sequences means that the adoption of oligo painting within this genus, and the legume family more generally, is both possible and to be expected.
Author Contributions
Conceptualization, E.L. and J.Ř.; writing, E.L.; review and editing, J.Ř. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Education, Youth and Sports of the Czech Republic (project No. MUNI/A/1522/2020).
Conflicts of Interest
The authors declare no conflict of interests.
References
- Zohary, M.; Heller, D. The Genus Trifolium, 1st ed.; Israel Academy of Sciences and Humanities: Jerusalem, Israel, 1984; pp. 1–610. [Google Scholar]
- Gillet, J.M.; Taylor, N.L. The World of Clovers, 1st ed.; Iowa State University Press: Ames, IA, USA, 2001; pp. 1–457. [Google Scholar]
- Ellison, N.W.; Liston, A.; Steiner, J.J.; Williams, W.M.; Taylor, N.L. Molecular phylogenetics of the clover genus (Trifolium-Leguminosae). Mol. Phylogenet. Evol. 2006, 39, 688–705. [Google Scholar] [CrossRef] [PubMed]
- Zohary, M. Origin and evolution in the genus Trifolium. Bot. Notiser. 1972, 125, 501–511. [Google Scholar]
- Taylor, N.L. Clovers Around the World. In Agronomy Monographs; Taylor, N.L., Ed.; Soil Science Society of America: Madison, WI, USA, 1985; Volume 25, pp. 1–6. [Google Scholar]
- Panitsa, M.; Trigas, P.; Iatrou, G.; Sfenthourakis, S. Factors affecting plant species richness and endemism on land-bridge islands—An example from the East Aegean Archipelago. Acta Oceol. 2010, 36, 431–437. [Google Scholar] [CrossRef]
- Scoppola, A.; Tirado, J.L.; Gutiérrez, F.M.; Magrini, S. The genus Trifolium (Fabaceae) in South Europe: A critical review on species richness and distribution. Nord. J. Bot. 2018, 36, e01723. [Google Scholar] [CrossRef]
- Boissier, E. Trifolium. In Flora Orientalis; H. Georg: Basileae, Switzerland, 1872; pp. 110–156. [Google Scholar]
- Hossain, M. A revision of Trifolium in the nearer East. Notes R. Bot. Gard. Edinb. 1961, 23, 387–481. [Google Scholar]
- Lavin, M.; Doyle, J.J.; Palmer, J.D. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 1990, 44, 390–402. [Google Scholar]
- Liston, A. Use of the polymerase chain reaction to survey for the loss of the inverted repeat in the legume chloroplast genome. In Advances in Legume Systematics; Crisp, M.D., Doyle, J.J., Eds.; Royal Botanic Gardens: Kew, UK, 1995; Volume 7, pp. 31–40. [Google Scholar]
- Watson, L.E.; Sayed-Ahmed, H.; Badr, A. Molecular phylogeny of Old World Trifolium (Fabaceae). Plant Syst. Evol. 2000, 224, 153–171. [Google Scholar] [CrossRef]
- Steele, K.; Wojciechowski, M. Phylogenetic analyses of tribes Trifolieae and Vicieae, based on sequences of the plastid gene matK (Papilionoideae: Leguminosae). Adv. Legume Syst. 2003, 1, 355–370. [Google Scholar]
- Sveinsson, S.; Cronk, Q. Evolutionary origin of highly repetitive plastid genomes within the clover genus (Trifolium). BMC Evol. Biol. 2014, 14, 228. [Google Scholar] [CrossRef]
- Kintl, A.; Elbl, J.; Lošák, T.; Vaverková, M.; Nedělník, J. Mixed intercropping of wheat and white clover to enhance the sustainability of the conventional cropping system: Effects on biomass production and leaching of mineral nitrogen. Sustainability 2018, 10, 3367. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.-K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Kim, J.-S.; Islam-Faridi, M.N.; Klein, P.E.; Stelly, D.M.; Price, H.J.; Klein, R.R.; Mullet, J.E. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: Distribution of euchromatin, heterochromatin, genes and recombination in comparison to cice. Genetics 2005, 171, 1963–1976. [Google Scholar] [CrossRef]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef]
- Schubert, I. Chromosome Evolution. Curr. Opin. Plant Biol. 2007, 10, 109–115. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.P.; Schwarzacher, T. Organisation of the plant genome in chromosomes: Organisation of the plant genome in chromosomes. Plant J. 2011, 66, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Liu, B.; Segal, G.; Abbo, S.; Levy, A.A.; Vega, J.M. Rapid elimination of low-copy dna sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 1997, 147, 1381–1387. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Hiscock, S.J. Hybrid speciation in plants: New insights from molecular studies. New Phytol. 2005, 165, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F.; Schnabel, A.; Seelanan, T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 1995, 92, 280–284. [Google Scholar] [CrossRef]
- Plant DNA C-Value Database. Available online: https://cvalues.science.kew.org/ (accessed on 20 August 2021).
- Taylor, N.L.; Quesenberry, K.H.; Anderson, M.K. Genetic system relationships in Trifolium. Econ. Bot. 1979, 33, 431–441. [Google Scholar] [CrossRef]
- Index to Plant Chromosome Numbers. Available online: http://legacy.tropicos.org/Project/IPCN (accessed on 3 June 2021).
- Salimpour, F.; Sharifnia, F.; Mostafavi, G.; Hajrasoliha, S.; Ukhneh, E. chromosome counts and determination of ploid levels in iranian species of Trifolium. Chromosome Bot. 2008, 3, 53–63. [Google Scholar] [CrossRef][Green Version]
- Uslu, E. Karyology of Nine Trifolium L. Taxa from Turkey. Caryologia 2012, 65, 304–310. [Google Scholar] [CrossRef]
- Chromosome Counts Database. Available online: http://ccdb.tau.ac.il/ (accessed on 20 August 2021).
- Vižintin, L.; Javornik, B.; Bohanec, B. Genetic characterization of selected Trifolium species as revealed by nuclear DNA content and ITS rDNA region analysis. Plant Sci. 2006, 170, 859–866. [Google Scholar] [CrossRef]
- Vozárová, R.; Macková, E.; Vlk, D.; Řepková, J. Variation in ribosomal DNA in the genus Trifolium (Fabaceae). Plants 2021, 10, 1771. [Google Scholar] [CrossRef]
- Lawrence, M. Population Genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Ann. Bot.-Lond. 2000, 85, 221–226. [Google Scholar] [CrossRef]
- Abberton, M.T. Interspecific hybridization in the genus Trifolium. Plant Breed. 2007, 126, 337–342. [Google Scholar] [CrossRef]
- Williams, W.M.; Ellison, N.W.; Ansari, H.A.; Verry, I.M.; Hussain, S. Experimental evidence for the ancestry of allotetraploid Trifolium repens and creation of synthetic forms with value for plant breeding. BMC Plant Biol. 2012, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Egan, L.M.; Hofmann, R.W.; Ghamkhar, K.; Hoyos-Villegas, V. Prospects for Trifolium improvement through germplasm characterisation and pre-breeding in New Zealand and beyond. Front. Plant Sci. 2021, 12, 653191. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.M.F. Inbreeding and fertility in egyptian clover, Trifolium alexandrinum. J. Pharmacogn. Phytother. 2012, 4, 16–25. [Google Scholar]
- Schifino, M.T.; Moreas-Fernandes, M.I.B. Cytological comparision of diploid and autotetraploid Trifolium riograndense Burkart (Leguminosae). Rev. Bras. Genet. IX 1986, 4, 637–643. [Google Scholar]
- Schifino-Wittmann, M.T.; Moraes-Fernandes, B. Chromosome numbers, karyotypes and meiotic behavior of populations of some Trifolium (Leguminosae) species. Rev. Brazil Genet. 1988, 11, 379–390. [Google Scholar]
- Sheidai, M.; Hamta, A.; Mofidabadi, A.J.; NooriDaloii, M.R. Karyotypic study of Trifolium species and cultivars in Iran. J. Sci. Islam. Repub. Iran 1998, 9, 215–222. [Google Scholar]
- Khatoon, S.; Ali, S.I. Chromosome numbers and polyploidy in the legumes of Pakistan. Pak. J. Bot. 2006, 38, 935–945. [Google Scholar]
- Conterato, I.F.; Schifino-Wittmann, M.T.; Dall′Agnol, M. Seed dimorphism, chromosome number and karyotype of the amphicarpic species Trifolium argentinense Speg. Genet. Resour. Crop. Evol. 2010, 57, 727–731. [Google Scholar] [CrossRef]
- Isobe, S.; Klimenko, I.; Ivashuta, S.; Gau, M.; Kozlov, N.N. First RFLP linkage map of red clover (Trifolium pratense L.) based on cDNA probes and its transferability to other red clover germplasm. Theor. Appl. Genet. 2003, 108, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Isobe, S.; Asamizu, E.; Ohmido, N.; Kataoka, R.; Nakamura, Y.; Kaneko, T.; Sakurai, N.; Okumura, K.; Klimenko, I.; et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res. 2005, 12, 301–364. [Google Scholar] [CrossRef]
- Isobe, S.; Kolliker, R.; Hisano, H.; Sasamoto, S.; Wada, T.; Klimenko, I.; Okumura, K.; Tabata, S. Construction of a consensus linkage map for red clover (Trifolium pratense L.). BMC Plant Biol. 2009, 9, 57. [Google Scholar] [CrossRef]
- Zhang, Y.; Sledge, M.K.; Bouton, J.H. Genome mapping of white clover (Trifolium repens L.) and comparative analysis within the Trifolieae using cross-species SSR markers. Theor. Appl. Genet. 2007, 114, 1367–1378. [Google Scholar] [CrossRef]
- Ghamkhar, K.; Isobe, S.; Nichols, P.G.H.; Faithfull, T.; Ryan, M.H.; Snowball, R.; Sato, S.; Appels, R. The first genetic maps for subterranean clover (Trifolium subterraneum L.) and comparative genomics with T. pratense L. and Medicago Truncatula Gaertn. to identify new molecular markers for breeding. Mol. Breed. 2012, 30, 213–226. [Google Scholar] [CrossRef]
- Isobe, S.N.; Hisano, H.; Sato, S.; Hirakawa, H.; Okumura, K.; Shirasawa, K.; Sasamoto, S.; Watanabe, A.; Wada, T.; Kishida, Y.; et al. Comparative genetic mapping and discovery of linkage disequilibrium across linkage groups in white clover (Trifolium repens L.). G3-Genes Genomes Genet. 2012, 2, 607–617. [Google Scholar] [CrossRef]
- Jones, E.S.; Hughes, L.J.; Drayton, M.C.; Abberton, M.T.; Michaelson-Yeates, T.P.T.; Bowen, C.; Forster, J.W. An SSR and AFLP molecular marker-based genetic map of white clover (Trifolium repens L.). Plant Sci. 2003, 165, 531–539. [Google Scholar] [CrossRef]
- Barrett, B.; Griffiths, A.; Schreiber, M.; Ellison, N.; Mercer, C.; Bouton, J.; Ong, B.; Forster, J.; Sawbridge, T.; Spangenberg, G.; et al. a microsatellite map of white clover. Theor. Appl. Genet. 2004, 109, 596–608. [Google Scholar] [CrossRef]
- Griffiths, A.G.; Barrett, B.A.; Simon, D.; Khan, A.K.; Bickerstaff, P.; Anderson, C.B.; Franzmayr, B.K.; Hancock, K.R.; Jones, C.S. An integrated genetic linkage map for white clover (Trifolium repens L.) with alignment to Medicago. BMC Genom. 2013, 14, 388. [Google Scholar] [CrossRef]
- Ištvánek, J.; Jaroš, M.; Křenek, A.; Řepková, J. Genome assembly and annotation for red clover (Trifolium pratense; Fabaceae). Am. J. Bot. 2014, 101, 327–337. [Google Scholar] [CrossRef]
- De Vega, J.J.; Ayling, S.; Hegarty, M.; Kudrna, D.; Goicoechea, J.L.; Ergon, Å.; Rognli, O.A.; Jones, C.; Swain, M.; Geurts, R.; et al. Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 2015, 5, 17394. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Dluhošová, J.; Ištvánek, J.; Nedělník, J.; Řepková, J. Red clover (Trifolium pratense) and zigzag clover (T. medium)—A picture of genomic similarities and differences. Front. Plant. Sci. 2018, 9, 724. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Kaur, P.; Shirasawa, K.; Nichols, P.; Nagano, S.; Appels, R.; Erskine, W.; Isobe, S.N. Draft genome sequence of subterranean clover, a reference for genus Trifolium. Sci. Rep. 2016, 6, 30358. [Google Scholar] [CrossRef]
- Griffiths, A.G.; Moraga, R.; Tausen, M.; Gupta, V.; Bilton, T.P.; Campbell, M.A.; Ashby, R.; Nagy, I.; Khan, A.; Larking, A.; et al. Breaking Free: The genomics of allopolyploidy-facilitated niche expansion in white clover. Plant Cell 2019, 31, 1466–1487. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M. Clovers. In Evolution of Crop Plants, 1st ed.; Simmonds, N.W., Ed.; Longman: London, UK, 1976; Volume 4, pp. 79–98. [Google Scholar]
- Ansari, H.A.; Ellison, N.W.; Williams, W.M. Molecular and cytogenetic evidence for an allotetraploid origin of Trifolium dubium (Leguminosae). Chromosoma 2008, 117, 159–167. [Google Scholar] [CrossRef]
- Neumann, P.; Navrátilová, A.; Schroeder-Reiter, E.; Koblížková, A.; Steinbauerová, V.; Chocholová, E.; Novák, P.; Wanner, G.; Macas, J. Stretching the rules: Monocentric chromosomes with multiple centromere domains. PLoS Genet. 2012, 8, e1002777. [Google Scholar] [CrossRef]
- Macas, J.; Novák, P.; Pellicer, J.; Čížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 2015, 10, e0143424. [Google Scholar] [CrossRef]
- Ávila Robledillo, L.; Koblížková, A.; Novák, P.; Böttinger, K.; Vrbová, I.; Neumann, P.; Schubert, I.; Macas, J. Satellite DNA in Vicia faba is characterized by remarkable diversity in its sequence composition, association with centromeres, and replication timing. Sci. Rep. 2018, 8, 5838. [Google Scholar] [CrossRef]
- Taylor, N.L.; Quesenberry, K.H. Red Clover Science; Kluwer Academic: Dordrecht, The Netherlands, 1996; Volume 28. [Google Scholar]
- Williams, W.M. Trifolium interspecific hybridisation: Widening the white clover gene pool. Crop Pasture Sci. 2014, 65, 1091. [Google Scholar] [CrossRef]
- Řepková, J.; Nedbálková, B.; Holub, J. Regeneration of plants from zygotic embryos after interspecific hybridization within the genus Trifolium and electrophoretic evaluation of hybrids. Sci. Stud. Res. Inst. Fodd. Plants 1991, 12, 7–14. [Google Scholar]
- Řepkova, J.; Jungmannová, B.; Jakešová, H. Identification of barriers to interspecific crosses in the genus Trifolium. Euphytica 2006, 151, 39–48. [Google Scholar] [CrossRef]
- Fuchs, J.; Strehl, S.; Brandes, A.; Schweizer, D.; Schubert, I. Molecular-cytogenetic characterization of the Vicia faba genome—Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res. 1998, 6, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Kühne, M.; Schubert, I. Assignment of linkage groups to pea chromosomes after karyotyping and gene mapping by fluorescent in situ hybridization. Chromosoma 1998, 107, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, R.; Hara, M.; Kato, S.; Isobe, S.; Sato, S.; Tabata, S.; Ohmido, N. Integration of linkage and chromosome maps of red clover (Trifolium pratense L.). Cytogenet. Genome Res. 2012, 137, 60–69. [Google Scholar] [CrossRef]
- De Oliveira Bustamante, F.; do Nascimento, T.H.; Montenegro, C.; Dias, S.; do Vale Martins, L.; Braz, G.T.; Benko-Iseppon, A.M.; Jiang, J.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Oligo-FISH barcode in beans: A new chromosome identification system. Theor. Appl. Genet. 2021, 134, 3675–3686. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, C.; do Vale Martins, L.; de Oliveira Bustamante, F.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Comparative cytogenomics reveals genome reshuffling and centromere repositioning in the legume tribe Phaseoleae. bioRxiv 2021. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef]
- Flavell, R.B. The structure and control of expression of ribosomal RNA genes. Oxf. Surv. Plant Mol. Cell Biol. 1986, 3, 251–274. [Google Scholar]
- Dvořák, J.; Zhang, H.-B.; Kota, R.S.; Lassner, M. Organization and evolution of the 5S ribosomal rna gene family in wheat and related species. Genome 1989, 32, 1003–1016. [Google Scholar] [CrossRef]
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148. [Google Scholar] [CrossRef]
- Raina, S.N.; Mukai, Y. Detection of a variable number of 18S-5.8S-26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome 1999, 42, 52–59. [Google Scholar] [CrossRef]
- Pedrosa-Harand, A.; de Almeida, C.C.S.; Mosiolek, M.; Blair, M.W.; Schweizer, D.; Guerra, M. Extensive ribosomal DNA amplification during andean common bean (Phaseolus vulgaris L.) evolution. Theor. Appl. Genet. 2006, 112, 924–933. [Google Scholar] [CrossRef]
- Chung, M.-C.; Lee, Y.-I.; Cheng, Y.-Y.; Chou, Y.-J.; Lu, C.-F. Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theor. Appl. Genet. 2008, 116, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Roa, F.; Guerra, M. Non-Random Distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet. Genome Res. 2015, 146, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Dluhošová, J.; Řepková, J.; Jakešová, H.; Nedělník, J. Impact of interspecific hybridization of T. pratense × T. medium and backcrossing on genetic variability of progeny. Czech J. Genet. Plant 2016, 52, 125–131. [Google Scholar] [CrossRef]
- Falistocco, E.; Marconi, G.; Falcinelli, M. Comparative cytogenetic study on Trifolium subterraneum (2n = 16) and Trifolium israeliticum (2n = 12). Genome 2013, 56, 307–313. [Google Scholar] [CrossRef]
- Ansari, H. Molecular cytogenetic organization of 5S and 18S-26S rDNA loci in white clover (Trifolium repens L.) and related species. Ann. Bot. Lond. 1999, 83, 199–206. [Google Scholar] [CrossRef]
- Vicient, C.M.; Suoniemi, A.; Anamthawat-Jonsson, K.; Tanskanen, J.; Beharav, A.; Nevo, E.; Schulman, A.H. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. The Plant Cell. 1999, 11, 1769. [Google Scholar] [CrossRef]
- Macas, J.; Neumann, P.; Navrátilová, A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom. 2007, 8, 427. [Google Scholar] [CrossRef]
- Macas, J.; Kejnovský, E.; Neumann, P.; Novák, P.; Koblížková, A.; Vyskot, B. Next generation sequencing-based analysis of repetitive DNA in the model dioecious [corrected] plant Silene latifolia. PLoS ONE 2011, 6, e27335. [Google Scholar] [CrossRef]
- Tenaillon, M.I.; Hufford, M.B.; Gaut, B.S.; Ross-Ibarra, J. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol. Evol. 2011, 3, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Piednoël, M.; Aberer, A.J.; Schneeweiss, G.M.; Macas, J.; Novak, P.; Gundlach, H.; Temsch, E.M.; Renner, S.S. Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol. Biol. Evol. 2012, 29, 3601–3611. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Hřibová, E.; Neumann, P.; Koblížková, A.; Doležel, J.; Macas, J. Genome-wide analysis of repeat diversity across the family Musaceae. PLoS ONE 2014, 9, e98918. [Google Scholar] [CrossRef]
- Neumann, P.; Koblížková, A.; Navrátilová, A.; Macas, J. Significant expansion of Vicia Pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 2006, 173, 1047–1056. [Google Scholar] [CrossRef]
- Kulikova, O.; Gualtieri, G.; Geurts, R.; Kim, D.-J.; Cook, D.; Huguet, T.; De Jong, J.H.; Fransz, P.F.; Bisseling, T. Integration of the FISH pachytene and genetic maps of Medicago truncatula: FISH pachytene and genetic maps of M. truncatula. Plant J. 2001, 27, 49–58. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Jacobus, B.H.; SanMiguel, P.; Walling, J.G.; Yuan, Y.; Shoemaker, R.C.; Young, N.D.; Jackson, S.A. Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 2005, 170, 1221–1230. [Google Scholar] [CrossRef]
- Jiang, J.; Birchler, J.A.; Parrott, W.A.; Kelly Dawe, R. A molecular view of plant centromeres. Trends Plant Sci. 2003, 8, 570–575. [Google Scholar] [CrossRef]
- Zhu, J.; Ellison, N.; Rowland, R. Chromosomal localization of a tandemly repeated DNA sequence in Trifolium repens L. Cell Res. 1996, 6, 39–46. [Google Scholar] [CrossRef]
- Bucknell, T.T. Comparative cytogenetics in the genus Trifolium section Trifolium (clover). Master’s Thesis, Massey University, Palmerston North, New Zealand, 1999. [Google Scholar]
- Ansari, H.A.; Ellison, N.W.; Griffiths, A.G.; Williams, W.M. A lineage-specific centromeric satellite sequence in the genus Trifolium. Chromosome Res. 2004, 12, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of Similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Galasso, I.; Schmidt, T.; Pignone, D.; Heslop-Harrison, J.S. The molecular cytogenetics of Vigna unguiculata (L.) Walp: The physical organization and characterization of 18S-5.8S-25S rRNA genes, 5S rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theoret. Appl. Genet. 1995, 91, 928–935. [Google Scholar] [CrossRef]
- Gill, N.; Findley, S.; Walling, J.G.; Hans, C.; Ma, J.; Doyle, J.; Stacey, G.; Jackson, S.A. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 2009, 151, 1167–1174. [Google Scholar] [CrossRef]
- Findley, S.D.; Cannon, S.; Varala, K.; Du, J.; Ma, J.; Hudson, M.E.; Birchler, J.A.; Stacey, G. A fluorescence in situ hybridization system for karyotyping soybean. Genetics 2010, 185, 727–744. [Google Scholar] [CrossRef]
- Tek, A.L.; Kashihara, K.; Murata, M.; Nagaki, K. Functional Centromeres in Soybean Include Two Distinct Tandem Repeats and a Retrotransposon. Chromosome Res. 2010, 18, 337–347. [Google Scholar] [CrossRef]
- Zatloukalová, P.; Hřibová, E.; Kubaláková, M.; Suchánková, P.; Šimková, H.; Adoración, C.; Kahl, G.; Millán, T.; Doležel, J. Integration of genetic and physical maps of the chickpea (Cicer arietinum L.) genome using flow-sorted chromosomes. Chromosome Res. 2011, 19, 729–739. [Google Scholar] [CrossRef]
- Iwata, A.; Tek, A.L.; Richard, M.M.S.; Abernathy, B.; Fonsêca, A.; Schmutz, J.; Chen, N.W.G.; Thareau, V.; Magdelenat, G.; Li, Y.; et al. Identification and characterization of functional centromeres of the common bean. Plant J. 2013, 76, 47–60. [Google Scholar] [CrossRef]
- Yu, F.; Dou, Q.; Liu, R.; Wang, H. A conserved repetitive DNA element located in the centromeres of chromosomes in Medicago genus. Genes Genom. 2017, 39, 903–911. [Google Scholar] [CrossRef]
- Karafiátová, M.; Bartoš, J.; Kopecký, D.; Ma, L.; Sato, K.; Houben, A.; Stein, N.; Doležel, J. Mapping nonrecombining regions in barley using multicolor FISH. Chromosome Res. 2013, 21, 739–751. [Google Scholar] [CrossRef]
- Danilova, T.V.; Friebe, B.; Gill, B.S. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor. Appl. Genet. 2014, 127, 715–730. [Google Scholar] [CrossRef]
- Ebeed, H.T. Omics approaches for developing abiotic stress tolerance in wheat. In Wheat Production in Changing Environments; Springer: Singapore, 2019. [Google Scholar]
- Simpson, P.R.; Newman, M.-A.; Davies, D.R. Detection of legumin gene DNA sequences in pea by in situ hybridization. Chromosoma 1988, 96, 454–458. [Google Scholar] [CrossRef]
- Schaff, D.A.; Koehler, S.M.; Matthews, B.F.; Bauchan, G.R. In situ hybridization of β-tubulin to alfalfa chromosomes. J. Hered. 1990, 81, 480–483. [Google Scholar] [CrossRef]
- Danilova, T.V.; Birchler, J.A. Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: Resolution, sensitivity, and banding paint development. Chromosoma 2008, 117, 345–356. [Google Scholar] [CrossRef]
- Jiang, J.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 4487–4491. [Google Scholar] [CrossRef] [PubMed]
- Lapitan, N.L.V.; Brown, S.E.; Kennard, W.; Stephens, J.L.; Knudson, D.L. FISH physical mapping with barley BAC clones. Plant J. 1997, 11, 149–156. [Google Scholar] [CrossRef]
- Peterson, D.G.; Lapitan, N.L.; Stack, S.M. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 1999, 152, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.-B.; Bodeau, J.; Fransz, P.F.; Williamson, V.M.; van Kammen, A.; de Jong, J.H.; Zabel, P. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively: Theor. Appl. Genet. 1999, 98, 365–370. [Google Scholar] [CrossRef]
- Islam-Faridi, M.N.; Childs, K.L.; Klein, P.E.; Hodnett, G.; Menz, M.A.; Klein, R.R.; Rooney, W.L.; Mullet, J.E.; Stelly, D.M.; Price, H.J. A molecular cytogenetic map of sorghum Chromosome 1. Fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 2002, 161, 345–353. [Google Scholar] [CrossRef]
- Lee, H.-R.; Eom, E.-M.; Lim, Y.-P.; Bang, J.-W.; Lee, D.-H. Construction of a garlic BAC library and chromosomal assignment of BAC clones using the FISH technique. Genome 2003, 46, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.; Kulikova, O.; Penmetsa, R.V.; Bisseling, T.; Cook, D.R.; Frugoli, J. An integrated physical, genetic and cytogenetic map around the sunn locus of Medicago truncatula. Genome 2003, 46, 665–672. [Google Scholar] [CrossRef]
- Pedrosa, A.; Sandal, N.; Stougaard, J.; Schweizer, D.; Bachmair, A. Chromosomal map of the model legume Lotus japonicus. Genetics 2002, 161, 1661–1672. [Google Scholar] [CrossRef]
- Pedrosa-Harand, A.; Kami, J.; Gepts, P.; Geffroy, V.; Schweizer, D. Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosome Res. 2009, 17, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Fonsêca, A.; Ferreira, J.; dos Santos, T.R.B.; Mosiolek, M.; Bellucci, E.; Kami, J.; Gepts, P.; Geffroy, V.; Schweizer, D.; dos Santos, K.G.B.; et al. Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosome Res. 2010, 18, 487–502. [Google Scholar] [CrossRef]
- Schubert, I.; Fransz, P.F.; Fuchs, J.; de Jong, J.H. Chromosome painting in plants. Method Cell Sci. 2001, 23, 57–69. [Google Scholar] [CrossRef]
- Lysak, M.A.; Fransz, P.F.; Ali, H.B.; Schubert, I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001, 28, 689–697. [Google Scholar] [CrossRef]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Betekhtin, A.; Jenkins, G.; Hasterok, R. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS ONE 2014, 9, e115108. [Google Scholar] [CrossRef]
- Iovene, M.; Wielgus, S.M.; Simon, P.W.; Buell, C.R.; Jiang, J. Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 2008, 180, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Amarillo, F.I.E.; Bass, H.W. A transgenomic cytogenetic sorghum (Sorghum propinquum) bacterial artificial chromosome fluorescence in situ hybridization map of maize (Zea mays L.) Pachytene chromosome 9, evidence for regions of genome hyperexpansion. Genetics 2007, 177, 1509–1526. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Braz, G.T.; He, L.; Zhao, H.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Torres, G.A.; Jiang, J. Comparative oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2018, 208, 513–523. [Google Scholar] [CrossRef]
- Qu, M.; Li, K.; Han, Y.; Chen, L.; Li, Z.; Han, Y. Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet. Genome Res. 2017, 153, 158–164. [Google Scholar] [CrossRef]
- Albert, P.S.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Kao, Y.-H.; Wang, C.-J.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sun, S.; Wu, Y.; Zhou, Y.; Gu, S.; Yu, H.; Yi, C.; Gu, M.; Jiang, J.; Liu, B.; et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 2020, 101, 112–121. [Google Scholar] [CrossRef]
- Šimoníková, D.; Němečková, A.; Karafiátová, M.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa spp.). Front. Plant Sci. 2019, 10, 1503. [Google Scholar] [CrossRef]
- Li, G.; Zhang, T.; Yu, Z.; Wang, H.; Yang, E.; Yang, Z. An efficient oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 2021, 105, 978–993. [Google Scholar] [CrossRef]
- Do Vale Martins, L.; de Oliveira Bustamante, F.; da Silva Oliveira, A.R.; da Costa, A.F.; de Lima Feitoza, L.; Liang, Q.; Zhao, H.; Benko-Iseppon, A.M.; Muñoz-Amatriaín, M.; Pedrosa-Harand, A.; et al. BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 2021, 130, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Cui, C.; Liu, H.; Fu, L.; Li, L.; Dai, X.; Qin, L.; Wang, S.; Han, S.; Xu, J.; et al. Development of an oligonucleotide dye solution facilitates high throughput and cost-efficient chromosome identification in peanut. Plant Methods 2019, 15, 69. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, E.M.; Fonsêca, A.; Almeida, C.; Dos Santos, K.G.B.; Pedrosa-Harand, A. Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor. Appl. Genet. 2012, 124, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).