Genome-Wide Analysis and the Expression Pattern of the MADS-Box Gene Family in Bletilla striata
Abstract
:1. Introduction
2. Results
2.1. Identification of BsMADS
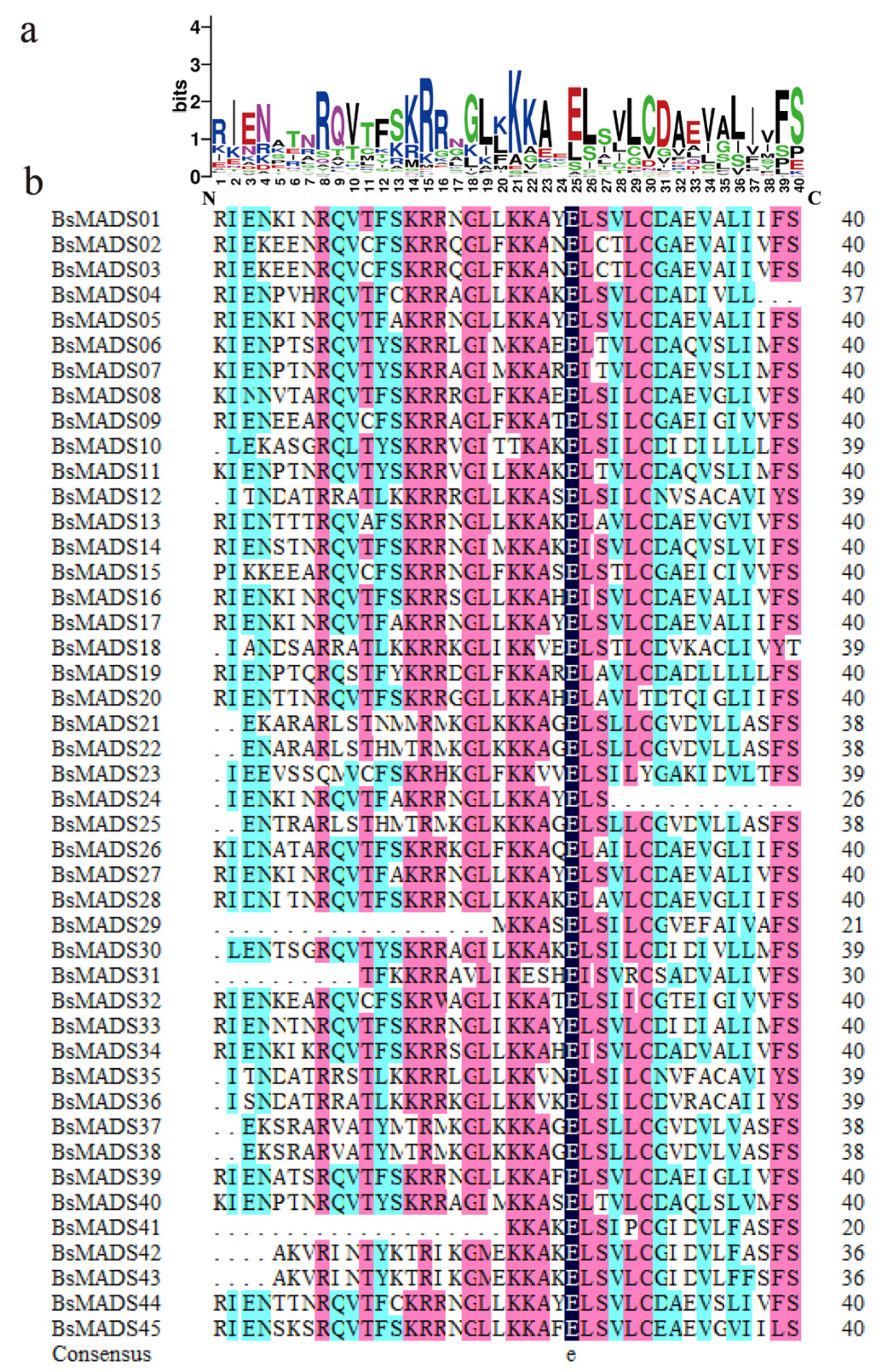
2.2. Analysis of Multiple Sequence Alignments and Cis-Acting Elements
2.3. Phylogenetic Analysis of the MADS-Box Family between B. striata and Arabidopsis
2.4. Gene Structure, Motif Composition, and Ka/Ks Analysis of BsMADSs
2.5. Synteny Analysis and Chromosomal Distribution of MADS Genes
2.6. Expression Profile of BsMADS Genes under Non-Stressed Growth Conditions
2.7. Analysis of Expression Patterns under Different Stress Treatments
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Identification of MADS-Box Gene in B. striata
4.3. Multiple Alignment and Phylogenetic Analysis
4.4. Prediction of Conserved Motifs and Gene Structure Analysis
4.5. Cis-Elements and Ka/Ks Analysis
4.6. RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Tu, Y.Y.; Cheng, X.J.; Zhang, L.L.; Meng, H.L.; Zhao, X.; Zhang, W.; He, B.; Amato, A. Genome-wide identification and expression profile of the MADS-box gene family in Erigeron breviscapus. PLoS ONE 2019, 14, e0226599. [Google Scholar] [CrossRef]
- Shchennikova, A.V.; Shulga, O.A.; Skryabin, K.G. Diversification of the homeotic AP3 clade MADS-box genes in asteraceae species Chrysanthemum morifolium L. and Helianthus annuus L. Dokl. Biochem. Biophys. 2018, 483, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.H.; Yu, J.Q.; Wen, L.Z.; Guo, Y.H.; Sun, X.; Hao, Y.J.; Hu, D.G.; Zheng, C.S. Chrysanthemum MADS-box transcription factor CmANR1 modulates lateral root development via homo-/heterodimerization to influence auxin accumulation in Arabidopsis. Plant Sci. 2017, 266, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Liu, S.; Somers, D.J. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Wu, Y.W.; Ke, Y.Z.; Wen, J.; Guo, P.C.; Ran, F.; Wang, M.M.; Liu, M.M.; Li, P.F.; Li, J.N.; Du, H.; et al. Evolution and expression analyses of the MADS-box gene family in Brassica napus. PLoS ONE 2018, 13, e0200762. [Google Scholar] [CrossRef]
- Koo, S.C.; Bracko, O.; Mi, S.P.; Schwab, R.; Chun, H.J.; Park, K.M.; Seo, J.S.; Grbic, V.; Balasubramanian, S.; Schmid, M.; et al. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J. 2010, 62, 807–816. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martinez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damveld, R.A.; Arentshorst, M.; Franken, A.; Vankuyk, P.A.; Klis, F.M.; Ram, A. The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol. Microbiol. 2010, 58, 305–319. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theissen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.; Carlsbecker, A.; Sundås-Larsson, A.; Vahala, T. APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta 2007, 225, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Passmore, S.; Maine, G.T.; Elble, R.; Christ, C.; Tye, B.K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988, 204, 593–606. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. Embo J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Bodt, S.D.; Raes, J.; Peer, Y.; TheiβEn, G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar] [CrossRef]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell. 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [Green Version]
- Becker, A.; TheiβEn, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Yang, Y.; Jack, T. Defining subdomains of the K domain important for protei–protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef]
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar] [CrossRef]
- Hou, X.J.; Liu, S.R.; Khan, M.R.G. Genome-wide identification, classification, expression profiling, and SSR marker development of the MADS-box gene family in citrus. Plant Mol. Biol. Rep. 2014, 32, 28–41. [Google Scholar] [CrossRef]
- Lu, S.J.; Wei, H.; Wang, Y.; Wang, H.M.; Yang, R.F.; Zhang, X.B.; Tu, J.M. Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 2012, 30, 1461–1469. [Google Scholar] [CrossRef]
- Xu, D.; Pan, Y.; Chen, J. Chemical constituents, pharmacologic properties, and clinical applications of Bletilla striata. Front Pharmacol. 2019, 10, 1168. [Google Scholar] [CrossRef]
- Hossain, M.M. Therapeutic orchids: Traditional uses and recent advances-an overview. Fitoterapia 2011, 82, 102–140. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.; Manorama; et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama; et al. Genome-Wide Identification and Characterization of the Brassinazole-resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Fang, Y.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-wide identifcation and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 2020, 93, 89–103. [Google Scholar] [CrossRef]
- Baruah, P.M.; Krishnatreya, D.B.; Bordoloi, K.S.; Gill, S.S.; Agarwala, N. Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum. Plant Physiol. Bioch. 2021, 162, 221–236. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, H.; Liu, J.; Du, Q.; Lu, S.; Liu, C. Genome-wide identification and functional characterization of natural antisense transcripts in Salvia miltiorrhiza. Sci. Rep. 2021, 11, 4769. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Zhou, W.; Xie, L.; Wang, L.; Zhang, Y.; Zhang, Q. Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genom. 2021, 22, 361. [Google Scholar]
- Burgeff, C.; Liljegren, S.J.; Tapia-López, R.; Yanofsky, M.F.; Alvarez-Buylla, E.R. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 2002, 214, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Dee, Z.P.; Wittich, P.; PE‘, M.E.; Rigola, D.; Buono, I.D.; Gorla, M.S.; Kater, M.M.; Colombo, L. OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev. Genet. 1999, 25, 237–244. [Google Scholar] [CrossRef]
- Prasad, K.; Parameswaran, S.; Vijayraghavan, U. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 2010, 43, 915–928. [Google Scholar] [CrossRef]
- Lin, C.S.; Hsu, C.T.; Liao, D.C.; Chang, W.J.; Chou, M.L.; Huang, Y.T.; Chen, J.W.; Ko, S.S.; Chan, M.T.; Shih, M.C. Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 2016, 14, 284–298. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, T.; Shi, J.L.; Chen, D.H.; Shen, D.L.; Feng, M. Cloning and characterization of a novel PI-like MADS-box gene in Phalaenopsis orchid. J. Biochem. Mol. Biol. 2008, 40, 845–852. [Google Scholar]
- Riechmann, J.L.; Meyerowitz, E.M. MADS domain proteins in plant development. Bio Chem. 1997, 378, 1079–1101. [Google Scholar]
- Yoshida, T.; Kawabe, A. Importance of gene duplication in the evolution of genomic imprinting revealed by molecular evolutionary analysis of the type I MADS-box gene family in Arabidopsis species. PLoS ONE 2013, 8, e73588. [Google Scholar] [CrossRef] [Green Version]
- Theien, G.; Kim, J.T.; Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef]
- Lydia, G.; Günter, T. Phylogenomics of MADS-box genes in plants-two opposing life styles in one gene family. Biology 2013, 2, 1150–1164. [Google Scholar]
- Tian, Y.; Dong, Q.L.; Ji, Z.R.; Chi, F.M.; Cong, P.H.; Zhou, Z.S. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant. 2019, 12, 489–505. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.T.; Hu, Z.L.; Deng, L.; Wang, Y.; Zhu, M.K.; Zhang, J.L.; Chen, G.P. A tomato MADS-box transcription factor, S1MADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Maekawa, M.; Miyao, A.; Hirochika, H.; Kyozuka, J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2009, 51, 47–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuer, S. The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiol. 2001, 127, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Li, X.Y.; Chen, W.J.; Peng, X.J.; Xiao, C.; Zhu, S.W.; Cheng, B.J. Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tiss Org. 2011, 105, 159–173. [Google Scholar] [CrossRef]
- Li, K.Y.; Anamika, S.; Crooks, D.R.; Dai, X.M.; Cong, Z.Z.; Liang, P.; Dung, H.; Rouault, T.A.; Tracey, A.; Deb, S. Expression of human frataxin is regulated by transcription factors SRF and TFAP2. PLoS ONE 2010, 5, e12286. [Google Scholar] [CrossRef]
- Lundon, D.J.; Boland, A.; Prencipe, M.; Hurley, G.; O’Neill, A.; Kay, E.; Aherne, S.T.; Doolan, P.; Madden, S.F.; Clynes, M.; et al. The prognostic utility of the transcription factor SRF in docetaxel-resistant prostate cancer: In-vitro discovery and in-vivo validation. BMC Cancer 2017, 17, 163. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, Y.; Liang, X.; Nie, S.S.; Chen, Y.L.; Liang, D.Y.; Sun, X.C.; Karanja, B.K.; Luo, X.B.; Liu, L.W. Genome-wide characterization of the MADS-box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef] [Green Version]
- Prachumwat, A.; Li, W.H. Gene number expansion and contraction in vertebrate genomes with respect to invertebrate genomes. Genome Res. 2008, 18, 221–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montiel, G.; Gaudet, M.; Laurans, F.; Rozenberg, P.; Simom, M.; Gantet, P.; Jay-Allemand, C.; Breton, C. Overexpression of MADS-box gene AGAMOUS-LIKE 12 activates root development in Juglans sp. and Arabidopsis thaliana. Plants 2020, 9, 444. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.A.; Yang, S.H.; Hair, A.; Galanis, A.; Sharrocks, A.D. Molecular characterization of a zebrafish TCF ETS-domain transcription factor. Front. Plant Sci. 2000, 18, 7985–7993. [Google Scholar] [CrossRef] [Green Version]
- Song, X.M.; Liu, T.K.; Duan, W.K.; Ma, Q.H.; Ren, J.; Wang, Z.; Li, Y.; Hou, X.L. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genomics 2014, 103, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.K.; Song, X.M.; Liu, T.K.; Huang, Z.N.; Ren, J.; Hou, X.L.; Ying, L. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef]
- Lu, W.J.; Chen, J.X.; Ren, X.C.; Yuan, J.J.; Han, X.Y.; Mao, L.C.; Ying, T.J.; Luo, Z.S. One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening. Sci. Hortic.-Amsterdam 2018, 227, 124–131. [Google Scholar] [CrossRef]
- Guo, J.; Shi, X.X.; Zhang, J.S.; Duan, Y.H.; Bai, P.F.; Guan, X.N.; Kang, Z.S. A type I MADS-box gene is differentially expressed in wheat in response to infection by the stripe rust fungus. Biol. Plantarum. 2013, 57, 540–546. [Google Scholar] [CrossRef]
- Tang, Y.H.; Wang, J.; Bao, X.X.; Wu, Q.; Yang, T.W.; Li, H.; Wang, W.X.; Zhang, Y.Z.; Bai, N.N.; Guan, Y.X.; et al. Genome-wide analysis of Jatropha curcas MADS-box gene family and functional characterization of the JcMADS40 gene in transgenic rice. BMC Genom. 2020, 21, 325. [Google Scholar] [CrossRef]
- Tanaka, Y.; Yamamura, T.; Terakawa, T. Identification and expression analysis of the Cyclamen persicum MADS-box gene family. Plant Biotechnol. 2011, 28, 167–172. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.T.; Zhao, P.C.; Cheng, L.Q.; Yuan, G.X.; Yang, W.G.; Liu, S.; Chen, S.Y.; Qi, D.M.; Liu, G.S.; Li, X.X. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Bio. 2018, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Zhang, R.Z.; Guo, J.J.; Liu, D.M.; Li, A.L.; Fan, R.C.; Zhang, X.Q. Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS ONE 2014, 9, e84781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.F.; Chen, X.Y.; Zhou, S.G.; Xie, Q.L.; Wang, Y.S.; Xiang, X.X.; Hu, Z.L.; Chen, G.P. Overexpression of SIMBP22 in tomato affects plant growth and enhances tolerance to drought stress. Plant Sci. 2020, 301, 110672. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Nadya, W.; Chris, M.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Magali, L.; Patrice, D.; Gert, T.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi, Z.-Y.; Zhao, Q.; Lu, C.; Zhang, Q.; Li, L.; Liu, S.; Wang, S.-Q.; Wang, Z.-Z.; Niu, J.-F. Genome-Wide Analysis and the Expression Pattern of the MADS-Box Gene Family in Bletilla striata. Plants 2021, 10, 2184. https://doi.org/10.3390/plants10102184
Mi Z-Y, Zhao Q, Lu C, Zhang Q, Li L, Liu S, Wang S-Q, Wang Z-Z, Niu J-F. Genome-Wide Analysis and the Expression Pattern of the MADS-Box Gene Family in Bletilla striata. Plants. 2021; 10(10):2184. https://doi.org/10.3390/plants10102184
Chicago/Turabian StyleMi, Ze-Yuan, Qian Zhao, Chan Lu, Qian Zhang, Lin Li, Shuai Liu, Shi-Qiang Wang, Zhe-Zhi Wang, and Jun-Feng Niu. 2021. "Genome-Wide Analysis and the Expression Pattern of the MADS-Box Gene Family in Bletilla striata" Plants 10, no. 10: 2184. https://doi.org/10.3390/plants10102184
APA StyleMi, Z.-Y., Zhao, Q., Lu, C., Zhang, Q., Li, L., Liu, S., Wang, S.-Q., Wang, Z.-Z., & Niu, J.-F. (2021). Genome-Wide Analysis and the Expression Pattern of the MADS-Box Gene Family in Bletilla striata. Plants, 10(10), 2184. https://doi.org/10.3390/plants10102184






