Gene Duplication and Differential Expression of Flower Symmetry Genes in Rhododendron (Ericaceae)
Abstract
:1. Introduction
2. Results
2.1. Characterization of CYC, RAD, and DIV Orthologs in Rhododendron
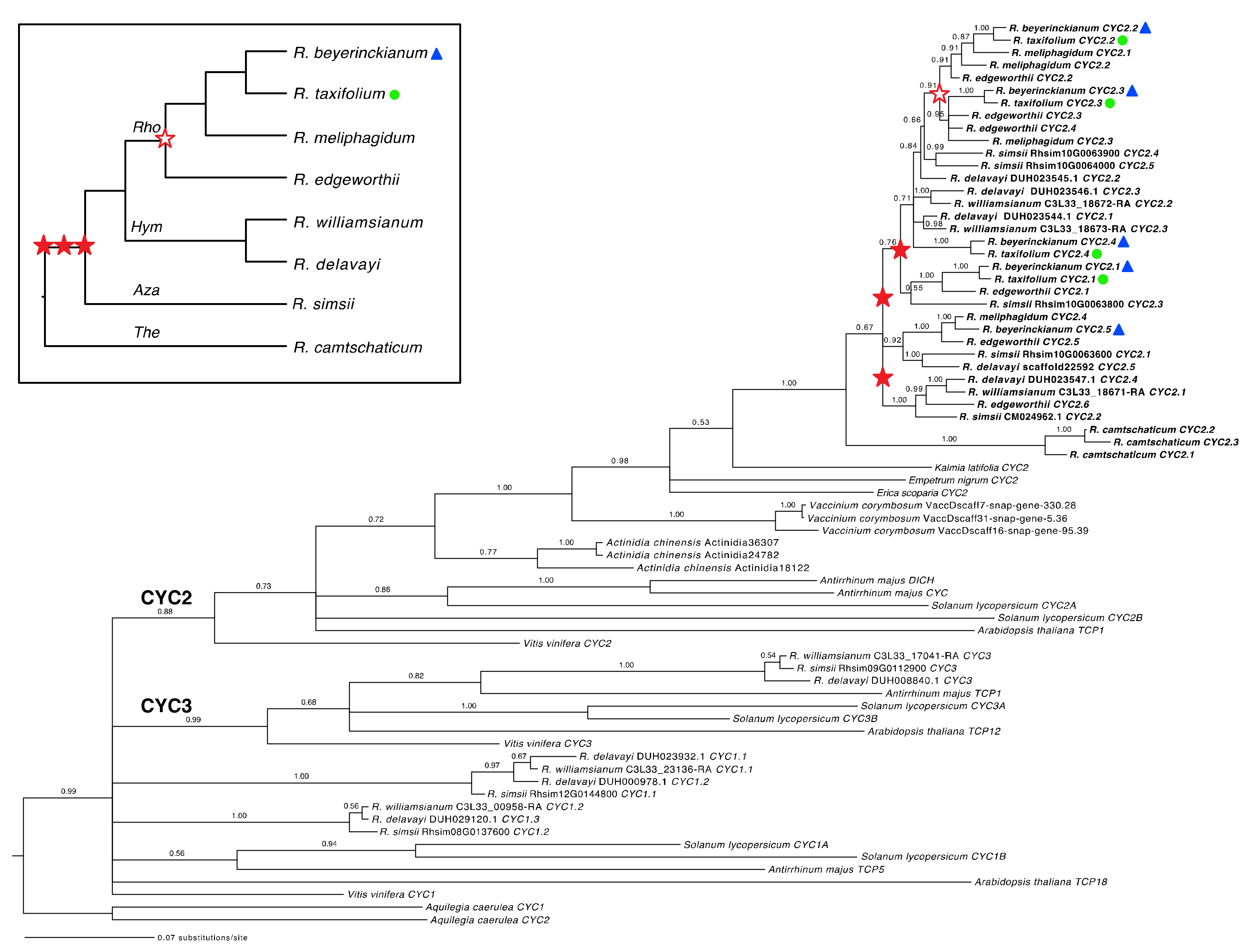
2.1.1. Both Shared and Independent Duplications and Losses in Rhododendron CYC Orthologs
2.1.2. Tandem Gene Duplication and Genetic Variation in Rhododendron CYC Orthologs
2.1.3. A Shared Duplication in Rhododendron RAD Orthologs Arose from an Ericalean Ancestor
2.1.4. Structural and Genetic Variation in Rhododendron RAD Orthologs
2.1.5. A Shared Duplication in Rhododendron DIV Orthologs Arose from an Ericalean Ancestor
2.1.6. Genetic Variation in Rhododendron DIV Orthologs
2.2. Comparative Expression Analyses of Flower Symmetry Genes in Rhododendron
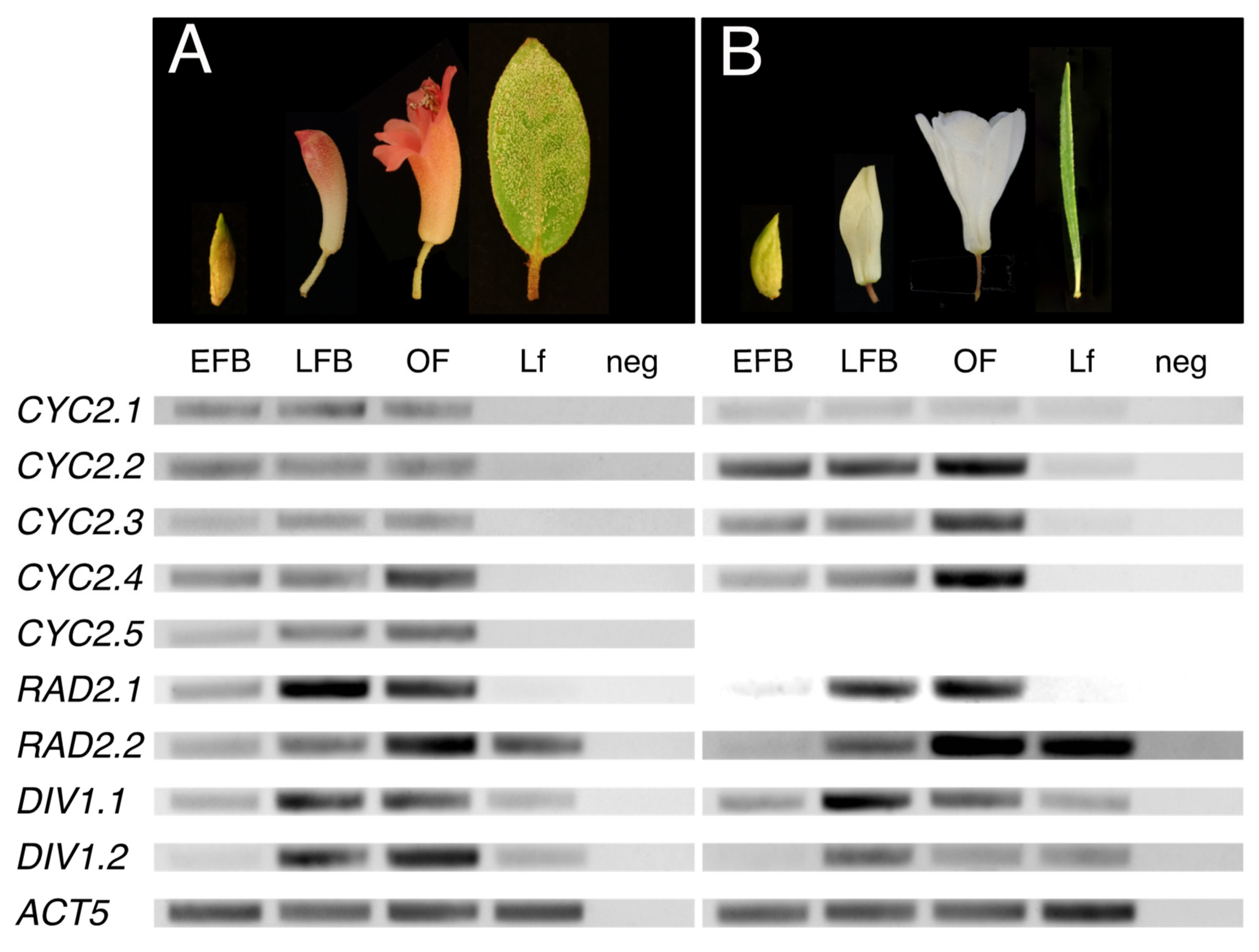
2.2.1. Uniform or Increasing Expression of CYC Orthologs across Rhododendron Flower Development
2.2.2. Increasing Expression of RAD Orthologs across Rhododendron Flower Development
2.2.3. Increasing Expression of DIV Orthologs across Rhododendron Flower Development
2.2.4. Dorsally Restricted Expression of CYC Orthologs in Bilateral Flowers versus Ubiquitous Expression in Radial Flowers
2.2.5. Ubiquitous Expression of RAD Orthologs in Bilateral and Radial Rhododendron Flowers
2.2.6. Uniform Expression of DIV Orthologs in Bilateral and Radial Rhododendron Flowers
3. Discussion
3.1. Evolution of Rhododendron CYC Orthologs Compared to Other Asterids
3.1.1. Tandem Gene Duplication Creates Diversity in Rhododendron CYC Orthologs
3.1.2. Conserved Patterns of Dorsally Restricted Expression of CYC Orthologs in Bilateral Rhododendron Flowers
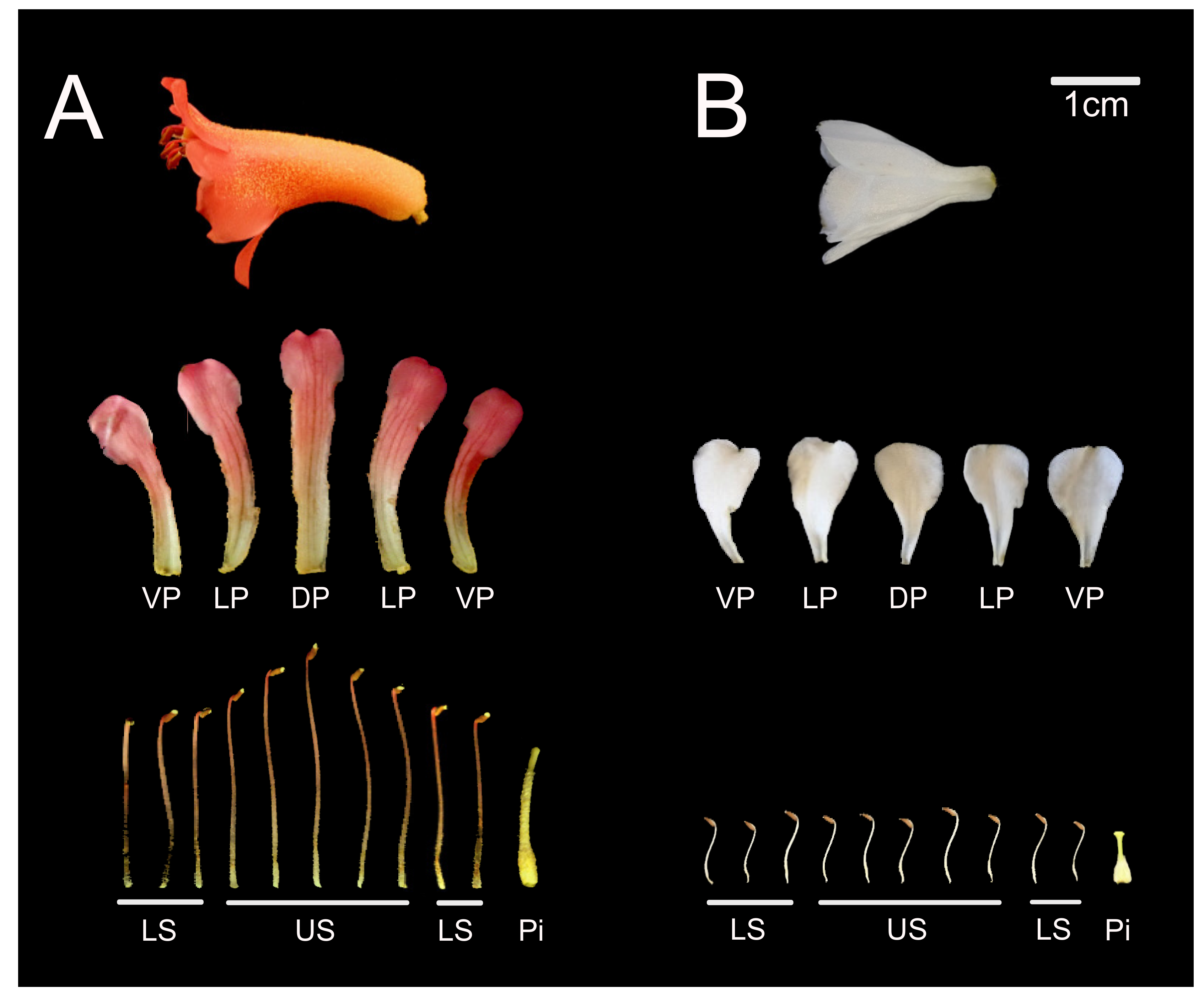
3.1.3. Differential Expression of CYC Orthologs in Bilateral Rhododendron Flowers Is Associated with Differential Growth in Petals and Stamens
3.1.4. CYC Orthologs Are Associated with Asymmetric Organ Growth in Bilateral Rhododendron Flowers
3.1.5. Multiple Conserved Expression Patterns of CYC Orthologs in Radial Rhododendron Flowers
3.2. Evolution of Rhododendron RAD and DIV Orthologs Compared to Other Asterids
3.2.1. Divergent Patterns of Gene Structure, Duplication, and Expression of Rhododendron RAD Orthologs
3.2.2. Conserved Ubiquitous Patterns of Expression of DIV Orthologs in Rhododendron Flowers
3.3. Future Directions in Rhododendron Floral Symmetry Research
4. Materials and Methods
4.1. Data Mining for CYC, RAD, and DIV Homologs in Rhododendron
4.2. Isolation of Rhododendron CYC, RAD, and DIV Orthologs
4.3. Phylogenetic Analyses of Rhododendron CYC, RAD, and DIV Orthologs
4.4. RNA Sample Collection
4.5. RNA Extraction and cDNA Synthesis
4.6. 3′RACE of Rhododendron CYC Orthologs
4.7. Expression Analyses of Rhododendron CYC, RAD, and DIV Orthologs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neal, P.R.; Dafni, A.; Giurfa, M. Floral symmetry and its role in plant-pollinator systems: Terminology, distribution, and hypotheses. Annu. Rev. Ecol. Syst. 1998, 29, 345–373. [Google Scholar] [CrossRef] [Green Version]
- Sargent, R.D. Floral symmetry affects speciation rates in angiosperms. Proc. R. Soc. B Boil Sci. 2004, 271, 603–608. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, B.C.; Smith, S.D.; Armbruster, W.S.; Harder, L.; Hardy, C.R.; Hileman, L.; Hufford, L.; Litt, A.; Magallón, S.; Smith, S.; et al. Non-equilibrium dynamics and floral trait interactions shape extant angiosperm diversity. Proc. R. Soc. B Boil Sci. 2016, 283, 20152304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citerne, H.; Jabbour, F.; Nadot, S.; Damerval, C. The evolution of floral symmetry. Adv. Bot. Res. 2010, 54, 85–137. [Google Scholar] [CrossRef]
- Reyes, E.; Sauquet, H.; Nadot, S. Perianth symmetry changed at least 199 times in angiosperm evolution. Taxon 2016, 65, 945–964. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nat. Cell Biol. 1996, 383, 794–799. [Google Scholar] [CrossRef]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Clark, J.; Coen, E. Control of organ asymmetry in flowers of antirrhinum. Cell 1999, 99, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Gübitz, T.; Caldwell, A.; Hudson, A. Rapid molecular evolution of Cycloidea-like genes in Antirrhinum and its relatives. Mol. Biol. Evol. 2003, 20, 1537–1544. [Google Scholar] [CrossRef] [Green Version]
- Hileman, L.C.; Baum, D.A. Why do paralogs persist? Molecular evolution of Cycloidea and related floral symmetry genes in Antirrhineae (Veronicaceae). Mol. Biol. Evol. 2003, 20, 591–600. [Google Scholar] [CrossRef]
- Li, M.; Zhang, D.; Gao, Q.; Luo, Y.; Zhang, H.; Ma, B.; Chen, C.; Whibley, A.; Zhang, Y.; Cao, Y.; et al. Genome structure and evolution of Antirrhinum majus L. Nat. Plants 2019, 5, 174–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galego, L. Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 2002, 16, 880–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corley, S.B.; Carpenter, R.; Copsey, L.; Coen, E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc. Natl. Acad. Sci. USA 2005, 102, 5068–5073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, M.M.; Fox, S.; Hanna, A.I.; Baxter, C.; Coen, E. Evolution of regulatory interactions controlling floral asymmetry. Development 2005, 132, 5093–5101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimundo, J.; Sobral, R.; Bailey, P.; Azevedo, H.; Galego, L.; Almeida, J.; Coen, E.; Costa, M.M.R. A subcellular tug of war involving three MYB-like proteins underlies a molecular antagonism in Antirrhinum flower asymmetry. Plant J. 2013, 75, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP Transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Howarth, D.G.; Donoghue, M.J. Duplications in CYC-like genes from dipsacales correlate with floral form. Int. J. Plant Sci. 2005, 166, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef] [Green Version]
- Madrigal, Y.; Alzate, J.F.; Pabón-Mora, N. Evolution and expression patterns of TCP genes in asparagales. Front. Plant Sci. 2017, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, A.; Zachgo, S. Flower symmetry evolution: Towards understanding the abominable mystery of angiosperm radiation. Bioessays 2009, 31, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Specht, C.; Howarth, D.G. Adaptation in flower form: A comparative evodevo approach. New Phytol. 2015, 206, 74–90. [Google Scholar] [CrossRef] [Green Version]
- Jabbour, F.; Cossard, G.; Le Guilloux, M.; Sannier, J.; Nadot, S.; Damerval, C. Specific duplication and dorsoventrally asymmetric expression patterns of cycloidea-like genes in zygomorphic species of Ranunculaceae. PLoS ONE 2014, 9, e95727. [Google Scholar] [CrossRef] [PubMed]
- Hileman, L.C. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Raimundo, J.; Sobral, R.; Laranjeira, S.; Costa, M.M.R. Successive domain rearrangements underlie the evolution of a regulatory module controlled by a small interfering peptide. Mol. Biol. Evol. 2018, 35, 2873–2885. [Google Scholar] [CrossRef] [PubMed]
- Howarth, D.G.; Donoghue, M.J. Duplications and expression of divaricata-like genes in dipsacales. Mol. Biol. Evol. 2009, 26, 1245–1258. [Google Scholar] [CrossRef] [Green Version]
- Boyden, G.S.; Donoghue, M.J.; Howarth, D.G. Duplications and expression of radialis-like genes in dipsacales. Int. J. Plant Sci. 2012, 173, 971–983. [Google Scholar] [CrossRef] [Green Version]
- Madrigal, Y.; Alzate, J.F.; González, F.; Pabón-Mora, N. Evolution of radialis and divaricata gene lineages in flowering plants with an expanded sampling in non-core eudicots. Am. J. Bot. 2019, 106, 334–351. [Google Scholar] [CrossRef]
- Gao, A.; Zhang, J.; Zhang, W. Evolution of rad- and div-like genes in plants. Int. J. Mol. Sci. 2017, 18, 1961. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.-R.; Wang, Y.-Z.; Smith, J.F.; Chen, R. Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol. 2008, 178, 532–543. [Google Scholar] [CrossRef]
- Preston, J.C.; Kost, M.A.; Hileman, L.C. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol. 2009, 182, 751–762. [Google Scholar] [CrossRef]
- Zhong, J.; Preston, J.C.; Hileman, L.C.; Kellogg, E.A. Repeated and diverse losses of corolla bilateral symmetry in the Lamiaceae. Ann. Bot. 2017, 119, 1211–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston, J.C.; Martinez, C.C.; Hileman, L. Gradual disintegration of the floral symmetry gene network is implicated in the evolution of a wind-pollination syndrome. Proc. Natl. Acad. Sci. USA 2011, 108, 2343–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reardon, W.; Gallagher, P.; Nolan, K.M.; Wright, H.; Rubio, M.D.L.C.C.; Bragalini, C.; Lee, C.; Fitzpatrick, D.; Corcoran, K.; Wolff, K.; et al. Different outcomes for the MYB floral symmetry genes divaricata and radialis during the evolution of derived actinomorphy in Plantago. New Phytol. 2014, 202, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Hsin, K.-T.; Wang, C.-N. Expression shifts of floral symmetry genes correlate to flower actinomorphy in East Asia endemic Conandron ramondioides (Gesneriaceae). Bot. Stud. 2018, 59, 24. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, J.; Yang, X.; Wang, Y. Regulatory pathways of CYC -like genes in patterning floral zygomorphy exemplified in Chirita pumila. J. Syst. Evol. 2020, 59, 567–580. [Google Scholar] [CrossRef]
- Zhong, J.; Kellogg, E.A. Stepwise evolution of corolla symmetry in cycloidea2 -like and radialis -like gene expression patterns in Lamiales. Am. J. Bot. 2015, 102, 1260–1267. [Google Scholar] [CrossRef] [Green Version]
- Baxter, C.E.L.; Costa, M.M.; Coen, E.S. Diversification and co-option of RAD-like genes in the evolution of floral asymmetry. Plant J. 2007, 52, 105–113. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, X.; Zuo, K.; Zhang, J.; Sun, X.; Tang, K. Molecular cloning and characterization of a novel Gossypium barbadense L. RAD-like gene. Plant Mol. Biol. Rep. 2011, 29, 324–333. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, X.; Zuo, K.; Sun, X.; Tang, K. Molecular cloning and expression analysis of a novel SANT/MYB gene from Gossypium barbadense. Mol. Biol. Rep. 2011, 38, 2329–2336. [Google Scholar] [CrossRef]
- Yang, B.; Song, Z.; Li, C.; Jiang, J.; Zhou, Y.; Wang, R.; Wang, Q.; Ni, C.; Liang, Q.; Chen, H.; et al. RSM1, an arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 2018, 14, e1007839. [Google Scholar] [CrossRef] [PubMed]
- Stevens, P.F.; Luteyn, J.; Oliver, E.G.H.; Bell, T.L.; Brown, E.A.; Crowden, R.K.; George, A.S.; Jordan, G.J.; Ladd, P.; Lemson, K.; et al. Ericaceae. In The Families and Genera of Vascular Plants: Flowering Plants, Dicotyledons, Celastrales, Oxalidales, Rosales, Cornales, Ericales; Springer: Berlin/Heidelberg, Germany, 2004; Volume 6, pp. 145–194. [Google Scholar]
- Byng, J.W.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Judd, W.S.; Mabberley, D.J.; Sennikov, A.N.; Soltis, D.E.; Soltis, P.S.; Stevens, P.F.; et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Goetsch, L.; Eckert, A.J.; Hall, B.D. The molecular systematics of Rhododendron (Ericaceae): A phylogeny based upon RPB2 gene sequences. Syst. Bot. 2005, 30, 616–626. [Google Scholar] [CrossRef]
- Shrestha, N.; Wang, Z.; Su, X.; Xu, X.; Lyu, L.; Liu, Y.; Dimitrov, D.; Kennedy, J.D.; Wang, Q.; Tang, Z.; et al. Global patterns of Rhododendron diversity: The role of evolutionary time and diversification rates. Glob. Ecol. Biogeogr. 2018, 27, 913–924. [Google Scholar] [CrossRef] [Green Version]
- Berry, E.; Sharma, S.K.; Pandit, M.K.; Geeta, R. Evolutionary correlation between floral monosymmetry and corolla pigmentation patterns in Rhododendron. Plant Syst. Evol. 2017, 304, 219–230. [Google Scholar] [CrossRef]
- Stevens, P.F. The altitudinal and geographical distributions of flower types in Rhododendron section Vireya, especially in the Papuasian species, and their significance. Bot. J. Linn. Soc. 1976, 73, 1–33. [Google Scholar] [CrossRef]
- Craven, L.; Dăneţ, F.; Veldkamp, J.; Goetsch, L.; Hall, B. Vireya Rhododendrons: Their monophyly and classification (Ericaceae, Rhododendron section Schistanthe). Blumea Biodivers. Evol. Biogeogr. Plants 2011, 56, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, P.; Cai, Y.; Ma, L.; Li, S.; Li, S.; Xie, W.; Song, J.; Peng, L.; Yan, H.; et al. The draft genome assembly of Rhododendron delavayi Franch. var. delavayi. GigaScience 2017, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Colle, M.; Leisner, C.P.; Wai, C.M.; Ou, S.; Bird, K.A.; Wang, J.; Wisecaver, J.H.; Yocca, A.E.; Alger, E.I.; Tang, H.; et al. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. GigaScience 2019, 8, giz012. [Google Scholar] [CrossRef] [Green Version]
- Soza, V.L.; Lindsley, D.; Waalkes, A.; Ramage, E.; Patwardhan, R.P.; Burton, J.N.; Adey, A.; Kumar, A.; Qiu, R.; Shendure, J.; et al. The Rhododendron genome and chromosomal organization provide insight into shared whole-genome duplications across the heath family (Ericaceae). Genome Biol. Evol. 2019, 11, 3353–3371. [Google Scholar] [CrossRef]
- Wu, H.; Ma, T.; Kang, M.; Ai, F.; Zhang, J.; Dong, G.; Liu, J. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic. Res. 2019, 6, 117–119. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.-S.; Nie, S.; Liu, H.; Shi, T.-L.; Tian, X.-C.; Zhou, S.-S.; Bao, Y.-T.; Jia, K.-H.; Guo, J.-F.; Zhao, W.; et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 2020, 11, 5269. [Google Scholar] [CrossRef] [PubMed]
- Ramage, E.; Soza, V.L.; Hall, B.D. Transcriptome Assemblies across the Genus Rhododendron (Ericaceae). University of Washington: Seattle, WA, USA, 2021; manuscript in preparation. [Google Scholar]
- Soza, V.L.; Kriebel, R.; Ramage, E.; Hall, B.D.; Twyford, A.D. The Symmetry Spectrum in a Hybridizing, Tropical Group of Rhododendrons. University of Washington: Seattle, WA, USA, 2021; manuscript in preparation. [Google Scholar]
- Gillespie, E.; Kron, K. Molecular phylogenetic relationships and a revised classification of the subfamily Ericoideae (Ericaceae). Mol. Phylogenetics Evol. 2010, 56, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.P.; Kleist, T.J.; Löfstrand, S.D.; Drew, B.T.; Schönenberger, J.; Sytsma, K.J. Phylogeny, historical biogeography, and diversification of angiosperm order Ericales suggest ancient Neotropical and East Asian connections. Mol. Phylogenetics Evol. 2018, 122, 59–79. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Leebens-Mack, J.H.; Burke, J.M. Positive selection and expression divergence following gene duplication in the sunflower Cycloidea gene family. Mol. Biol. Evol. 2008, 25, 1260–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaut, B.S.; Wright, S.I.; Rizzon, C.; Dvorak, J.; Anderson, L.K. Recombination: An underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 2007, 8, 77–84. [Google Scholar] [CrossRef]
- Gao, Q.; Tao, J.-H.; Yan, D.; Wang, Y.-Z.; Li, Z.-Y. Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev. Genes Evol. 2008, 218, 341–351. [Google Scholar] [CrossRef]
- Song, C.-F.; Lin, Q.-B.; Liang, R.-H.; Wang, Y.-Z. Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae). BMC Evol. Biol. 2009, 9, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howarth, D.G.; Martins, T.; Chimney, E.; Donoghue, M.J. Diversification of Cycloidea expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot. 2011, 107, 1521–1532. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Pang, H.-B.; Liu, B.-L.; Qiu, Z.-J.; Gao, Q.; Wei, L.; Dong, Y.; Wang, Y.-Z. Evolution of double positive autoregulatory feedback loops in Cycloidea 2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 2012, 24, 1834–1847. [Google Scholar] [CrossRef] [Green Version]
- Zhong, J.; Kellogg, E. Duplication and expression of CYC2-like genes in the origin and maintenance of corolla zygomorphy in Lamiales. New Phytol. 2015, 205, 852–868. [Google Scholar] [CrossRef]
- Hsin, K.-T.; Lu, J.-Y.; Möller, M.; Wang, C.-N. Gene duplication and relaxation from selective constraints of GCYC genes correlated with various floral symmetry patterns in Asiatic Gesneriaceae tribe Trichosporeae. PLoS ONE 2019, 14, e0210054. [Google Scholar] [CrossRef]
- Garcês, H.M.P.; Spencer, V.M.; Kim, M. Control of floret symmetry by RAY3, SvDIV1B, and SvRAD in the capitulum of Senecio vulgaris. Plant Physiol. 2016, 171, 2055–2068. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.-C.; Wang, C.-N.; Liang, C.-H.; Wang, C.-C.; Kuo, Y.-F. Association between petal form variation and CYC2-like genotype in a hybrid line of Sinningia speciosa. Front. Plant Sci. 2017, 8, 558. [Google Scholar] [CrossRef] [Green Version]
- Broholm, S.K.; Tähtiharju, S.; Laitinen, R.A.E.; Albert, V.A.; Teeri, T.; Elomaa, P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 9117–9122. [Google Scholar] [CrossRef] [Green Version]
- Reardon, W.; Fitzpatrick, D.; Fares, M.A.; Nugent, J.M. Evolution of flower shape in Plantago lanceolata. Plant Mol. Biol. 2009, 71, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Pang, H.-B.; Sun, Q.-W.; He, S.-Z.; Wang, Y.-Z. Expression pattern of CYC-like genes relating to a dorsalized actinomorphic flower in Tengia (Gesneriaceae). J. Syst. Evol. 2010, 48, 309–317. [Google Scholar] [CrossRef]
- Larson, D.A.; Walker, J.F.; Vargas, O.M.; Smith, S.A. A consensus phylogenomic approach highlights paleopolyploid and rapid radiation in the history of Ericales. Am. J. Bot. 2020, 107, 773–789. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.70. Available online: http://www.mesquiteproject.org (accessed on 16 August 2021).
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salminen, M.; Carr, J.K.; Burke, D.S.; McCutchan, F.E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 1995, 11, 1423–1425. [Google Scholar] [CrossRef]
- Martin, D.; Posada, D.; Crandall, K.; Williamson, C. A Modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 2005, 21, 98–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Citerne, H.L.; Le Guilloux, M.; Sannier, J.; Nadot, S.; Damerval, C. Combining phylogenetic and syntenic analyses for understanding the evolution of TCP ECE genes in eudicots. PLoS ONE 2013, 8, e74803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. In Selected Papers of Hirotugu Akaike; Springer: New York, NY, USA, 1974; pp. 215–222. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Rannala, B. Bayesian phylogenetic inference using DNA sequences: A markov chain Monte Carlo method. Mol. Biol. Evol. 1997, 14, 717–724. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree Version 1.4.3. 2016. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 16 August 2021).
- De Keyser, E.; Desmet, L.; Van Bockstaele, E.; De Riek, J. How to perform RT-qPCR accurately in plant species? A case study on flower colour gene expression in an azalea (Rhododendron simsii hybrids) mapping population. BMC Mol. Biol. 2013, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- Loh, E. Anchored PCR: Amplification with single-sided specificity. Methods 1991, 2, 11–19. [Google Scholar] [CrossRef]
- Scotto–Lavino, E.; Du, G.; Frohman, M.A. 3′ End cDNA amplification using classic RACE. Nat. Protoc. 2006, 1, 2742–2745. [Google Scholar] [CrossRef]
- Yi, S.; Qian, Y.; Han, L.; Sun, Z.; Fan, C.; Liu, J.; Ju, G. Selection of reliable reference genes for gene expression studies in Rhododendron micranthum Turcz. Sci. Hortic. 2012, 138, 128–133. [Google Scholar] [CrossRef]
- Xiao, Z.; Sun, X.; Liu, X.; Li, C.; He, L.; Chen, S.; Su, J. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Front. Plant Sci. 2016, 7, 1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer 3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramage, E.; Soza, V.L.; Yi, J.; Deal, H.; Chudgar, V.; Hall, B.D.; Di Stilio, V.S. Gene Duplication and Differential Expression of Flower Symmetry Genes in Rhododendron (Ericaceae). Plants 2021, 10, 1994. https://doi.org/10.3390/plants10101994
Ramage E, Soza VL, Yi J, Deal H, Chudgar V, Hall BD, Di Stilio VS. Gene Duplication and Differential Expression of Flower Symmetry Genes in Rhododendron (Ericaceae). Plants. 2021; 10(10):1994. https://doi.org/10.3390/plants10101994
Chicago/Turabian StyleRamage, Elizabeth, Valerie L. Soza, Jing Yi, Haley Deal, Vaidehi Chudgar, Benjamin D. Hall, and Verónica S. Di Stilio. 2021. "Gene Duplication and Differential Expression of Flower Symmetry Genes in Rhododendron (Ericaceae)" Plants 10, no. 10: 1994. https://doi.org/10.3390/plants10101994
APA StyleRamage, E., Soza, V. L., Yi, J., Deal, H., Chudgar, V., Hall, B. D., & Di Stilio, V. S. (2021). Gene Duplication and Differential Expression of Flower Symmetry Genes in Rhododendron (Ericaceae). Plants, 10(10), 1994. https://doi.org/10.3390/plants10101994







