RNA-Based Technologies for Engineering Plant Virus Resistance
Abstract
1. Introduction
2. Non-Coding RNAs and Plant Antiviral Defense
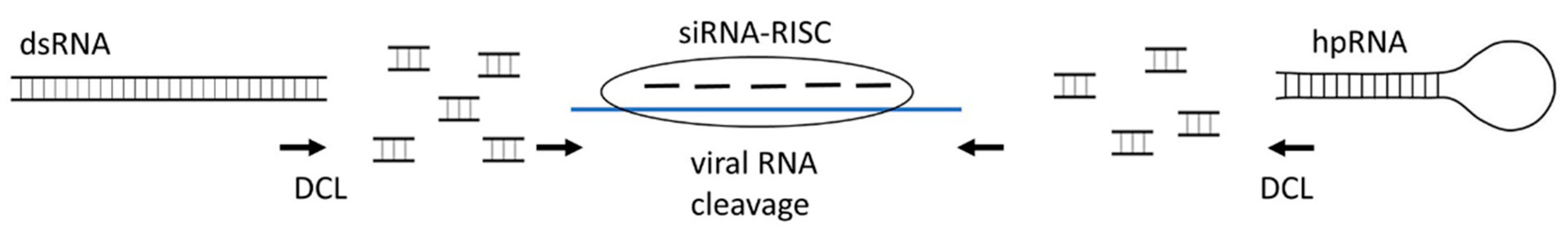
2.1. siRNAs
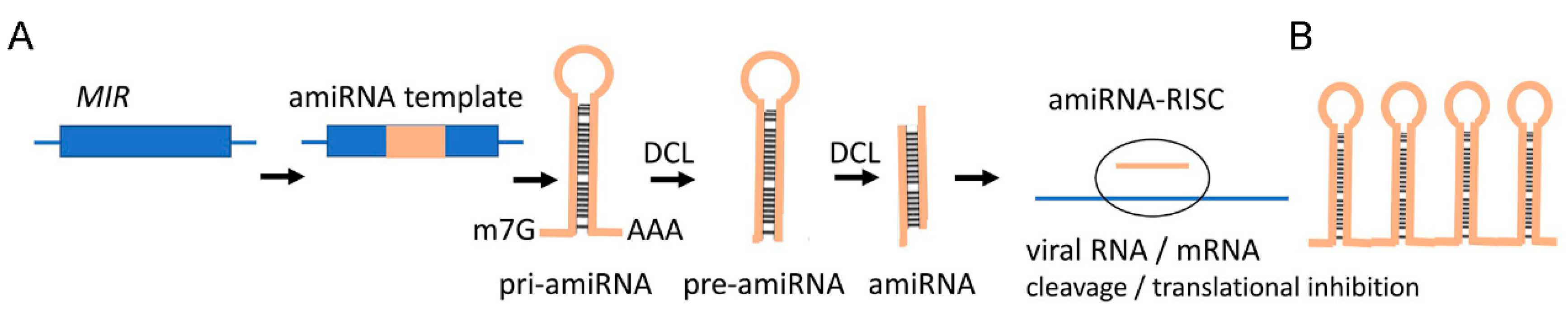
2.2. miRNAs
2.3. LncRNAs
2.4. Small Peptides in lncRNAs
2.5. Other Non-Coding RNAs
3. RNA-Based Tools for Engineering Viral Resistance
3.1. Host Induced Gene Silencing
3.2. Exogenous dsRNA (hpRNA)-Induced Gene Silencing
3.3. Host Gene Targets for SIGS
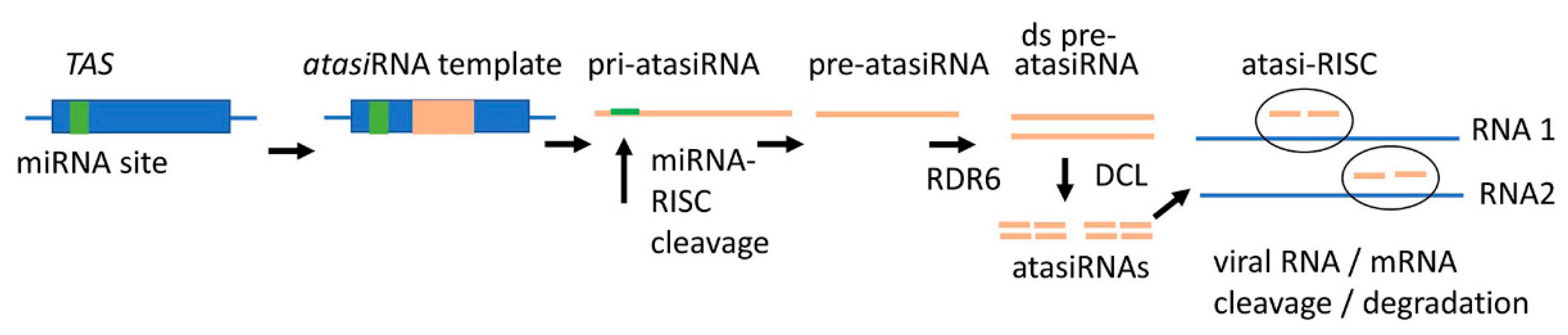
3.4. New RNA Tools for SIGS
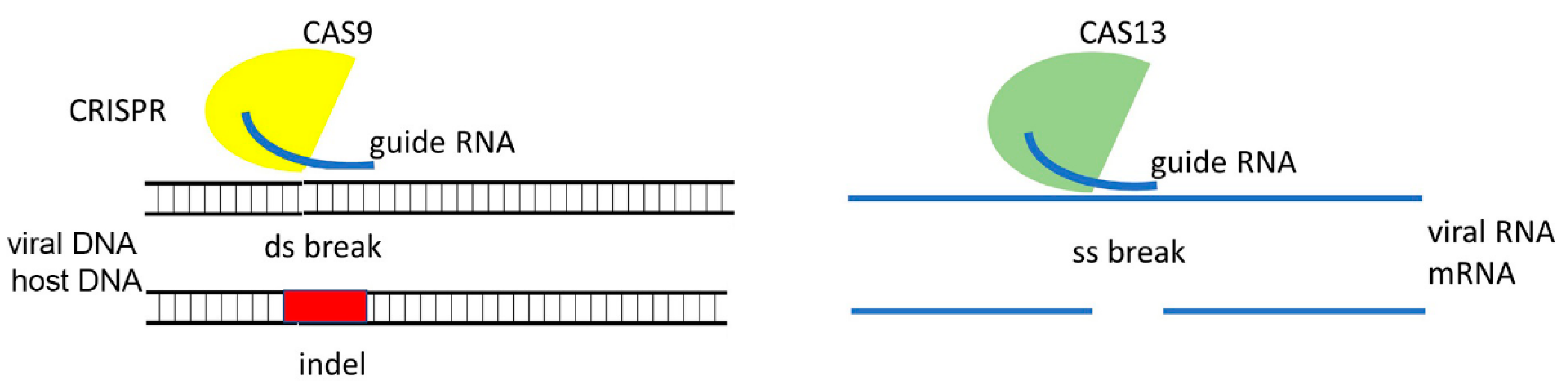
3.5. RNA Guided CRISPR-Cas System
3.6. RNAi Versus CRISPR
3.7. Stability and Uptake of RNA Molecules by Plant Cells
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dormatey, R.; Sun, C.; Ali, K.; Coulter, J.A.; Bi, Z.; Bai, J. Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses. Agronomy 2020, 10, 1255. [Google Scholar] [CrossRef]
- Cillo, F.; Palukaitis, P. Transgenic Resistance. Adv. Virus Res. 2014, 90, 35–146. [Google Scholar] [CrossRef]
- Mello, C.C.; Conte, D. Revealing the world of RNA interference. Nature 2004, 431, 338–342. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing. Trends Biochem. Sci. 2005, 30, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-X.; Li, Y.; Ding, S.-W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Li, J.; Zhang, K.-Q. Structure, Regulation, and Function of Linear and Circular Long Non-Coding RNAs. Front. Genet. 2020, 11, 150. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Jørgensen, H.J.L.; Lund, O.S.; Lyngkjær, M.F. Engineering Pathogen Resistance in Crop Plants: Current Trends and Future Prospects. Annu. Rev. Phytopathol. 2010, 48, 269–291. [Google Scholar] [CrossRef]
- Thompson, J.R.; Tepfer, M. Assessment of the Benefits and Risks for Engineered Virus Resistance. Adv. Virus Res. 2010, 76, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-B.; Masuta, C.; Smith, N.A.; Shimura, H. RNA Silencing and Plant Viral Diseases. Mol. Plant-Microbe Interact. 2012, 25, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Morozov, S.Y.; Solovyev, A.G.; Kalinina, N.O.; Taliansky, M.E. Double-Stranded RNAs in Plant Protection Against Pathogenic Organisms and Viruses in Agriculture. Acta Nat. 2019, 11, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, N.O.; Khromov, A.; Love, A.J.; Talianksy, M.E. CRISPR Applications in Plant Virology: Virus Resistance and Beyond. Phytopathology 2020, 110, 18–28. [Google Scholar] [CrossRef]
- Cao, Y.; Zhou, H.; Zhou, X.; Li, F. Control of Plant Viruses by CRISPR/Cas System-Mediated Adaptive Immunity. Front. Microbiol. 2020, 11, 593700. [Google Scholar] [CrossRef]
- Hadidi, A.; Flores, R.; Candresse, T.; Barba, M. Next-Generation Sequencing and Genome Editing in Plant Virology. Front. Microbiol. 2016, 7, 1325. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 6164–6169. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Severinov, K.; Regev, A.; Lander, E.S.; Koonin, E.V.; Zhang, F.; Konermann, S.M.; Joung, J.; Slaymaker, I.M.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Gaffar, F.Y.; Koch, A. Catch Me If You Can! RNA Silencing-Based Improvement of Antiviral Plant Immunity. Viruses 2019, 11, 673. [Google Scholar] [CrossRef]
- Zhang, B.; Li, W.; Zhang, J.; Wang, L.; Wu, J. Roles of Small RNAs in Virus-Plant Interactions. Viruses 2019, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Djami-Tchatchou, A.T.; Sanan-Mishra, N.; Ntushelo, K.; Dubery, I.A. Functional Roles of microRNAs in Agronomically Important Plants—Potential as Targets for Crop Improvement and Protection. Front. Plant Sci. 2017, 8, 378. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Zhang, Q.; Yasir, M.; Liu, F. Small RNA Based Genetic Engineering for Plant Viral Resistance: Application in Crop Protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2019, 18, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Mezzetti, B.; Sabbadini, S.; Sorteberg, H.; Sweet, J.; Ventura, V.; et al. RNA-based biocontrol compounds: Current status and perspectives to reach the market. Pest Manag. Sci. 2020, 76, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Xu, Z.P.; Carroll, B.J. Induction of virus resistance by exogenous application of double-stranded RNA. Curr. Opin. Virol. 2017, 26, 49–55. [Google Scholar] [CrossRef]
- Saha, D.; Dey, P. New Dimensions of RNA Based Technologies in Plant Functional Genomics. Indian J. Nat. Sci. 2020, 10, 23286–23294. [Google Scholar]
- Cisneros, A.E.; Carbonell, A. Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants. Plants 2020, 9, 669. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K. Exogenous RNAs for Gene Regulation and Plant Resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef]
- Baulcombe, D.C. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, V.; Scholthof, H.B. Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin. Cell Dev. Biol. 2009, 20, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, Q.; Smith, N.A.; Liang, G.; Wang, M.-B. RNA Silencing in Plants: Mechanisms, Technologies and Applications in Horticultural Crops. Curr. Genom. 2016, 17, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.-S.; De Alba, A.E.M.; Vaucheret, H. The origin and effect of small RNA signaling in plants. Front. Plant Sci. 2012, 3, 179. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and Secondary siRNAs in Geminivirus-induced Gene Silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyán, J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 85–103. [Google Scholar] [CrossRef]
- Li, F.; Ding, S.-W. Virus Counterdefense: Diverse Strategies for Evading the RNA-Silencing Immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Sun, G. MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 2011, 80, 17–36. [Google Scholar] [CrossRef]
- Du, Z.; Chen, A.; Chen, W.; Westwood, J.H.; Baulcombe, D.C.; Carr, J.P. Using a Viral Vector to Reveal the Role of MicroRNA159 in Disease Symptom Induction by a Severe Strain of Cucumber mosaic virus. Plant Physiol. 2014, 164, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.; Yuan, Q.; Wang, S.; Peng, J.; Lu, Y.; Zheng, H.; Lin, L.; Chen, H.; Gong, Y.; Chen, J.; et al. Altered accumulation of osa-miR171b contributes to rice stripe virus infection by regulating disease symptoms. J. Exp. Bot. 2017, 68, 4357–4367. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.A.; Eamens, A.L.; Wang, M.-B. Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants. PLoS Pathog. 2011, 7, e1002022. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jia, M.-A.; Yang, Y.; Zhan, L.; Cheng, X.; Cai, J.; Zhang, J.; Yang, J.; Liu, T.; Fu, Q.; et al. Integrated analysis of tobacco miRNA and mRNA expression profiles under PVY infection provids insight into tobacco-PVY interactions. Sci. Rep. 2017, 7, 4895. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A Signaling Cascade from miR444 to RDR1 in Rice Antiviral RNA Silencing Pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.-M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A MicroRNA Superfamily Regulates Nucleotide Binding Site–Leucine-Rich Repeats and Other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Liu, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional Regulation of miR528 by OsSPL9 Orchestrates Antiviral Response in Rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.; Zhao, S.; Yang, Z.; et al. Suppression of Jasmonic Acid-Mediated Defense by Viral-Inducible MicroRNA319 Facilitates Virus Infection in Rice. Mol. Plant 2016, 9, 1302–1314. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, X.; Li, L.; He, Y.; Hong, G.; Li, J.; Lin, L.; Cheng, Y.; Yan, F.; Chen, J.; et al. Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol. Plant Pathol. 2019, 20, 1093–1104. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs inArabidopsis thalianaby integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef]
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef]
- Wu, H.; Yang, L.; Chen, L.-L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Markenscoff-Papadimitriou, E.; Kuhl, D.; Bito, H.; Worley, P.F.; Kreiman, G.; Greenberg, M.E.; Gray, J.M.; Costa, A.M.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Karlik, E.; Ari, S.; Gözükirmizi, N. LncRNAs: Genetic and epigenetic effects in plants. Biotechnol. Biotechnol. Equip. 2019, 33, 429–439. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Q.-H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ding, B.; Fei, Z.; Wang, Y. Comprehensive transcriptome analyses reveal tomato plant responses to tobacco rattle virus-based gene silencing vectors. Sci. Rep. 2017, 7, 9771. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cho, W.K.; Byun, H.-S.; Chavan, V.; Kil, E.-J.; Lee, S.; Hong, S.-W. Genome-wide identification of long non-coding RNAs in tomato plants irradiated by neutrons followed by infection withTomato yellow leaf curl virus. PeerJ 2019, 7, e6286. [Google Scholar] [CrossRef]
- Seo, J.S.; Diloknawarit, P.; Park, B.S.; Chua, N.-H. ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol. 2018, 221, 2067–2079. [Google Scholar] [CrossRef]
- Taliansky, M.; Brown, J.; Rajamäki, M.-L.; Valkonen, J.P.T.; Kalinina, N.O. Involvement of the Plant Nucleolus in Virus and Viroid Infections. Adv. Virus Res. 2010, 77, 119–158. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Makarova, S.; Makhotenko, A.; Love, A.J.; Talianksy, M.E. The Multiple Functions of the Nucleolus in Plant Development, Disease and Stress Responses. Front. Plant Sci. 2018, 9, 132. [Google Scholar] [CrossRef]
- Kang, S.H.; Sun, Y.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A Long Non-Coding RNA of Citrus tristeza virus: Role in the Virus Interplay with the Host Immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef]
- Carr, J.P.; Lewsey, M.G.; Palukaitis, P. Signaling in Induced Resistance. Adv. Virus Res. 2010, 76, 57–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, S.; Chen, M. Characterization and Function of Circular RNAs in Plants. Front. Mol. Biosci. 2020, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Bujarski, J.J. Long Noncoding RNAs in Plant Viroids and Viruses: A Review. Pathogens 2020, 9, 765. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-W.; Kim, H.-W.; Nam, J.-W. The small peptide world in long noncoding RNAs. Briefings Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef]
- Wang, S.; Tian, L.; Liu, H.; Li, X.; Zhang, J.; Chen, X.; Jia, X.; Zheng, X.; Wu, S.; Chen, Y.; et al. Large-Scale Discovery of Non-conventional Peptides in Maize and Arabidopsis through an Integrated Peptidogenomic Pipeline. Mol. Plant 2020, 13, 1078–1093. [Google Scholar] [CrossRef]
- Couso, J.-P.; Patraquim, P. Classification and Function of Small Open Reading Frames. Nat. Rev. Mol. Cell Biol. 2017, 18, 575–589. [Google Scholar] [CrossRef]
- Fesenko, I.A.; Kirov, I.; Kniazev, A.; Khazigaleeva, R.; Lazarev, V.; Kharlampieva, D.; Grafskaia, E.; Zgoda, V.; Butenko, I.; Arapidi, G.; et al. Distinct types of short open reading frames are translated in plant cells. Genome Res. 2019, 29, 1464–1477. [Google Scholar] [CrossRef]
- Lauressergues, D.; Couzigou, J.-M.; Clemente, H.S.; Martinez, Y.; Dunand, C.; Bécard, G.; Combier, J.-P. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015, 520, 90–93. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Ryazantsev, D.Y.; Erokhina, T.N. Bioinformatics Analysis of the Novel Conserved Micropeptides Encoded by the Plants of Family Brassicaceae. J. Bioinform. Syst. Biol. 2019, 2, 66–77. [Google Scholar] [CrossRef][Green Version]
- Jackson, R.; Kroehling, L.; Harman, C.C.D.; Chang, L.; Bielecki, P.; Solis, A.G.; Steach, H.R.; Slavoff, S.; Flavell, R.A.; Khitun, A.; et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature 2018, 564, 434–438. [Google Scholar] [CrossRef]
- Razooky, B.S.; Obermayer, B.; O’May, J.B.; Tarakhovsky, A. Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs. Genes 2017, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009, 139, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Regulation of Ribosomal Proteins on Viral Infection. Cells 2019, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Abel, P.P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 1986, 232, 738–743. [Google Scholar] [CrossRef]
- Tenllado, F.; Díaz-Ruíz, J.R. Double-Stranded RNA-Mediated Interference with Plant Virus Infection. J. Virol. 2001, 75, 12288–12297. [Google Scholar] [CrossRef]
- Tenllado, F.; Martínez, B.; Vargas, M.; Díaz-Ruíz, J.R. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 2003, 3, 3. [Google Scholar] [CrossRef]
- Carbonell, A.; De Alba, A.E.M.; Flores, R.; Gago, S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008, 371, 44–53. [Google Scholar] [CrossRef]
- Yin, G.; Sun, Z.; Liu, N.; Zhang, L.; Song, Y.; Zhu, C.; Wen, F. Production of double-stranded RNA for interference with TMV infection utilizing a bacterial prokaryotic expression system. Appl. Microbiol. Biotechnol. 2009, 84, 323–333. [Google Scholar] [CrossRef]
- Sun, Z.-N.; Song, Y.-Z.; Yin, G.-H.; Zhu, C.-X.; Wen, F.-J. HpRNAs Derived from Different Regions of the NIb Gene Have Different Abilities to Protect Tobacco from Infection withPotato Virus Y. J. Phytopathol. 2009, 158, 566–568. [Google Scholar] [CrossRef]
- Sun, Z.-N.; Yin, G.-H.; Song, Y.-Z.; An, H.-L.; Zhu, C.-X.; Wen, F.-J. Bacterially Expressed Double-Stranded RNAs against Hot-Spot Sequences of Tobacco Mosaic Virus or Potato Virus Y Genome Have Different Ability to Protect Tobacco from Viral Infection. Appl. Biochem. Biotechnol. 2010, 162, 1901–1914. [Google Scholar] [CrossRef]
- Gan, D.; Zhang, J.; Jiang, H.; Jiang, T.; Zhu, S.; Cheng, B. Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 2010, 29, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Yang, G.; Chen, Y.; Yan, P.; Tuo, D.; Li, X.; Zhou, P. Resistance of non-transgenic papaya plants to papaya ringspot virus (PRSV) mediated by intron-containing hairpin dsRNAs expressed in bacteria. Acta Virol. 2014, 58, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.E.; Mazumdar, P.; Hee, T.W.; Song, A.L.A.; Othman, R.Y.; Harikrishna, J.A. Crude extracts of bacterially-expressed dsRNA protect orchid plants against Cymbidium mosaic virus during transplantation from in vitro culture. J. Hortic. Sci. Biotechnol. 2014, 89, 569–576. [Google Scholar] [CrossRef]
- Šafářová, D.; Brazda, P.; Navrátil, M. Effect of artificial dsRNA on infection of pea plants by Pea seed-borne mosaic virus. Czech J. Genet. Plant Breed. 2014, 50, 105–108. [Google Scholar] [CrossRef]
- Konakalla, N.C.; Kaldis, A.; Berbati, M.; Masarapu, H.; Voloudakis, A. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 2016, 244, 961–969. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.M.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Kaldis, A.; Berbati, M.; Melita, O.; Reppa, C.; Holeva, M.; Otten, P.; Voloudakis, A. Exogenously applied dsRNA molecules deriving from theZucchini yellow mosaic virus(ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol. Plant Pathol. 2017, 19, 883–895. [Google Scholar] [CrossRef]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef]
- Worrall, E.A.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.P.; Mitter, N. Exogenous Application of RNAi-Inducing Double-Stranded RNA Inhibits Aphid-Mediated Transmission of a Plant Virus. Front. Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef]
- Namgial, T.; Kaldis, A.; Chakraborty, S.; Voloudakis, A. Topical application of double-stranded RNA molecules containing sequences of Tomato leaf curl virus and Cucumber mosaic virus confers protection against the cognate viruses. Physiol. Mol. Plant Pathol. 2019, 108. [Google Scholar] [CrossRef]
- Vadlamudi, T.; Patil, B.L.; Kaldis, A.; Gopal, D.V.R.S.; Mishra, R.; Berbati, M.; Voloudakis, A. DsRNA-mediated protection against two isolates of Papaya ringspot virus through topical application of dsRNA in papaya. J. Virol. Methods 2019, 275, 113750. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Heinlein, M. Perception of double-stranded RNA in plant antiviral immunity. Mol. Plant Pathol. 2019, 20, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNA s induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Sanfaçon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef]
- Wang, X.; Kohalmi, S.E.; Svircev, A.; Wang, A.; Sanfaçon, H.; Tian, L. Silencing of the Host Factor eIF(iso)4E Gene Confers Plum Pox Virus Resistance in Plum. PLoS ONE 2013, 8, e50627. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Love, A.J.; Makarova, S.S.; Kalinina, N.O.; Harrison, B.D.; Talianksy, M.E. Coilin, the signature protein of Cajal bodies, differentially modulates the interactions of plants with viruses in widely different taxa. Nucleus 2014, 5, 85–94. [Google Scholar] [CrossRef]
- Carbonell, A.; Rubio, C.; Daròs, J.-A. Multi-targeting of viral RNAs with synthetic trans -acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019, 100, 720–737. [Google Scholar] [CrossRef]
- Meng, Y.; Shao, C.; Wang, H.; Jin, Y. Target mimics: An embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genom. 2012, 13, 197. [Google Scholar] [CrossRef]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR /Cas9 system. Plant Biotechnol. J. 2019, 17, 1004–1006. [Google Scholar] [CrossRef]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.-E.-A.; Mahfouz, M.M. CRISPR/Cas9-Mediated Immunity to Geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016, 6, 26912. [Google Scholar] [CrossRef]
- Liu, H.; Soyars, C.L.; Li, J.; Fei, Q.; He, G.; Peterson, B.A.; Meyers, B.C.; Nimchuk, Z.L.; Wang, X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct 2018, 2, e00047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; et al. Establishing CRISPR /Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185–1187. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M.M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1–9. [Google Scholar] [CrossRef]
- Ran, Y.; Liang, Z.; Gao, C. Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 2017, 60, 490–505. [Google Scholar] [CrossRef]
- Makhotenko, A.V.; Khromov, A.V.; Snigir, E.A.; Makarova, S.S.; Makarov, V.V.; Suprunova, T.P.; Kalinina, N.O.; Taliansky, M.E. Functional Analysis of Coilin in Virus Resistance and Stress Tolerance of Potato Solanum tuberosum using CRISPR-Cas9 Editing. Dokl. Biochem. Biophys. 2019, 484, 88–91. [Google Scholar] [CrossRef]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.-R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1–23. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The Formation and Function of Plant Cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Oparka, K.J.; Turgeon, R. Sieve Elements and Companion Cells—Traffic Control Centers of the Phloem. Plant Cell 1999, 11, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Popat, A.; Hartono, S.B.; Stahr, F.; Liu, J.; Qiao, S.; Lu, G.Q. Mesoporous silica nanoparticles for bioadsorption, enzyme immobilisation, and delivery carriers. Nanoscale 2011, 3, 2801–2818. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.J.; Ellington, A.D.; Finkelstein, I.J. Retrons and their applications in genome engineering. Nucleic Acids Res. 2019, 47, 11007–11019. [Google Scholar] [CrossRef]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial Retrons Function In Anti-Phage Defense. Cell 2020, 183, 1551–1561.e12. [Google Scholar] [CrossRef]
- Boettcher, M.; McManus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef]
| Virus/Viroid | RNA Production Method | Delivery Method | Host Plant/Crop | Reference |
|---|---|---|---|---|
| PMMoV, TEV, AMV | In vitro synthesized dsRNAs | Mechanical inoculation | Nicotiana tabacum Capsicum chinense (pepper), Nicotiana benthamiana | [85] |
| PMMoV, TMV, PPV | Bacterially expressed dsRNAs (crude extracts) | Mechanical inoculation or spraying | N. benthamiana | [86] |
| PSTVd, CEVd, CChMVd | In vitro synthesized dsRNAs | Mechanical inoculation | Solanum lycopersicum (tomato), Gynura aurantiaca (gynura), Dendranthema grandiflora (chrysanthemum) | [87] |
| TMV | Bacterially expressed dsRNA (crude extracts) | Mechanical inoculation | N. tabacum | [88] |
| PVY | Bacterially expressed hpRNAs (crude extracts) | Mechanical inoculation | N. tabacum | [89] |
| TMV, PVY | Bacterially expressed hpRNAs (crude extracts) | Mechanical inoculation | N. tabacum | [90] |
| SCMV | Bacterially expressed hpRNA (crude extracts) | Spraying | Zea mays (maize) | [91] |
| PRSV | Bacterially expressed hpRNA (crude extracts) | Mechanical inoculation | Carica papaya (papaya) | [92] |
| CymMV | Bacterially expressed dsRNAs and ssRNAs (crude extracts) | Mechanical inoculation | Brassolaeliocattleya hybrida (orchid) | [93] |
| PSbMV | In vitro synthesized dsRNA | Spraying | Pisum sativum (pea) | [94] |
| TMV | In vitro synthesized dsRNA | Mechanical inoculation | N. tabacum | [95] |
| CMV, PMMoV | In vitro synthesized dsRNAs or bacterially expressed dsRNAs (crude extracts); applied directly or loaded into LDH | Spraying | N. tabacum, Vigna unguiculata (cowpea) | [96] |
| ZYMV | In vitro synthesized dsRNAs | Mechanical inoculation | Citrulus lanatus (watermelon), Cucurbita pepo (squash), Cucumis sativus (cucumber) | [97] |
| TMV | Bacterially expressed or in vitro synthesized dsRNA | Mechanical inoculation, spraying | N. benthamiana | [98] |
| BCMV | Chemically synthesized dsRNA applied directly or loaded into LDH | Spraying | N. benthamiana, Vigna unguiculata (cowpea) | [99] |
| ToLCV, CMV | In vitro synthesized dsRNAs | Mechanical inoculation | S. lycopersicum (tomato), N. tabacum | [100] |
| PRSV | In vitro synthesized dsRNA | Mechanical inoculation | Carica papaya | [101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-Based Technologies for Engineering Plant Virus Resistance. Plants 2021, 10, 82. https://doi.org/10.3390/plants10010082
Taliansky M, Samarskaya V, Zavriev SK, Fesenko I, Kalinina NO, Love AJ. RNA-Based Technologies for Engineering Plant Virus Resistance. Plants. 2021; 10(1):82. https://doi.org/10.3390/plants10010082
Chicago/Turabian StyleTaliansky, Michael, Viktoria Samarskaya, Sergey K. Zavriev, Igor Fesenko, Natalia O. Kalinina, and Andrew J. Love. 2021. "RNA-Based Technologies for Engineering Plant Virus Resistance" Plants 10, no. 1: 82. https://doi.org/10.3390/plants10010082
APA StyleTaliansky, M., Samarskaya, V., Zavriev, S. K., Fesenko, I., Kalinina, N. O., & Love, A. J. (2021). RNA-Based Technologies for Engineering Plant Virus Resistance. Plants, 10(1), 82. https://doi.org/10.3390/plants10010082





