Targeted Mutagenesis of the Female-Suppressor SyGI Gene in Tetraploid Kiwifruit by CRISPR/CAS9
Abstract
1. Introduction
2. Results
2.1. SyGI Target Identification
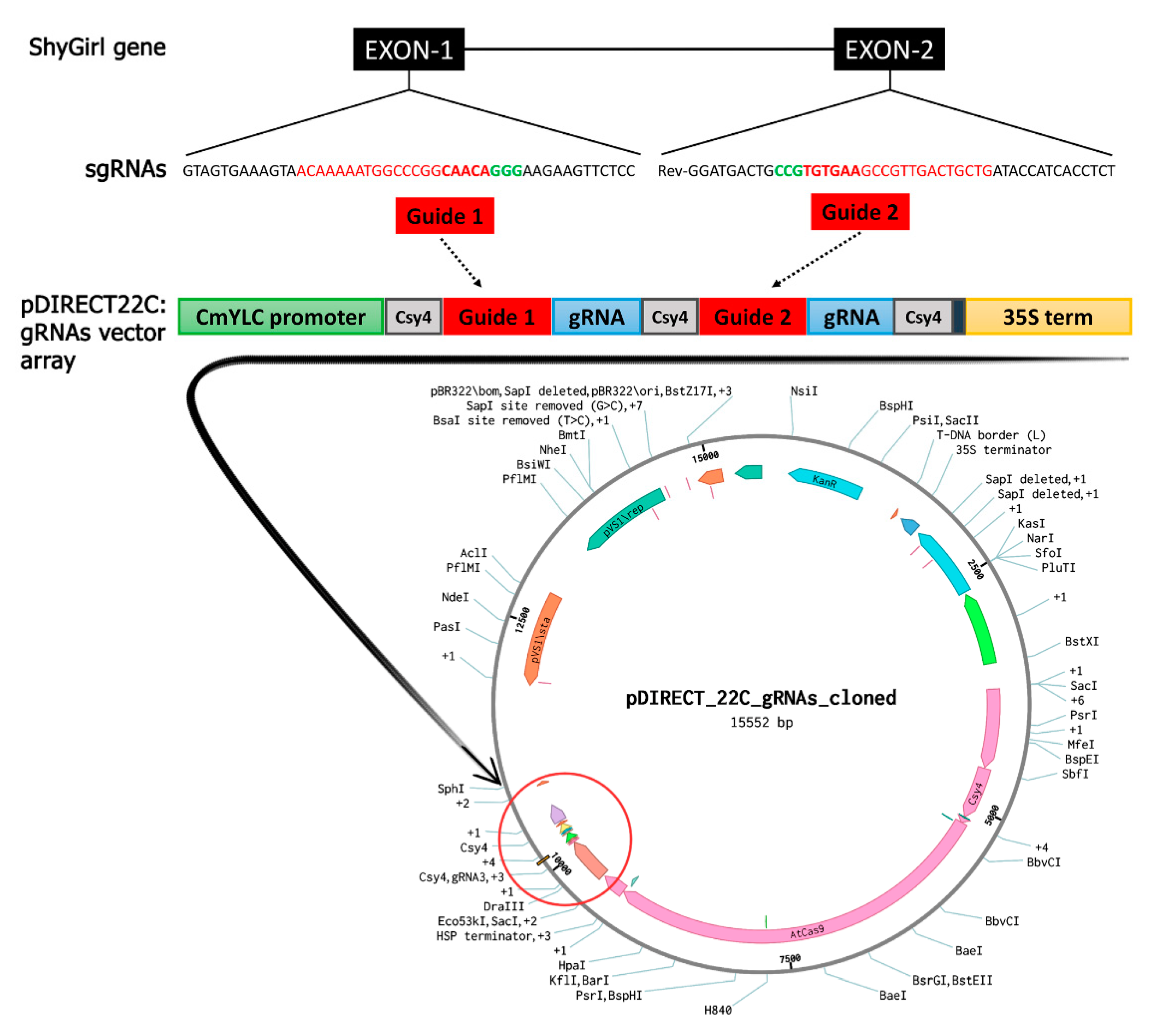
2.2. CRISPR/Cas9 T-DNA Vector Assembly
2.3. Actinidia Transformation
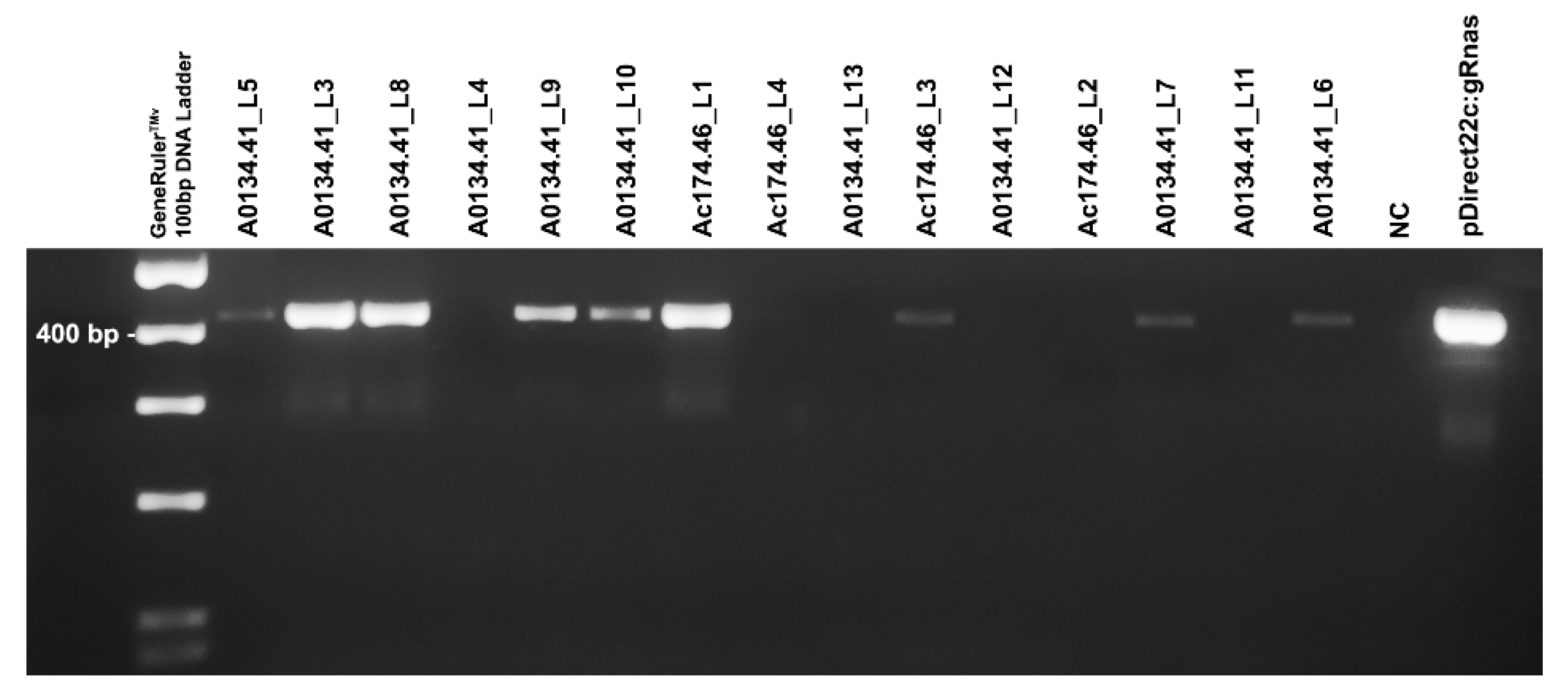
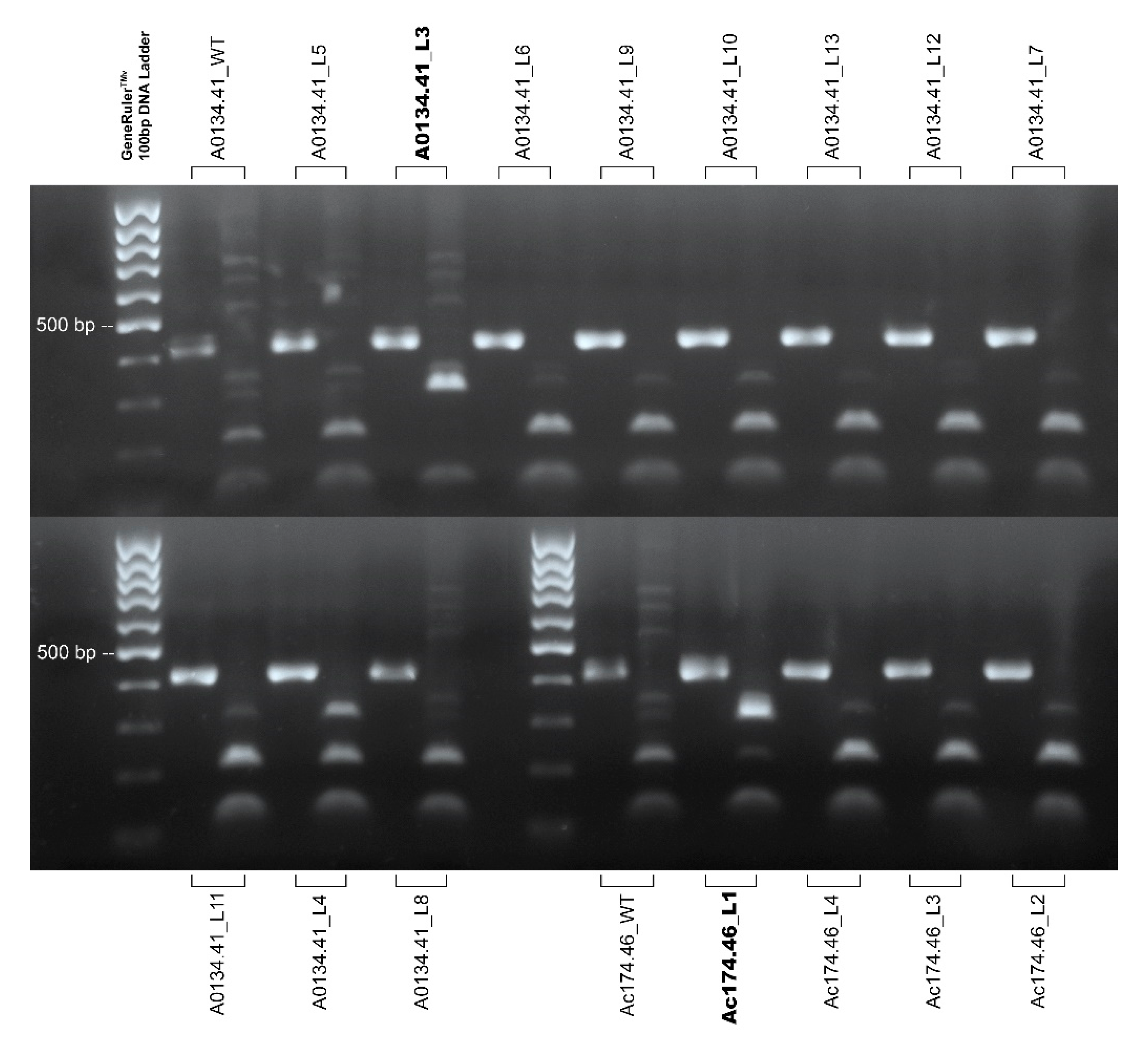
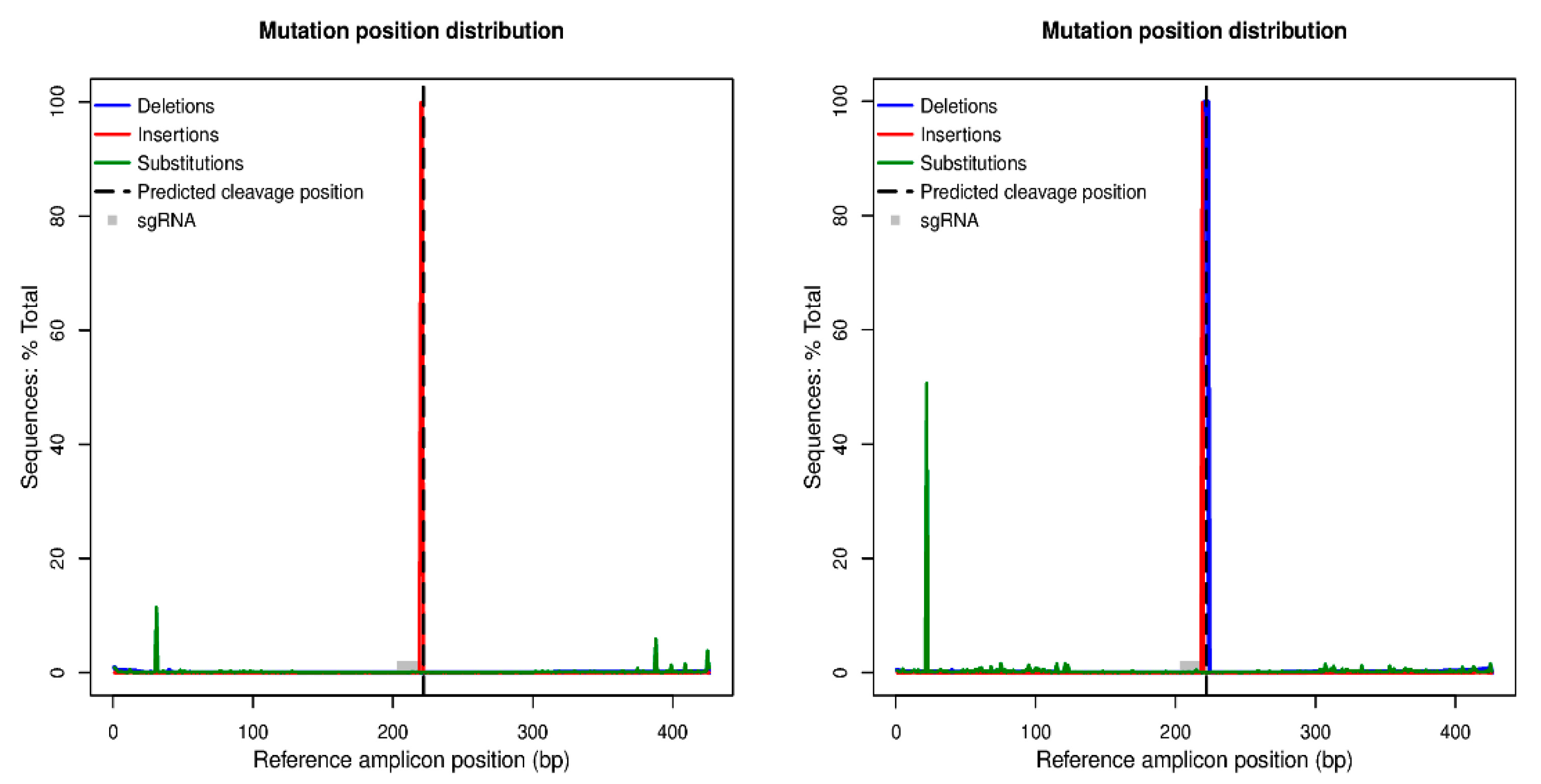
2.4. Evaluation of Mutation Induced by Editing
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Target Identification and CRISPR/Cas9 T-DNA Vector Assembly
4.3. Final CRISPR/Cas9 T-DNA Vector Purification and Sequencing
4.4. Actinidia Transformation
4.5. Mutation Detection
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, C.; Ferguson, A.R. Actinidia; Feng, K.-M., Ed.; Floria Reipublicae Pop. Sin; Science Press: Beijing, China, 1984; Volume 49/2, pp. 196–268, 309–324. [Google Scholar]
- Huang, H. Kiwifruit: The Genus ACTINIDIA; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 1–334. ISBN 9780128030660. [Google Scholar]
- McNeilage, M.A. Gender variation in Actinidia deliciosa, the kiwifruit. Sex. Plant Reprod. 1991, 4, 267–273. [Google Scholar] [CrossRef]
- Testolin, R.; Cipriani, G.; Costa, G. Sex segregation ratio and gender expression in the genus Actinidia. Sex. Plant Reprod. 1995, 8, 129–132. [Google Scholar] [CrossRef]
- Harvey, C.F.; Gill, G.P.; Fraser, L.G.; Mcneilage, M.A. Sex determination in Actinidia. 1. Sex-linked markers and progeny sex ratio in diploid A. chinensis. Sex. Plant Reprod. 1997, 10, 149–154. [Google Scholar] [CrossRef]
- Kawagoe, T.; Suzuki, N. Cryptic dioecy in Actinidia polygama: A test of the pollinator attraction hypothesis. Can. J. Bot. 2004, 82, 214–218. [Google Scholar] [CrossRef]
- McNeilage, M.A. Sex expression in fruiting male vines of kiwifruit. Sex. Plant Reprod. 1991, 8, 129–132. [Google Scholar] [CrossRef]
- Marsh, H.D.; Paterson, T.; Seal, A.G.; McNeilage, M.A. Heritability estimates in kiwifruit. Acta Hortic. 2003, 622, 221–229. [Google Scholar] [CrossRef]
- Testolin, R. Kiwifruit breeding: From the phenotypic analysis of parents to the genomic estimation of their breeding value (GEBV). Acta Hortic. 2011, 913, 123–130. [Google Scholar] [CrossRef]
- Van Nocker, S.; Gardiner, S.E. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 22. [Google Scholar] [CrossRef]
- Mcneilage, M.A.; Steinhagen, S. Flower and fruit characters in a kiwifruit hermaphrodite. Euphytica 1998, 101, 69–72. [Google Scholar] [CrossRef]
- Testolin, R. Male density and arrangement in kiwifruit orchards. Sci. Hortic. 1991, 48, 41–50. [Google Scholar] [CrossRef]
- Goodwin, R.; McBrydie, H.; Taylor, M. Wind and honey bee pollination of kiwifruit (Actinidia chinensis‘HORT16A’). N. Z. J. Bot. 2013, 51, 229–240. [Google Scholar] [CrossRef]
- Stefani, E.; Giovanardi, D. Dissemination of Pseudomonas syringae pv. Actinidiae through pollen and its epiphytic life on leaves and fruits. Phytopathol. Mediterr. 2011, 50, 489–496. [Google Scholar] [CrossRef]
- Testolin, R.; Cipriani, G.; Messina, R. Sex control in Actinidia is monofactorial and remains so in polyploids. In Sex Determination in Plants; Ainsworth, C.C., Ed.; BIOS Scientific Publisher Limited: Milton Park, UK, 1999; pp. 173–181. [Google Scholar]
- Pilkington, S.M.; Tahir, J.; Hilario, E.; Gardiner, S.E.; Chagné, D.; Catanach, A.; McCallum, J.; Jesson, L.; Fraser, L.G.; Mcneilage, M.A.; et al. Genetic and cytological analyses reveal the recombination landscape of a partially differentiated plant sex chromosome in kiwifruit. BMC Plant Biol. 2019, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Henry, I.M.; Ohtani, H.; Morimoto, T.; Beppu, K.; Kataoka, I.; Tao, R. A Y-Encoded Suppressor of Feminization Arose via Lineage-Specific Duplication of a Cytokinin Response Regulator in Kiwifruit. Plant Cell 2018, 30, 780–795. [Google Scholar] [CrossRef]
- Akagi, T.; Pilkington, S.M.; Varkonyi-Gasic, E.; Henry, I.M.; Sugano, S.S.; Sonoda, M.; Firl, A.; Mcneilage, M.A.; Douglas, M.J.; Wang, T.; et al. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 2019, 5, 801–809. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Baltes, N.J.; Voytas, D.F. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 2015, 33, 120–131. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.; Li, D.; Zhang, Q.; Li, L.; Zhong, C.; Liu, Y.; Huang, H. Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 2018, 16, 1424–1433. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wang, T.; Voogd, C.; Jeon, S.; Drummond, R.S.M.; Gleave, A.P.; Allan, A.C. Mutagenesis of kiwifruit CENTRORADIALIS -like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 2019, 17, 869–880. [Google Scholar] [CrossRef]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.G.; Tsang, G.K.; Datson, P.M.; De Silva, H.N.; Harvey, C.F.; Gill, G.P.; Crowhurst, R.N.; Mcneilage, M.A. A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genom. 2009, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Testolin, R. Kiwifruit Yield Efficiency, Plant Density, and Bud Number per Surface Unit. J. Am. Soc. Hortic. Sci. 1990, 115, 704–707. [Google Scholar] [CrossRef]
- Ferguson, A.R.; Huang, H. Genetic Resources of Kiwifruit: Domestication and Breeding. Hortic. Rev. 2007, 33, 1–122. [Google Scholar]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Schindele, A.; Dorn, A.; Puchta, H. CRISPR/Cas brings plant biology and breeding into the fast lane. Curr. Opin. Biotechnol. 2020, 61, 7–14. [Google Scholar] [CrossRef]
- Pinto, C.; Testolin, R.; Cipriani, G. Analysis of components of genetic variance and breeding value of kiwifruit (Actinidia chinensis Planch.) using best linear unbiased prediction (BLUP) and pedigree information. Acta Hortic. 2018, 1218, 73–80. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. Carpel development genes in Arabidopsis. Flower. Newsl. 1997, 23, 12–17. [Google Scholar]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef]
- Walton, E.F.; Fowke, P.J.; Weis, K.; McLeay, P.L. Shoot Axillary Bud Morphogenesis in Kiwifruit (Actinidia deliciosa). Ann. Bot. 1997, 80, 13–21. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Moss, S.M.; Voogd, C.; Wu, R.; Lough, R.H.; Wang, Y.-Y.; Hellens, R.P. Identification and characterization of flowering genes in kiwifruit: Sequence conservation and role in kiwifruit flower development. BMC Plant Biol. 2011, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Gill, G.P.; Harvey, C.F.; Gardner, R.C.; Fraser, L.G. Development of sex-linked PCR markers for gender identification in Actinidia. Theor. Appl. Genet. 1998, 97, 439–445. [Google Scholar] [CrossRef]
- Fraser, L.G.; McNeilage, M.A. Reproductive Biology. In The kiwifruit genome; Springer: Cham, Switzerland, 2016; pp. 65–84. [Google Scholar] [CrossRef]
- Müller, N.A.; Kersten, B.; Leite Montalvão, A.P.; Mähler, N.; Bernhardsson, C.; Bräutigam, K.; Carracedo Lorenzo, Z.; Hoenicka, H.; Kumar, V.; Mader, M.; et al. A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 2020, 6, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Park, J.; Bae, S.; Kim, J.-S. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 2015, 31, 4014–4016. [Google Scholar] [CrossRef]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 1–19. [Google Scholar] [CrossRef]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, K.; Unit, P.; et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Del Fabbro, C.; Scalabrin, S.; Morgante, M.; Giorgi, F.M. An Extensive Evaluation of Read Trimming Effects on Illumina NGS Data Analysis. PLoS ONE 2013, 8, e85024. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Sutton, G.G.; Jones, S.J.M.; Holt, R.A.; Bateman, A. Assembling millions of short DNA sequences using SSAKE. Bioinformatics 2007, 23, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Wang, T.; Lin-Wang, K. High throughput transformation of Actinidia: A platform for kiwifruit functional genomics and molecular breeding. Transgenic Plant J. 2007, 1, 173–184. [Google Scholar]
- Wang, T.; Karunairetnam, S.; Wu, R.; Wang, Y.Y.; Gleave, A.P. High efficiency transformation platforms for kiwifruit (Actinidia spp.) functional genomics. Acta Hortic. 2012, 929, 143–148. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J. Isolation of Plant DNA from Fresh Tissue. Focus (Madison) 1990, 12, 13–15. [Google Scholar]
- Andrews, S. Fast QC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 29 December 2020).
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Project, G.; et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Magoč, T.; Magoč, M.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Pinello, L.; Canver, M.C.; Hoban, M.D.; Orkin, S.H.; Kohn, D.B.; Bauer, D.E.; Yuan, G.-C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016, 34, 695–697. [Google Scholar] [CrossRef] [PubMed]



| Genotype | No. of Leaf Discs | No. of Petioles | Kanamycin-Resistant Calli | No. of Putative Transgenic Shoots | No. of Tested Plantlets | |
|---|---|---|---|---|---|---|
| No. of Leaf Discs | No. of Petioles | |||||
| A0134.41 | 40 | 35 | 2 | 30 | 44 | 11 |
| Ac174.46 | 50 | 33 | 3 | 20 | 61 | 4 |
| Genotype | Line Code | Ti integration * | BslI Restriction Assay ** | Illumina Sequencing |
|---|---|---|---|---|
| A0134.41 | A0134.41_L5 | Yes | Site-specific digestion and wild-type patterns | 0.0038% edited reads |
| A0134.41 | A0134.41_L6 | Yes | Site-specific digestion and wild-type patterns | 0.20% edited reads |
| A0134.41 | A0134.41_L3 | Yes | Site specific digestion | 99.85% edited reads |
| A0134.41 | A0134.41_L8 | Yes | Wild-type digestion pattern | 0.00% edited reads |
| A0134.41 | A0134.41_L4 | No | Site-specific digestion and wild-type patterns | 0.091% edited reads |
| A0134.41 | A0134.41_L9 | Yes | Wild-type digestion pattern | 0.00% edited reads |
| A0134.41 | A0134.41_L10 | Yes | Site-specific digestion and wild-type patterns | 0.31 % edited reads |
| Ac174.46 | Ac174.46_L1 | Yes | Site specific digestion | 99.76% edited reads |
| Ac174.46 | Ac174.46_L4 | No | Wild-type digestion pattern | Not sequenced |
| A0134.41 | A0134.41_L13 | No | Wild-type digestion pattern | Not sequenced |
| Ac174.46 | Ac174.46_L3 | Yes | Wild-type digestion pattern | Not sequenced |
| A0134.41 | A0134.41_L12 | No | Wild-type digestion pattern | Not sequenced |
| Ac174.46 | Ac174.46_L2 | No | Wild-type digestion pattern | Not sequenced |
| A0134.41 | A0134.41_L7 | Yes | Wild-type digestion pattern | Not sequenced |
| A0134.41 | A0134.41_L11 | No | Wild-type digestion pattern | Not sequenced |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Mori, G.; Zaina, G.; Franco-Orozco, B.; Testolin, R.; De Paoli, E.; Cipriani, G. Targeted Mutagenesis of the Female-Suppressor SyGI Gene in Tetraploid Kiwifruit by CRISPR/CAS9. Plants 2021, 10, 62. https://doi.org/10.3390/plants10010062
De Mori G, Zaina G, Franco-Orozco B, Testolin R, De Paoli E, Cipriani G. Targeted Mutagenesis of the Female-Suppressor SyGI Gene in Tetraploid Kiwifruit by CRISPR/CAS9. Plants. 2021; 10(1):62. https://doi.org/10.3390/plants10010062
Chicago/Turabian StyleDe Mori, Gloria, Giusi Zaina, Barbara Franco-Orozco, Raffaele Testolin, Emanuele De Paoli, and Guido Cipriani. 2021. "Targeted Mutagenesis of the Female-Suppressor SyGI Gene in Tetraploid Kiwifruit by CRISPR/CAS9" Plants 10, no. 1: 62. https://doi.org/10.3390/plants10010062
APA StyleDe Mori, G., Zaina, G., Franco-Orozco, B., Testolin, R., De Paoli, E., & Cipriani, G. (2021). Targeted Mutagenesis of the Female-Suppressor SyGI Gene in Tetraploid Kiwifruit by CRISPR/CAS9. Plants, 10(1), 62. https://doi.org/10.3390/plants10010062







