Assessment of the Current Status of Potyviruses in Watermelon and Pumpkin Crops in Spain: Epidemiological Impact of Cultivated Plants and Mixed Infections
Abstract
1. Introduction
2. Materials & Methods
2.1. Sample Surveys
2.2. Cucurbit Virus Detection
2.3. Full-Length MWMV and WMV Genome Amplification and Construction of Infectious Clones
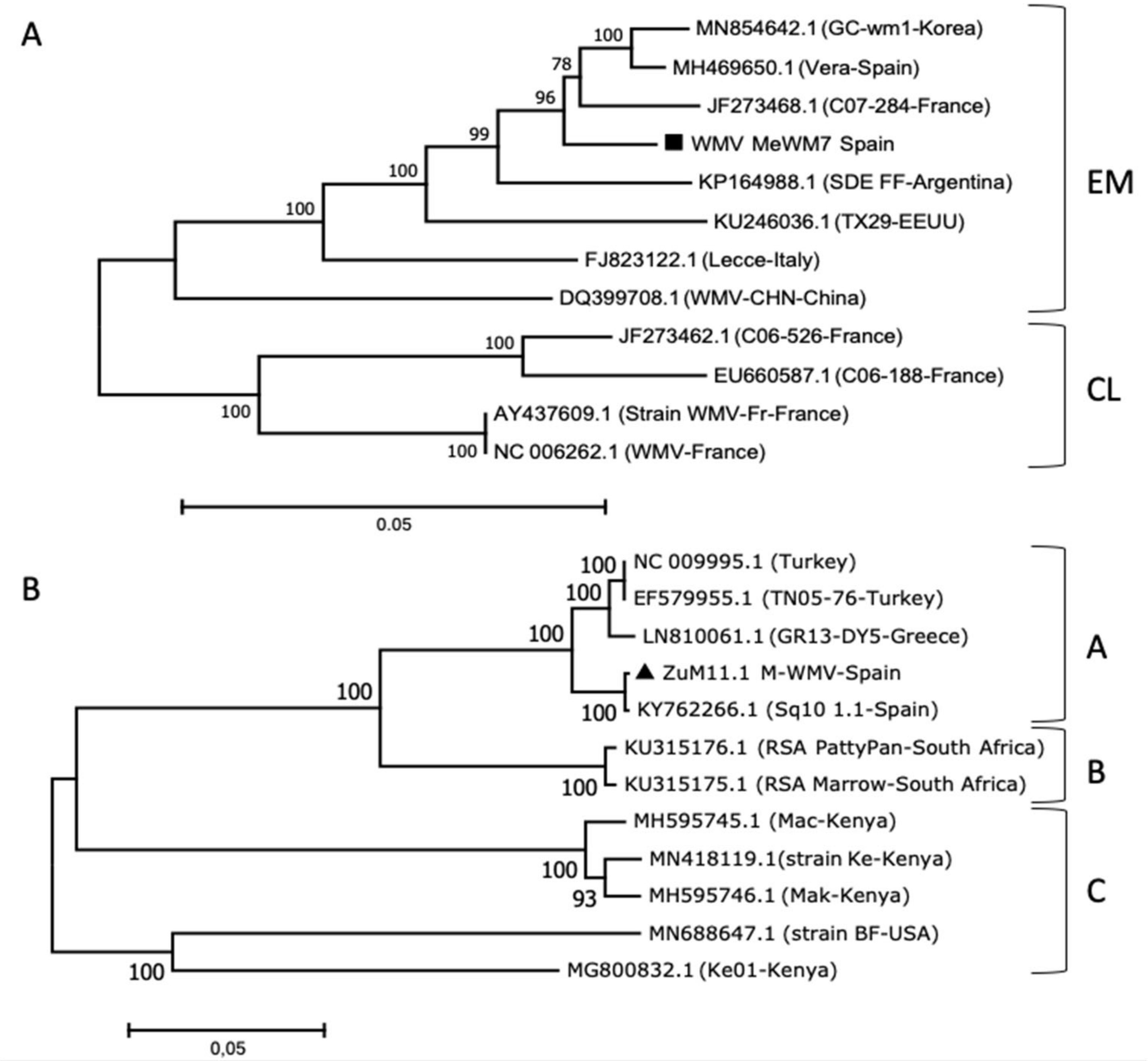
2.4. Sequencing and Phylogenetic Relationships of MWMV and WMV Isolates
2.5. Agro-Inoculation of Cucurbit Plants Species
2.6. Viral Load Quantification
2.7. Statistical Analyses
2.8. Nucleotide Sequence Accession Numbers
3. Results
3.1. Cucurbit Aphid-Borne Mosaic Viruses’ Occurrence in Watermelon and Pumpkin
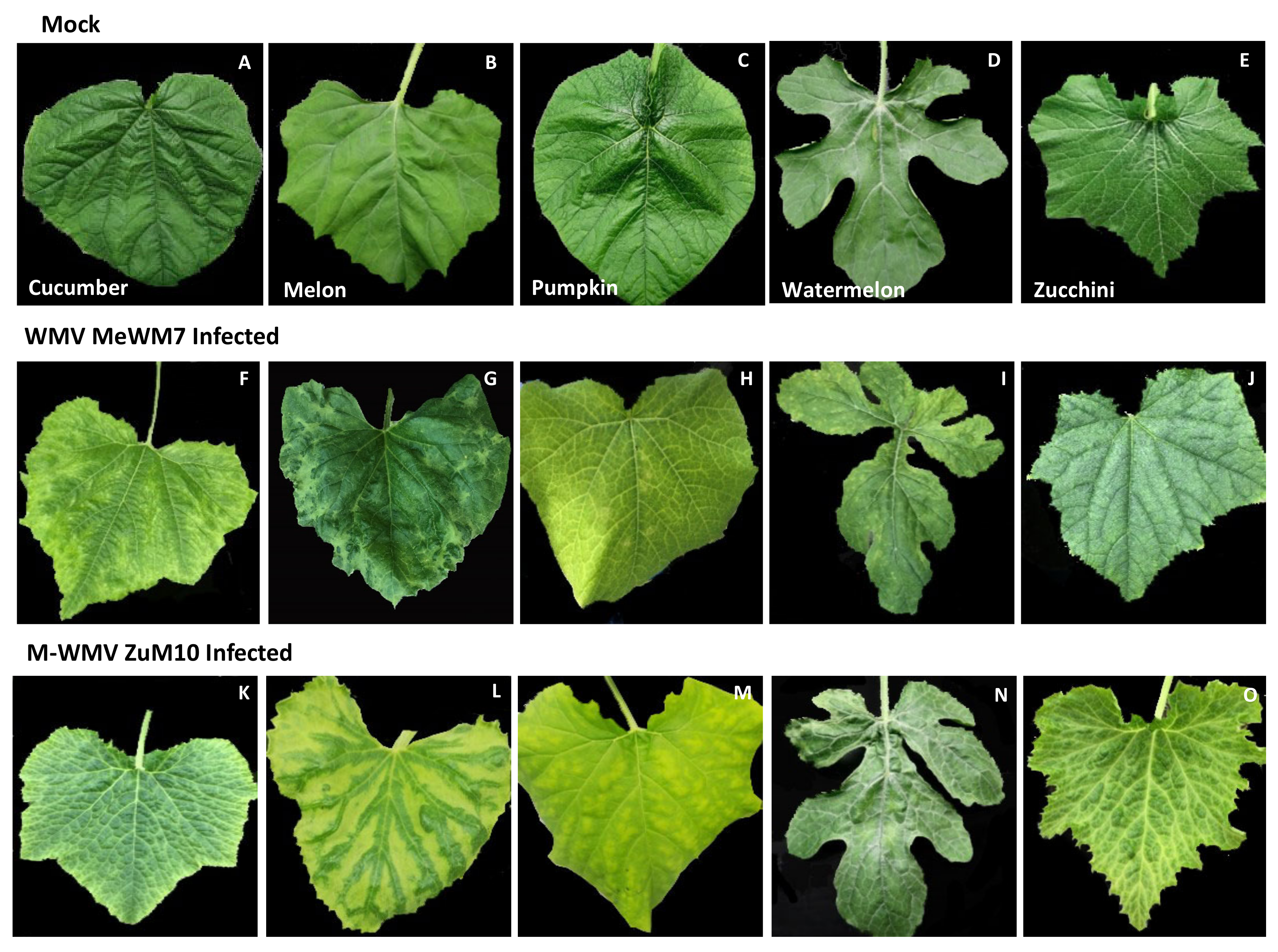
3.2. Genetic Analysis and Symptom Expression of the WMV and MWMV Clones
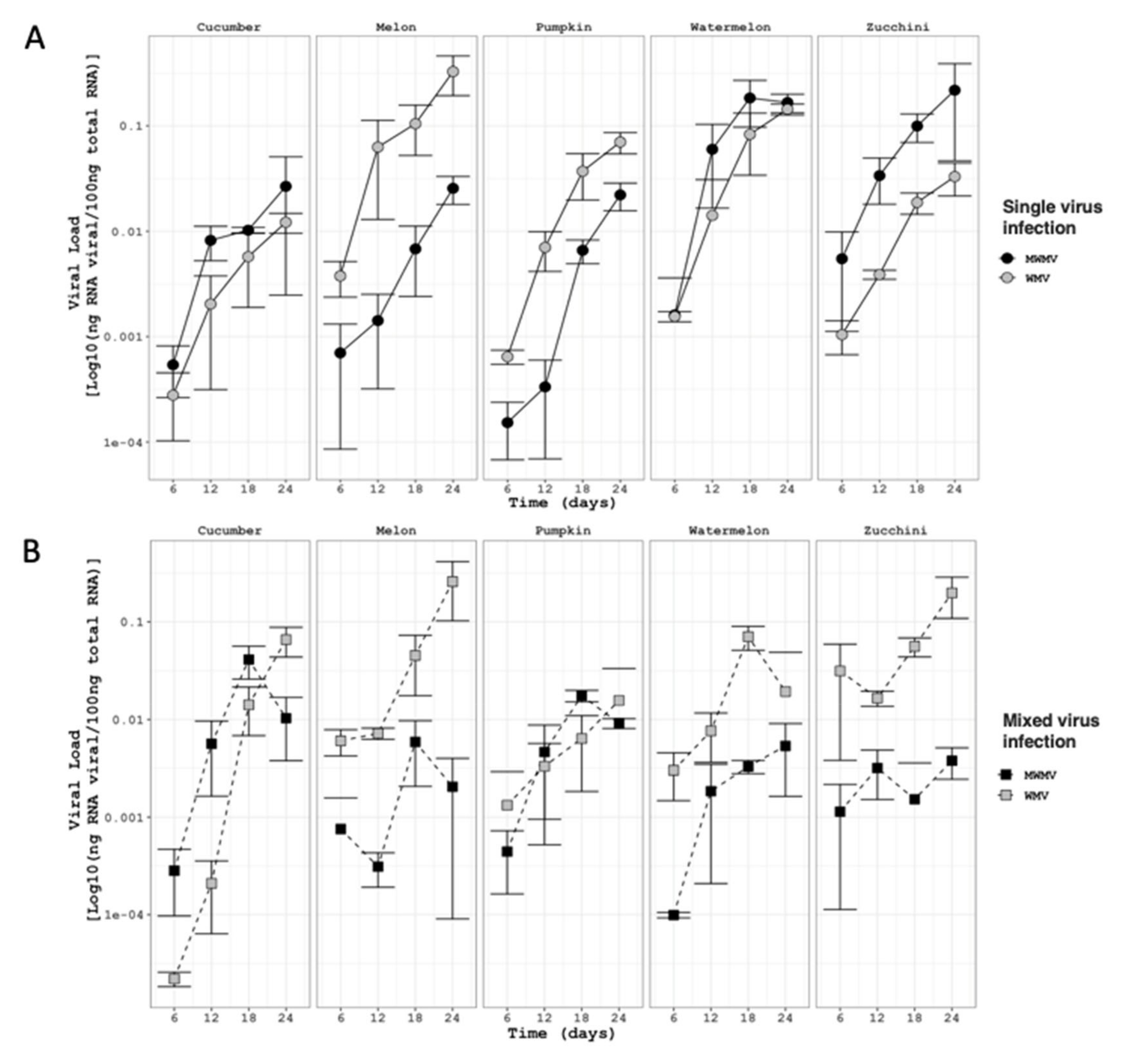
3.3. Virus Accumulation of WMV-MeWM7 and MWMV-ZuM10 Clones in Single and Mixed Infections
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Romay, G.; Lecoq, H.; Desbiez, C. Cucurbit crops and their viral diseases in latin america and the caribbean islands: A review. J. Plant Pathol. 2014, 96, 227–242. [Google Scholar] [CrossRef]
- Lecoq, H.; Katis, N. Control of cucurbit viruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2014; Volume 90, ISBN 9780128012468. [Google Scholar]
- Moreno, A.B.; López-Moya, J.J. When viruses play team sports: Mixed infections in plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, C.; Rabadán, M.P.; Moreno-Pérez, M.G.; Gómez, P. Implications of mixed viral infections on plant disease ecology and evolution. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2020; Volume 106, pp. 145–169. ISBN 9780128207543. [Google Scholar]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Lecoq, H.; Desbiez, C. Viruses of Cucurbit Crops in the Mediterranean Region. An Ever-Changing Picture. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2012; Volume 84, ISBN 9780123943149. [Google Scholar]
- Kassem, M.; Juárez, M.; Gómez, P.; Mengual, C.M.; Sempere, R.N.; Plaza, M.; Elena, S.F.; Moreno, A.; Fereres, A.; Aranda, M. Genetic diversity and potential vectors and reservoirs of Cucurbit aphid-borne yellows virus in Southeastern Spain. Phytopathology 2013, 103, 1188–1197. [Google Scholar] [CrossRef]
- Mnari-Hattab, M.; Gauthier, N.; Zouba, A. Biological and molecular characterization of the Cucurbit aphid-borne yellows virus affecting cucurbits in Tunisia. Plant Dis. 2009, 93, 1065–1072. [Google Scholar] [CrossRef]
- Abeykoon, A.M.S.K.; Basnayake, B.M.V.S.; Salim, N. First report of Cucurbit aphid-borne yellow virus infecting bitter gourd (Momordica charantia) and spiny gourd (Momordica dioica) in Sri Lanka. New Dis. Rep. 2018, 38, 22. [Google Scholar] [CrossRef]
- Choi, S.K.; Yoon, J.Y.; Choi, G.S. Biological and molecular characterization of a Korean isolate of Cucurbit aphid-borne yellows virus infecting cucumis species in Korea. Plant Pathol. J. 2015, 31, 371–378. [Google Scholar] [CrossRef]
- Harth, J.E.; Ferrari, M.J.; Helms, A.M.; Tooker, J.F.; Stephenson, A.G. Zucchini Yellow mosaic virus infection limits establishment and severity of powdery mildew in wild populations of Cucurbita pepo. Front. Plant Sci. 2018, 9, 792. [Google Scholar] [CrossRef]
- Juárez, M.; Legua, P.; Mengual, C.M.; Kassem, M.A.; Sempere, R.N.; Gómez, P.; Truniger, V.; Aranda, M.A. Relative incidence, spatial distribution and genetic diversity of cucurbit viruses in eastern Spain. Ann. Appl. Biol. 2013, 162, 362–370. [Google Scholar] [CrossRef]
- Desbiez, C.; Costa, C.; Wipf-Scheibel, C.; Girard, M.; Lecoq, H. Serological and molecular variability of watermelon mosaic virus (genus Potyvirus). Arch. Virol. 2007, 152, 775–781. [Google Scholar] [CrossRef]
- Bertin, S.; Manglli, A.; McLeish, M.; Tomassoli, L. Genetic variability of watermelon mosaic virus isolates infecting cucurbit crops in Italy. Arch. Virol. 2020, 165, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Vučurović, A.; Bulajić, A.; Stanković, I.; Ristić, D.; Berenji, J.; Jović, J.; Krstić, B. Non-persistently aphid-borne viruses infecting pumpkin and squash in Serbia and partial characterization of Zucchini yellow mosaic virus isolates. Eur. J. Plant Pathol. 2012, 133, 935–947. [Google Scholar] [CrossRef]
- Cohen, S.; Nitzany, F.E. Identity of viruses affecting cucurbits in Israel. Phytopathology 1963, 53, 193–196. [Google Scholar]
- Purcifull, D.E. Serological Distinction of Watermelon Mosaic Virus Isolates. Phytopathology 1979, 69, 112. [Google Scholar] [CrossRef]
- Purcifull, D.; Hiebert, E.; Edwardson, J.R. CMI/AAB Descriptions of Plant Viruses. Assoc. Appl. Biol. 1984, 7. [Google Scholar]
- Alonso-Prados, J.L.; Fraile, A.; García-Arenal, F. Impact of cucumber mosaic virus and watermelon mosaic virus 2 infection on melon production in central Spain. J. Plant Pathol. 1997, 79, 131–134. [Google Scholar]
- Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. High-throughput sequencing facilitates discovery of new plant viruses in Poland. Plants 2020, 9, 820. [Google Scholar] [CrossRef]
- Lecoq, H.; Desbiez, C. Watermelon mosaic virus. In Encyclopedia Virol, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 433–440. [Google Scholar]
- Hajizadeh, M.; Bahrampour, H.; Abdollahzadeh, J. Genetic diversity and population structure of Watermelon mosaic virus. J. Plant Dis. Prot. 2017, 124, 601–610. [Google Scholar] [CrossRef]
- Desbiez, C.; Joannon, B.; Wipf-Scheibel, C.; Chandeysson, C.; Lecoq, H. Emergence of new strains of Watermelon mosaic virus in South-eastern France: Evidence for limited spread but rapid local population shift. Virus Res. 2009, 141, 201–208. [Google Scholar] [CrossRef]
- Kassem, M.A.; Sempere, R.N.; Juárez, M.; Aranda, M.A.; Truniger, V. Cucurbit aphid-borne yellows virus Is Prevalent in Field-Grown Cucurbit Crops of Southeastern Spain. Plant Dis. 2007, 91, 232–238. [Google Scholar] [CrossRef]
- Luis-Arteaga, M.; Alvarez, J.M.; Alonso-Prados, J.L.; Bernal, J.J.; Garcia-Arenal, F.; Lavina, A.; Batlle, A.; Moriones, E. Occurrence, distribution, and relative incidence of mosaic viruses infecting field-grown melon in Spain. Plant Dis. 1998, 82, 979–982. [Google Scholar] [CrossRef]
- Juárez, M.; Tovar, R.; Fiallo-Olivé, E.; Aranda, M.A.; Gosálvez, B.; Castillo, P.; Moriones, E.; Navas-Castillo, J. First Detection of Tomato leaf curl New Delhi virus Infecting Zucchini in Spain. Plant Dis. 2013, 98, 857. [Google Scholar] [CrossRef] [PubMed]
- Moreno, I.M.; Malpica, J.M.; Díaz-Pendón, J.A.; Moriones, E.; Fraile Garcia-Arenal, F.A. Variability and genetic structure of the population of watermelon mosaic virus infecting melon in Spain. Virology 2004, 318, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Miras, M.; Juárez, M.; Aranda, M.A. Resistance to the emerging Moroccan watermelon mosaic virus in squash. Phytopathology 2019, 109, 895–903. [Google Scholar] [CrossRef] [PubMed]
- McKern, N.M.; Strike, P.M.; Barnett, O.W.; Ward, C.W.; Shukla, D.D. Watermelon mosaic virus-Morocco is a distinct potyvirus. Arch. Virol. 1993, 131, 467–473. [Google Scholar] [CrossRef]
- Yakoubi, S.; Desbiez, C.; Fakhfakh, H.; Wipf-Scheibel, C.; Marrakchi, M.; Lecoq, H. Biological characterization and complete nucleotide sequence of a Tunisian isolate of Moroccan watermelon mosaic virus. Arch Virol 2008, 153, 117–125. [Google Scholar] [CrossRef]
- Chatzivassiliou, E.K.; Papapanagiotou, A.P.; Mpenardis, P.D.; Perdikis, D.C.; Menexes, G. Transmission of Moroccan watermelon mosaic virus (MWMV) by Aphids in Greece. Plant Dis. 2016, 100, 601–606. [Google Scholar] [CrossRef]
- Ibaba, J.D.; Laing, M.D.; Gubba, A. Genome sequence analysis of two South African isolates of Moroccan watermelon mosaic virus infecting cucurbits. Virus Genes 2016, 52, 896–899. [Google Scholar] [CrossRef]
- Malandraki, I.; Vassilakos, N.; Xanthis, C.; Kontosfiris, G.; Katis, N.I.; Varveri, C. First Report of Moroccan watermelon mosaic virus in Zucchini Crops in Greece. Plant Dis. 2013, 98, 702. [Google Scholar] [CrossRef]
- Lecoq, H.; Justafré, I.; Wipf-Scheibel, C.; Desbiez, C. Moroccan watermelon mosaic virus newly reported on zucchini squash in France. Plant Pathol. 2008, 57, 766. [Google Scholar] [CrossRef]
- Juárez, M.; Rabadan, M.; Diaz Martinez, L.; Tayahi, M.; Grande-Perez, A.; Gómez, P. Natural Hosts and Genetic Diversity of the Emerging Tomato Leaf Curl New Delhi Virus in Spain. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Valverde, S.; Vidiella, B.; Montañez, R.; Fraile, A.; Sacristán, S.; García-Arenal, F. Coexistence of nestedness and modularity in host–pathogen infection networks. Nat. Ecol. Evol. 2020, 4, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Sallinen, S.; Norberg, A.; Susi, H.; Laine, A.-L. Intraspecific host variation plays a key role in virus community assembly. Nat. Commun. 2020, 11, 5610. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Rousseau, C.M.; Birditt, B.A.; McKay, A.R.; Stoddard, J.N.; Lee, T.C.; McLaughlin, S.; Moore, S.W.; Shindo, N.; Learn, G.H.; Korber, B.T.; et al. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J. Virol. Methods 2006, 136, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Dilernia, D.A.; Chien, J.-T.; Monaco, D.C.; Brown, M.P.S.; Ende, Z.; Deymier, M.J.; Yue, L.; Paxinos, E.E.; Allen, S.; Tirado-Ramos, A.; et al. Multiplexed highly-accurate DNA sequencing of closely-related HIV-1 variants using continuous long reads from single molecule, real-time sequencing. Nucleic Acids Res. 2015, 43, e129. [Google Scholar] [CrossRef] [PubMed]
- Blawid, R.; Nagata, T. Construction of an infectious clone of a plant RNA virus in a binary vector using one-step Gibson Assembly. J. Virol. Methods 2015, 222, 11–15. [Google Scholar] [CrossRef]
- Tuo, D.; Shen, W.; Yan, P.; Li, X.; Zhou, P. Rapid Construction of Stable Infectious Full-Length cDNA Clone of Papaya Leaf Distortion Mosaic Virus Using In-Fusion Cloning. Viruses 2015, 7, 6241–6250. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Sempere, R.N.; Gómez, P.; Truniger, V.; Aranda, M.A. Development of expression vectors based on pepino mosaic virus. Plant Methods 2011, 7, 1–14. [Google Scholar] [CrossRef]
- MAPAMA Statistic Datasets from the Ministry of Agriculture, Fisheries and Food in Spain. Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/agricultura/superficies-producciones-anuales-cultivos/2019 (accessed on 16 November 2020).
- Desbiez, C.; Wipf-Scheibel, C.; Millot, P.; Berthier, K.; Girardot, G.; Gognalons, P.; Hirsch, J.; Moury, B.; Nozeran, K.; Piry, S.; et al. Distribution and evolution of the major viruses infecting cucurbitaceous and solanaceous crops in the French Mediterranean area. Virus Res. 2020, 286, 198042. [Google Scholar] [CrossRef]
- Verma, R.K.; Mishra, M.; Marwal, A.; Gaur, R.K. Identification, genetic diversity and recombination analysis of Watermelon Mosaic Virus isolates. 3 Biotech 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lecoq, H.; Dafalla, G.; Desbiez, C.; Wipf-Scheibel, C.; Delécolle, B.; Lanina, T.; Ullah, Z.; Grumet, R. Biological and molecular characterization of Moroccan watermelon mosaic virus and a potyvirus isolate from Eastern Sudan. Plant Dis. 2001, 85, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Lecoq, H.; Fabre, F.; Joannon, B.; Wipf-Scheibel, C.; Chandeysson, C.; Schoeny, A.; Desbiez, C. Search for factors involved in the rapid shift in Watermelon mosaic virus (WMV) populations in South-eastern France. Virus Res. 2011, 159, 115–123. [Google Scholar] [CrossRef]
- Desbiez, C.; Lecoq, H. Evidence for multiple intraspecific recombinants in natural populations of Watermelon mosaic virus (WMV, Potyvirus). Arch. Virol. 2008, 153, 1749–1754. [Google Scholar] [CrossRef] [PubMed]
- Gadhave, K.R.; Dutta, B.; Coolong, T.; Srinivasan, R.A. non-persistent aphid-transmitted Potyvirus differentially alters the vector and non-vector biology through host plant quality manipulation. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Mauck, K.E.; Chesnais, Q.; Shapiro, L.R. Evolutionary determinants of host and vector manipulation by plant viruses. Adv Virus Res. 2018, 101, 189–250. [Google Scholar] [CrossRef]
- Tollenaere, C.; Susi, H.; Laine, A.-L. Evolutionary and epidemiological implications of multiple infection in plants. Trends Plant Sci. 2016, 21, 80–90. [Google Scholar] [CrossRef]
- Desbiez, C.; Joannon, B.; Wipf-Scheibel, C.; Chandeysson, C.; Lecoq, H. Recombination in natural populations of watermelon mosaic virus: New agronomic threat or damp squib? J. Gen. Virol. 2011, 92, 1939–1948. [Google Scholar] [CrossRef]
- Quiot-Douine, L.; Lecoq, H.; Quiot, J.B.; Pitrat, M.; Labonne, G. Serological and biological variability of virus isolates related to strains of papaya ringspot virus. Phytopathology 1990, 80, 256–263. [Google Scholar] [CrossRef]
- Domingo-Calap, M.L.; Moreno, A.B.; Pendón, J.A.D.; Moreno, A.; Fereres, A.; López-Moya, J.J. Assessing the impact on virus transmission and insect vector behavior of a viral mixed infection in melon. Phytopathology 2020, 110, 174–186. [Google Scholar] [CrossRef]
- Syller, J. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Syller, J.; Grupa, A. Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef]
- Alcaide, C.; Rabadán;, M.P.; Juárez, M.; Gómez, P. Long-Term Cocirculation of Two Strains of Pepino Mosaic Virus in Tomato Crops and Its Effect on Population Genetic Variability. Phytopathology 2020, 110, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Peláez, A.; McLeish, M.J.; Paswan, R.R.; Dubay, B.; Fraile, A.; García-Arenal, F. Ecological fitting is the forerunner to diversification in a plant virus with broad host range. J. Evol. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Valli, A.A.; Gallo, A.; Rodamilans, B.; López-Moya, J.J.; García, J.A. The HCPro from the Potyviridae family: An enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763. [Google Scholar] [CrossRef]
- Salvaudon, L.; De Moraes, C.M.; Mescher, M.C. Outcomes of co-infection by two potyviruses: Implications for the evolution of manipulative strategies. Proc. R. Soc. B Biol. Sci. 2013, 280. [Google Scholar] [CrossRef]

| Positive Samples for Virus Tested | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cucurbit Plant | Locality | Year | N° Plots | N° Samples Analyzed | WMV | M-WMV | PRSV | ZYMV | CMV |
| Watermelon | Murcia | 2018 | 3 | 12 | 6 | - | - | - | - |
| 2019 | 5 | 20 | 12 | - | - | - | - | ||
| 2020 | 2 | 8 | 4 | - | - | - | - | ||
| TOTAL | 10 | 40 | 22 (55%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Alicante | 2018 | 4 | 16 | 10 | 2 | - | - | - | |
| 2019 | 2 | 8 | - | - | - | - | - | ||
| 2020 | 3 | 12 | 6 | - | - | - | - | ||
| TOTAL | 9 | 36 | 16 (44%) | 2 (5%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| C. La-Mancha | 2018 | 3 | 12 | 4 | 4 | - | - | - | |
| 2019 | 5 | 20 | 8 | - | - | - | - | ||
| 2020 | 1 | 4 | 4 | - | - | - | - | ||
| TOTAL | 9 | 36 | 16 (44%) | 4 (11%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Pumpkin | Murcia | 2018 | 2 | 8 | - | 1 | - | - | - |
| 2019 | 1 | 4 | 4 | - | - | 2 | - | ||
| 2020 | 3 | 12 | 12 | - | - | 4 | - | ||
| TOTAL | 6 | 24 | 16 (66.6%) | 1 (4.1%) | 0 (0%) | 6 (25%) | 0 (0%) | ||
| Alicante | 2018 | 6 | 24 | 12 | 5 | - | - | - | |
| 2019 | 4 | 16 | 12 | - | 6 | - | - | ||
| 2020 | 1 | 4 | 4 | - | - | - | - | ||
| TOTAL | 11 | 44 | 28 (63.6%) | 5 (11.3%) | 6 (13.6%) | 0 (0%) | 0 (0%) | ||
| C. La-Mancha | 2018 | 1 | 4 | 4 | - | 2 | - | - | |
| 2019 | 3 | 12 | 2 | - | - | - | - | ||
| 2020 | 0 | 0 | - | - | - | - | - | ||
| TOTAL | 4 | 16 | 6 (37.5%) | 0 (0%) | 2 (12.5%) | 0 (0%) | 0 (0%) | ||
| TOTAL | 49 | 196 | 104 (53%) | 12 (6.1%) | 8 (4%) | 6 (3%) | 0 (0%) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Moya-Ruiz, C.; Rabadán, P.; Juárez, M.; Gómez, P. Assessment of the Current Status of Potyviruses in Watermelon and Pumpkin Crops in Spain: Epidemiological Impact of Cultivated Plants and Mixed Infections. Plants 2021, 10, 138. https://doi.org/10.3390/plants10010138
De Moya-Ruiz C, Rabadán P, Juárez M, Gómez P. Assessment of the Current Status of Potyviruses in Watermelon and Pumpkin Crops in Spain: Epidemiological Impact of Cultivated Plants and Mixed Infections. Plants. 2021; 10(1):138. https://doi.org/10.3390/plants10010138
Chicago/Turabian StyleDe Moya-Ruiz, Celia, Pilar Rabadán, Miguel Juárez, and Pedro Gómez. 2021. "Assessment of the Current Status of Potyviruses in Watermelon and Pumpkin Crops in Spain: Epidemiological Impact of Cultivated Plants and Mixed Infections" Plants 10, no. 1: 138. https://doi.org/10.3390/plants10010138
APA StyleDe Moya-Ruiz, C., Rabadán, P., Juárez, M., & Gómez, P. (2021). Assessment of the Current Status of Potyviruses in Watermelon and Pumpkin Crops in Spain: Epidemiological Impact of Cultivated Plants and Mixed Infections. Plants, 10(1), 138. https://doi.org/10.3390/plants10010138






