Genetic and Pharmacological Inhibition of Autophagy Increases the Monoubiquitination of Non-Photosynthetic Phosphoenolpyruvate Carboxylase
Abstract
1. Introduction
2. Results
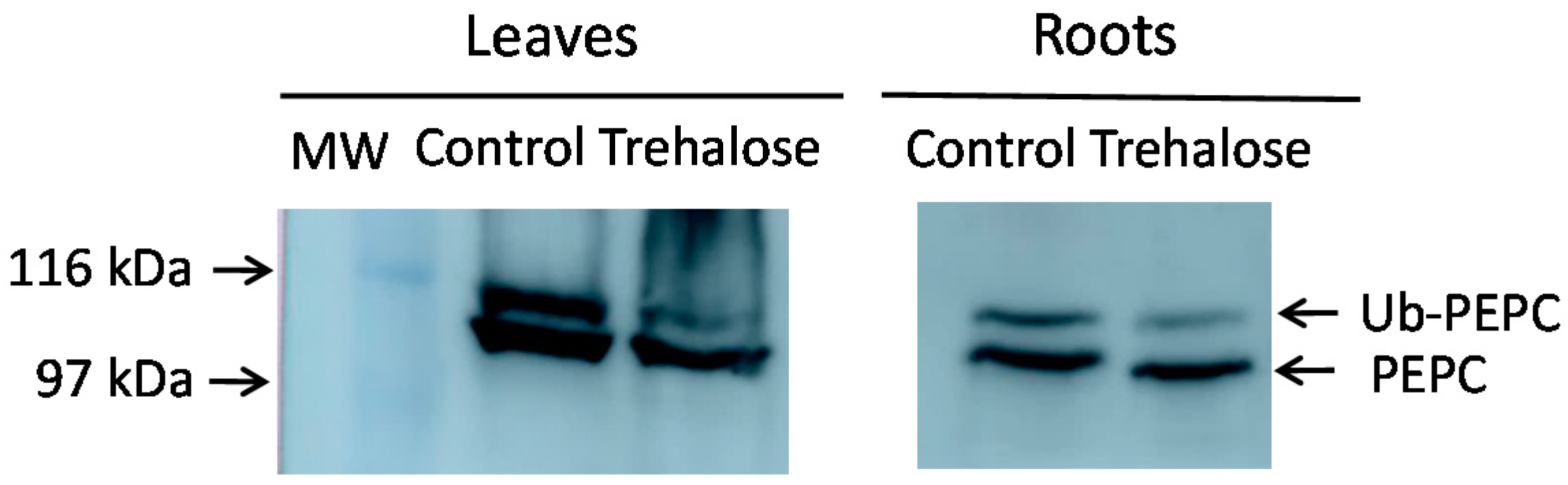
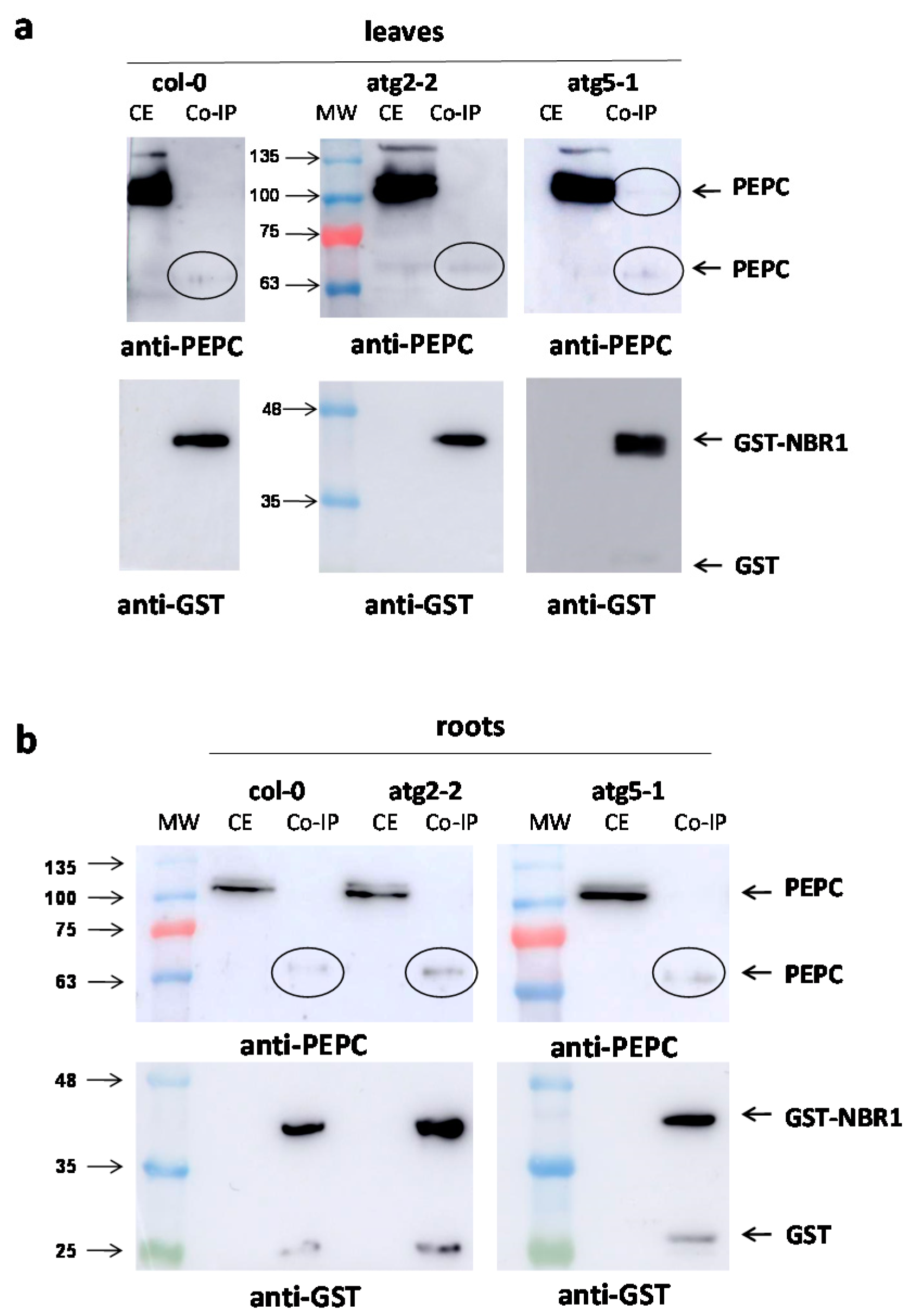
2.1. Increased Monoubiquitinated PEPC in Arabidopsis Mutants and Defective Autophagy
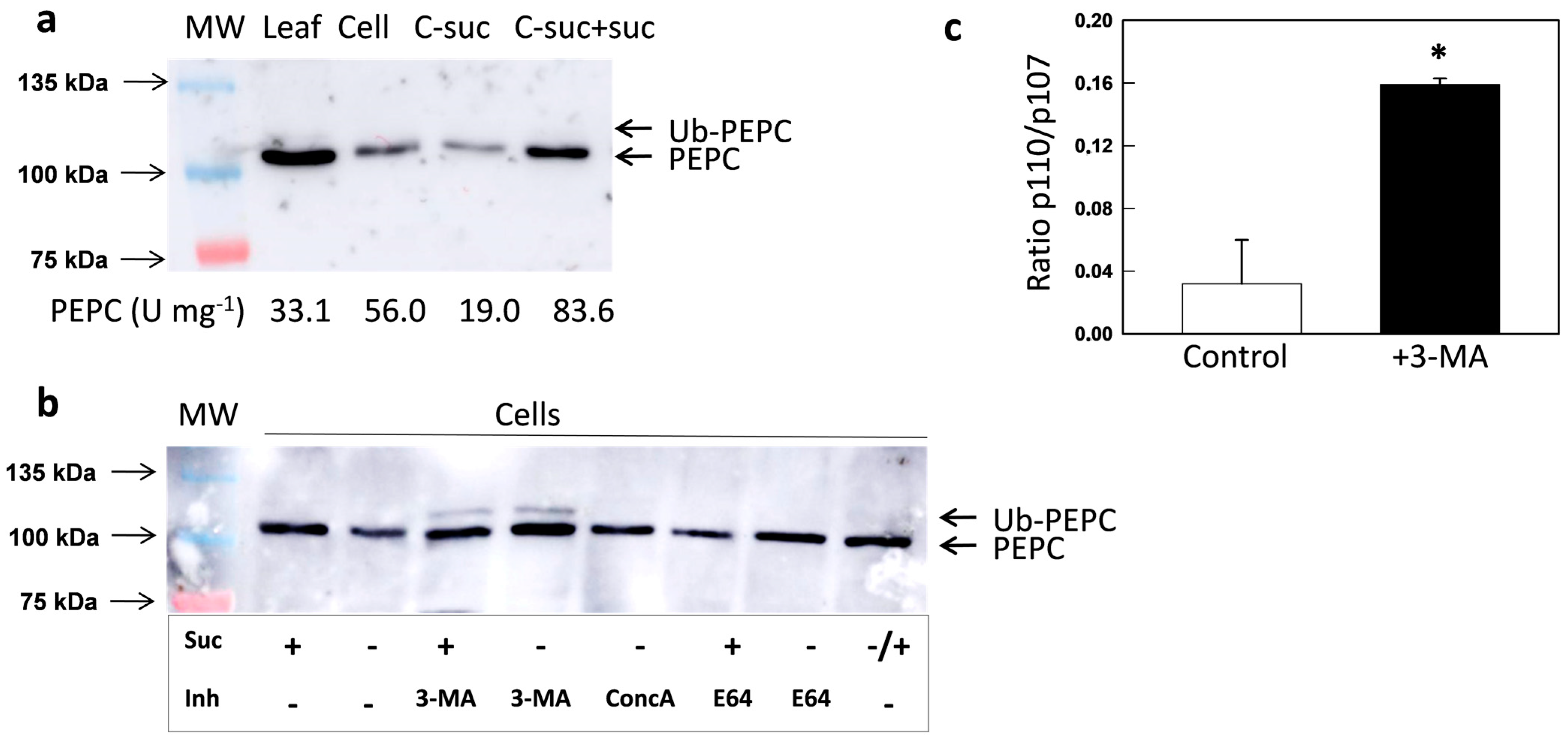
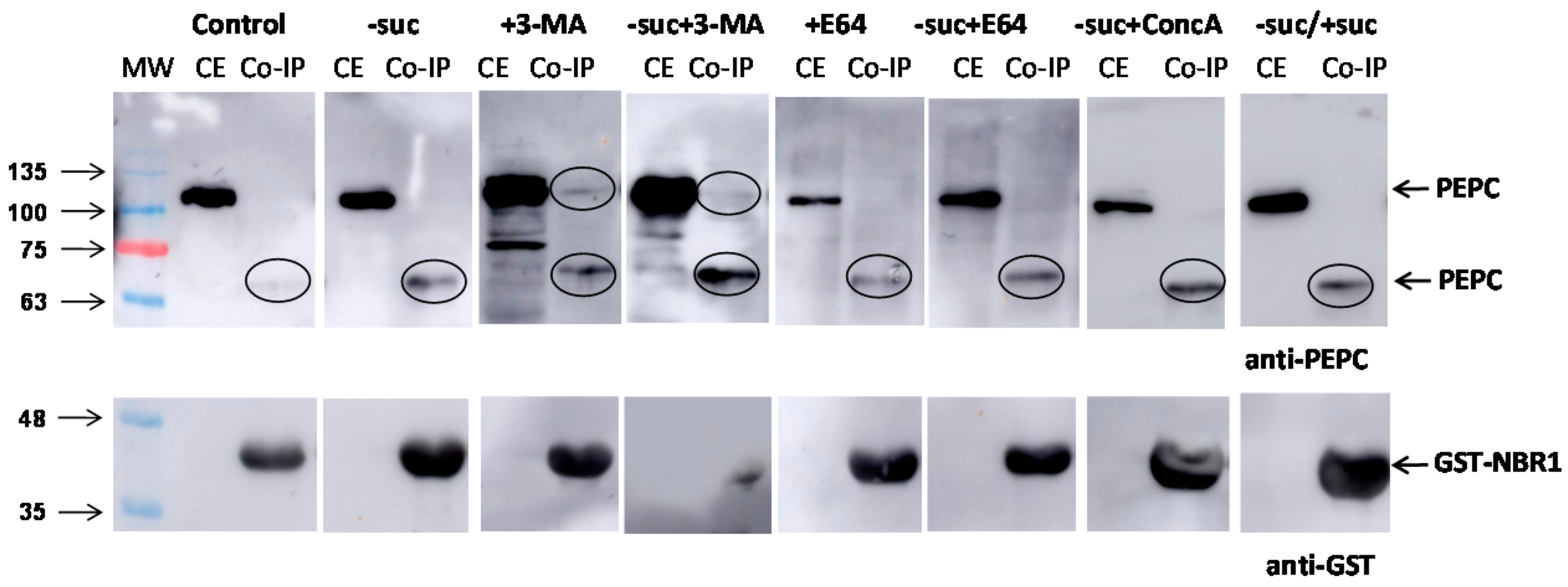
2.2. Chemical Inhibition of Autophagy in Cultured N. benthamiana Cells Increased the Amount of Monoubiquitinated PEPC
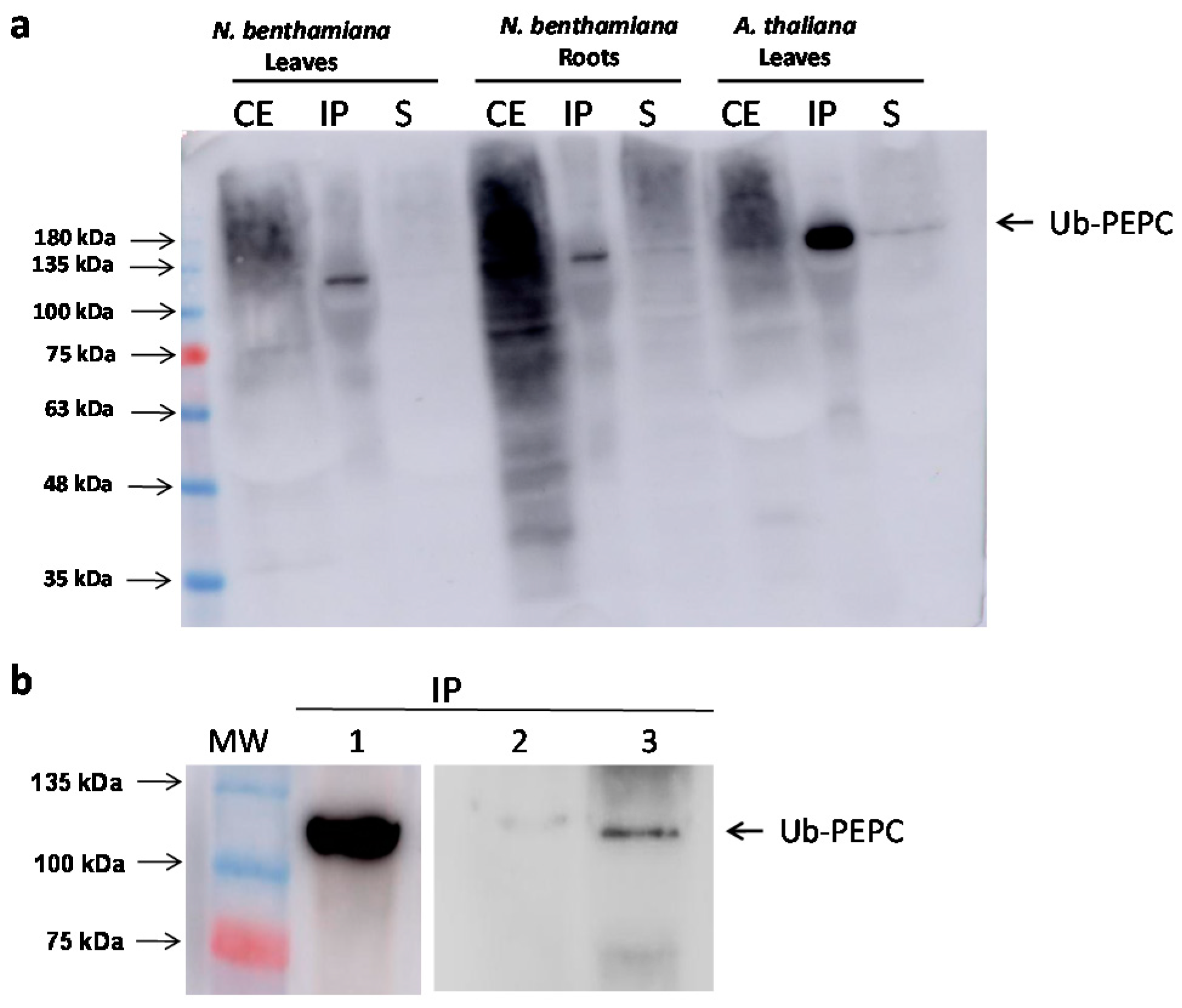
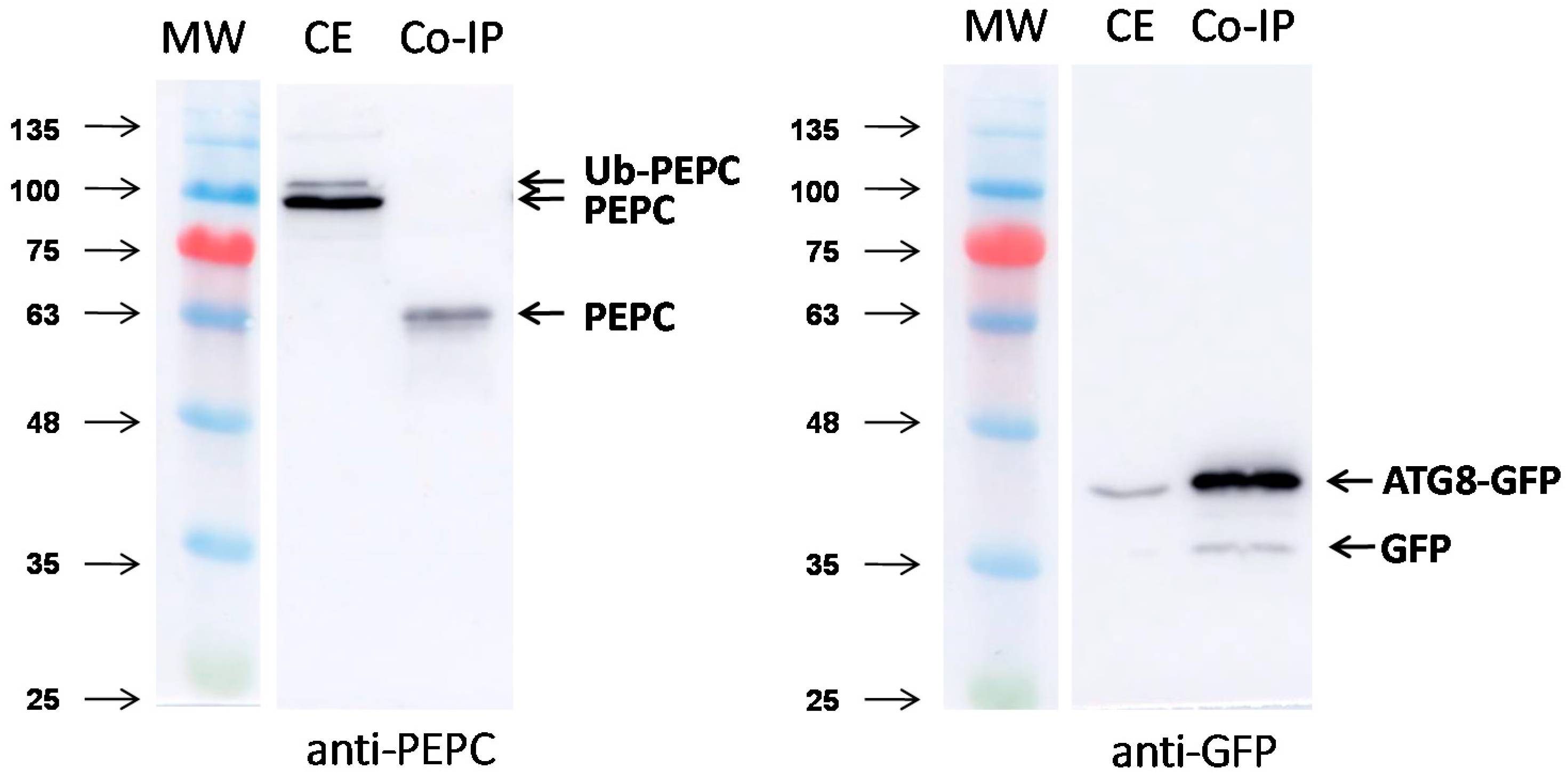
2.3. Pull-Down with GST-NBR1 and GFP-ATG8a
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. N. benthamiana Cell Cultures
4.3. Determination of Enzyme Activity and Protein Quantification
4.4. Antibodies
4.5. Electrophoresis and Protein Gel Blot Analysis
4.6. Recombinant Protein Expression and Purification
4.7. Pull-Down Experiments
4.8. Protein Databases Searches, Alignment, and Phylogenetic Analysis
4.9. Protein Digestion and Mass Spectrometry Identification
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chollet, R.; Vidal, J.; O’Leary, M.H. Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1996, 47, 273–298. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Park, J.; Plaxton, W.C. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): Recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 2011, 436, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Nimmo, H.G. The regulation of PEPC in CAM plants. Trends Plant. Sci. 2000, 5, 75–80. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Sánchez, R.; Cejudo, F.J. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 2003, 132, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yi, K.; Liu, Y.; Xie, L.; Zhou, Z.; Chen, Y.; Hu, Z.; Zheng, T.; Liu, R.; Chen, Y.; et al. Phosphoenolpyruvate carboxylase in Arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiol. 2015, 167, 671–681. [Google Scholar] [CrossRef]
- Sánchez, R.; Flores, A.; Cejudo, F.J. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 2006, 223, 901–909. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef]
- Echevarría, C.; Vidal, J. The unique phosphoenolpyruvate carboxylase kinase. Plant Physiol. Biochem. 2003, 41, 541–547. [Google Scholar] [CrossRef]
- Ruiz-Ballesta, I.; Feria, A.B.; Ni, H.; She, Y.M.; Plaxton, W.C.; Echevarría, C. In Vivo monoubiquitination of anaplerotic phosphoenolpyruvate carboxylase occurs at Lys624 in germinating sorghum seeds. J. Exp. Bot. 2014, 65, 443–451. [Google Scholar] [CrossRef]
- Ruiz-Ballesta, I.; Baena, G.; Gandullo, J.; Wang, L.; She, Y.M.; Plaxton, W.C.; Echevarría, C. New insights into the post-translational modification of multiple phosphoenolpyruvate carboxylase isoenzymes by phosphorylation and monoubiquitination during sorghum seed development and germination. J. Exp. Bot. 2016, 67, 3523–3536. [Google Scholar] [CrossRef] [PubMed]
- Arias-Baldrich, C.; de la Osa, C.; Bosch, N.; Ruiz-Ballesta, I.; Monreal, J.A.; García-Mauriño, S. Enzymatic activity, gene expression and posttranslational modifications of photosynthetic and non-photosynthetic phosphoenolpyruvate carboxylase in ammonium-stressed sorghum plants. J. Plant Physiol. 2017, 214, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Baena, G.; Feria, A.B.; Echevarría, C.; Monreal, J.A.; García-Mauriño, S. Salinity promotes opposite patterns of carbonylation and nitrosylation of C4 phosphoenolpyruvate carboxylase in sorghum leaves. Planta 2017, 246, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, R.G.; Schläpfer, P.; Roschitzki, B.; Hirsch-Hoffmann, M.; Gruissem, W. Diurnal changes in concerted plant protein phosphorylation and acetylation in Arabidopsis organs and seedlings. Plant J. 2019, 99, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Echevarría, C.; Vidal, J.; Jiao, J.A.; Chollet, R. Reversible light activation of phosphoenolpyruvate carboxylase protein-serine kinase in maize leaves. FEBS Lett. 1990, 275, 25–28. [Google Scholar] [CrossRef]
- Boxall, S.F.; Dever, L.V.; Knrová, J.; Gould, P.D.; Hartwell, J. Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate Crassulacean Acid Metabolism species Kalanchoë fedtschenkoi. Plant Cell 2017, 29, 2519–2536. [Google Scholar] [CrossRef]
- Pickart, C.; Eddins, M. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Uhrig, R.G.; She, Y.M.; Leach, C.A.; Plaxton, W.C. Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J. Biol. Chem. 2008, 283, 29650–29657. [Google Scholar] [CrossRef]
- Shane, M.W.; Fedosejevs, E.T.; Plaxton, W.C. Reciprocal control of anaplerotic phosphoenolpyruvate carboxylase by in vivo monoubiquitination and phosphorylation in developing proteoid roots of phosphate deficient harsh hakea. Plant Physiol. 2013, 161, 1634–1644. [Google Scholar] [CrossRef]
- Figueroa, C.; Feil, R.; Ishihara, H.; Watanabe, M.; Kölling, K.; Krause, U.; Höhne, M.; Encke, B.; Plaxton, W.C.; Zeeman, S.C.; et al. Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 2016, 85, 410–423. [Google Scholar] [CrossRef]
- Bassham, D.C. Plant autophagy-more than starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of ATG proteins in autophagosome formation. Annu. Rev. Cell. Dev. Biol. 2011, 27, 107–312. [Google Scholar] [CrossRef] [PubMed]
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell 2011, 2, 784–791. [Google Scholar] [CrossRef]
- Ren, C.; Liu, J.; Gong, Q. Functions of autophagy in plant carbon and nitrogen metabolism. Front. Plant Sci. 2014, 5, 301. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Chen, Q.; Havé, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 2017, 39, 8–17. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant. Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataillé, M.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef]
- Chen, Q.; Soulay, F.; Saudemont, B.; Elmayan, T.; Marmagne, A.; Masclaux-Daubresse, C. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 2019, 60, 343–352. [Google Scholar] [CrossRef]
- Yu, J.; Zhen, X.; Li, X.; Li, N.; Xu, F. Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 2019, 10, 584. [Google Scholar] [CrossRef]
- Wang, P.; Mugume, Y.; Bassham, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell. Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Honig, A.; Galili, G. Variations on a theme, plant autophagy in comparison to yeast and mammals. Protoplasma 2012, 249, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.F.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M.Y. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef]
- Kellner, R.; De la Concepcion, J.C.; Maqbool, A.; Kamoun, S.; Dagdas, Y.F. ATG8 expansion: A driver of selective autophagy diversification? Trends Plant Sci. 2017, 22, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.P.; Promponas, V.J. iLIR. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef]
- Xie, Q.; Tzfadia, O.; Levy, M.; Weithorn, E.; Peled-Zehavi, H.; Van Parys, T.; Vande Peer, Y.; Galili, G. HfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 2016, 12, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 2019, 177, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzynska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158. [Google Scholar] [CrossRef]
- Sarkar, S.; Davies, J.E.; Huang, Z.; Tunnacliffe, A.; Rubinsztein, D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J. Biol. Chem. 2007, 282, 5641–5652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Song, L.; Tang, Y.; Shen, Y.; Jia, L. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy 2014, 10, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Njaci, I.; Moghaddam, L.; Long, H.; Dickman, M.B.; Zhang, X.; Mundree, S. Trehalose accumulation triggers autophagy during plant desiccation. PLoS Genet. 2015, 11, e1005705. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, A.; Peter, M. Substrate recognition in selective autophagy and the ubiquitin–proteasome system. Biochim. Biophys. Acta 2014, 1843, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Seglen, P.O.; Gordon, P.B. 3-methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 1982, 79, 1889–1892. [Google Scholar] [CrossRef]
- Matsuoka, K.; Higuchi, T.; Maeshima, M.; Nakamura, K. A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 1997, 9, 533–546. [Google Scholar] [CrossRef]
- Takatsuka, C.; Inoue, Y.; Higuchi, T.; Hillmer, S.; Robinson, D.G.; Moriyasu, Y. Autophagy in tobacco BY-2 cells cultured under sucrose starvation conditions: Isolation of the autolysosome and its characterization. Plant Cell Physiol. 2011, 52, 2074–2087. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef]
- Osuna, L.; Pierre, J.N.; González, M.C.; Alvarez, R.; Cejudo, F.J.; EchevarrÞa, C.; Vidal, J. Evidence for a slow-turnover form of the Ca2+-independent phosphoenolpyruvate carboxylase kinase in the aleurone-endosperm tissue of germinating barley seeds. Plant Physiol. 1999, 119, 511–520. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feria, A.B.; Bosch, N.; Sánchez, A.; Nieto-Ingelmo, A.I.; de la Osa, C.; Echevarría, C.; García-Mauriño, S.; Monreal, J.A. Phosphoenolpyruvate carboxylase (PEPC) and PEPC-kinase (PEPC-k) isoenzymes in Arabidopsis thaliana: Role in control and abiotic stress conditions. Planta 2016, 244, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.S. Photosynthetic efficiency and carbon concentration in terrestrial plants: The C4 and CAM solutions. J. Exp. Bot. 2014, 65, 3323–3325. [Google Scholar] [CrossRef] [PubMed]
- Begum, H.; Osaki, M.; Watanabe, T.; Shinano, T. Mechanisms of aluminum tolerance in phosphoenolpyruvate carboxylase transgenic rice. J. Plant Nutr. 2009, 32, 84–96. [Google Scholar] [CrossRef]
- Willick, I.R.; Plaxton, W.C.; Lolle, S.J.; Macfie, S.M. Transcriptional and post-translational upregulation of phosphoenolpyruvate carboxylase in Arabidopsis thaliana (L. Heynh) under cadmium stress. Environ. Exp. Bot. 2019, 164, 29–39. [Google Scholar] [CrossRef]
- Monreal, J.A.; Arias-Baldrich, C.; Pérez-Montaño, F.; Gandullo, J.; Echevarría, C.; García-Mauriño, S. Factors involved in the rise of phosphoenolpyruvate carboxylase-kinase activity caused by salinity in sorghum leaves. Planta 2013, 237, 1401–1413. [Google Scholar] [CrossRef]
- Schulz, M.; Klockenbring, T.; Hunte, C.; Schnabl, H. Involvement of ubiquitin in phosphoenolpyruvate carboxylase degradation. Bot. Acta 1993, 106, 143–145. [Google Scholar] [CrossRef]
- Agetsuma, M.; Furumoto, T.; Yanagisawa, S.; Izui, K. The ubiquitin-proteasome pathway is involved in rapid degradation of phosphoenolpyruvate carboxylase kinase for C4 photosynthesis. Plant Cell Physiol. 2005, 46, 389–398. [Google Scholar] [CrossRef]
- Wang, Y.; Nishimura, M.T.; Zhao, T.; Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011, 68, 74–87. [Google Scholar] [CrossRef]
- McLoughlin, F.; Augustine, R.C.; Marshall, R.S.; Li, F.; Kirkpatrick, L.D.; Otegui, M.S.; Vierstra, R.D. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat. Plants 2018, 4, 1056–1070. [Google Scholar] [CrossRef]
- Gandullo, J.; Monreal, J.A.; Álvarez, R.; Díaz, I.; García-Mauriño, S.; Echevarría, C. Anionic phospholipids induce conformational changes in phosphoenolpyruvate carboxylase to increase sensitivity to cathepsin proteases. Front. Plant Sci. 2019, 10, 582. [Google Scholar] [CrossRef] [PubMed]
- Echevarría, C.; Pacquit, V.; Bakrim, N.; Osuna, L.; Delgado, B.; Arrio-Dupont, M.; Vidal, J. The effect of pH on the covalent and metabolic controls of C4 phosphoenolpyruvate carboxylase from Sorghum leaf. Arch. Biochem. Biophys. 1994, 315, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Pacquit, V.; Giglioli, N.; Crétin, C.; Pierre, J.N.; Vidal, J.; Echevarría, C. Regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase from Sorghum. An immunological study using specific anti-phosphorylation site antibodies. Photosynth. Res. 1995, 43, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: A multiple sequence alignment method with reduced time and space complexity. Biomed. Cent. Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular evolutionary genetics anaysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Miller, M.; Holder, M.; Vos, R.P.; Midford, P.; Liebowitz, T.; Chan, L.; Hoover, P.; Warnow, T. The CIPRES Portals. 2009. Available online: http://www.phylo.org/portal2/ (accessed on 15 October 2019).
- Stamatakis, A. RA×ML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-Rex: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef]
- Maguilla, E.; Escudero, M.; Waterway, M.J.; Hipp, A.L.; Luceño, M. Phylogeny, systematics, and trait evolution of Carex section Glareosae. Am. J. Bot. 2015, 102, 1128–1144. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Dayhoff, M.; Schwartz, R.; Orcutt, B. A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure; Dayhoff, M., Ed.; National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5, pp. 345–352. [Google Scholar]
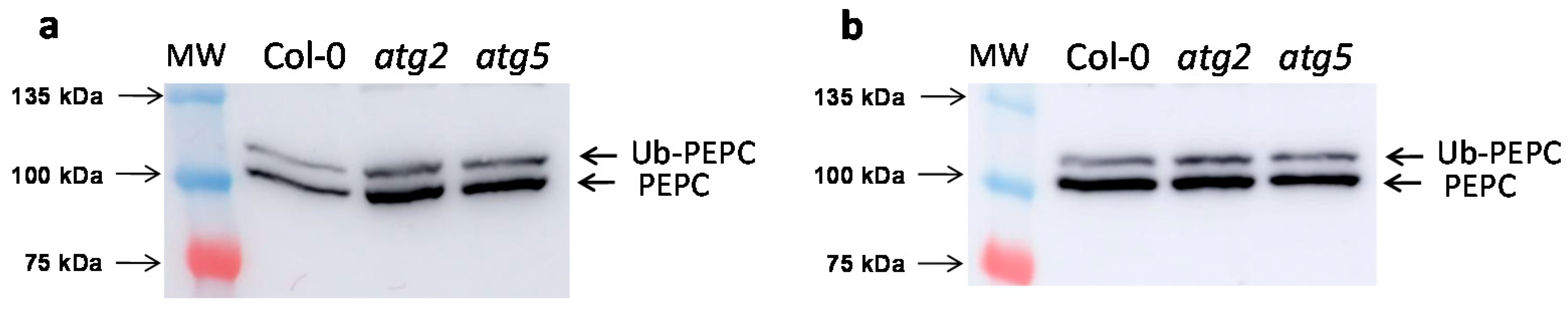


| Ratio of p110/p107 | ||
|---|---|---|
| Leaves | Roots | |
| Col-0 | 0.34 ± 0.04 | 0.11 ± 0.02 |
| atg2 | 0.53 ± 0.04 | 0.18 ± 0.02 |
| atg5 | 0.48 ± 0.03 | 0.16 ± 0.02 |
| nbr1 | 0.58 ± 0.16 | 0.38 ± 0.19 |
| atg18a | 0.50 ± 0.07 | 0.19 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baena, G.; Feria, A.B.; Hernández-Huertas, L.; Gandullo, J.; Echevarría, C.; Monreal, J.A.; García-Mauriño, S. Genetic and Pharmacological Inhibition of Autophagy Increases the Monoubiquitination of Non-Photosynthetic Phosphoenolpyruvate Carboxylase. Plants 2021, 10, 12. https://doi.org/10.3390/plants10010012
Baena G, Feria AB, Hernández-Huertas L, Gandullo J, Echevarría C, Monreal JA, García-Mauriño S. Genetic and Pharmacological Inhibition of Autophagy Increases the Monoubiquitination of Non-Photosynthetic Phosphoenolpyruvate Carboxylase. Plants. 2021; 10(1):12. https://doi.org/10.3390/plants10010012
Chicago/Turabian StyleBaena, Guillermo, Ana B. Feria, Luis Hernández-Huertas, Jacinto Gandullo, Cristina Echevarría, José A. Monreal, and Sofía García-Mauriño. 2021. "Genetic and Pharmacological Inhibition of Autophagy Increases the Monoubiquitination of Non-Photosynthetic Phosphoenolpyruvate Carboxylase" Plants 10, no. 1: 12. https://doi.org/10.3390/plants10010012
APA StyleBaena, G., Feria, A. B., Hernández-Huertas, L., Gandullo, J., Echevarría, C., Monreal, J. A., & García-Mauriño, S. (2021). Genetic and Pharmacological Inhibition of Autophagy Increases the Monoubiquitination of Non-Photosynthetic Phosphoenolpyruvate Carboxylase. Plants, 10(1), 12. https://doi.org/10.3390/plants10010012








