Molecular Characterization of HOXA2 and HOXA3 Binding Properties
Abstract
:1. Introduction
2. Material and Methods
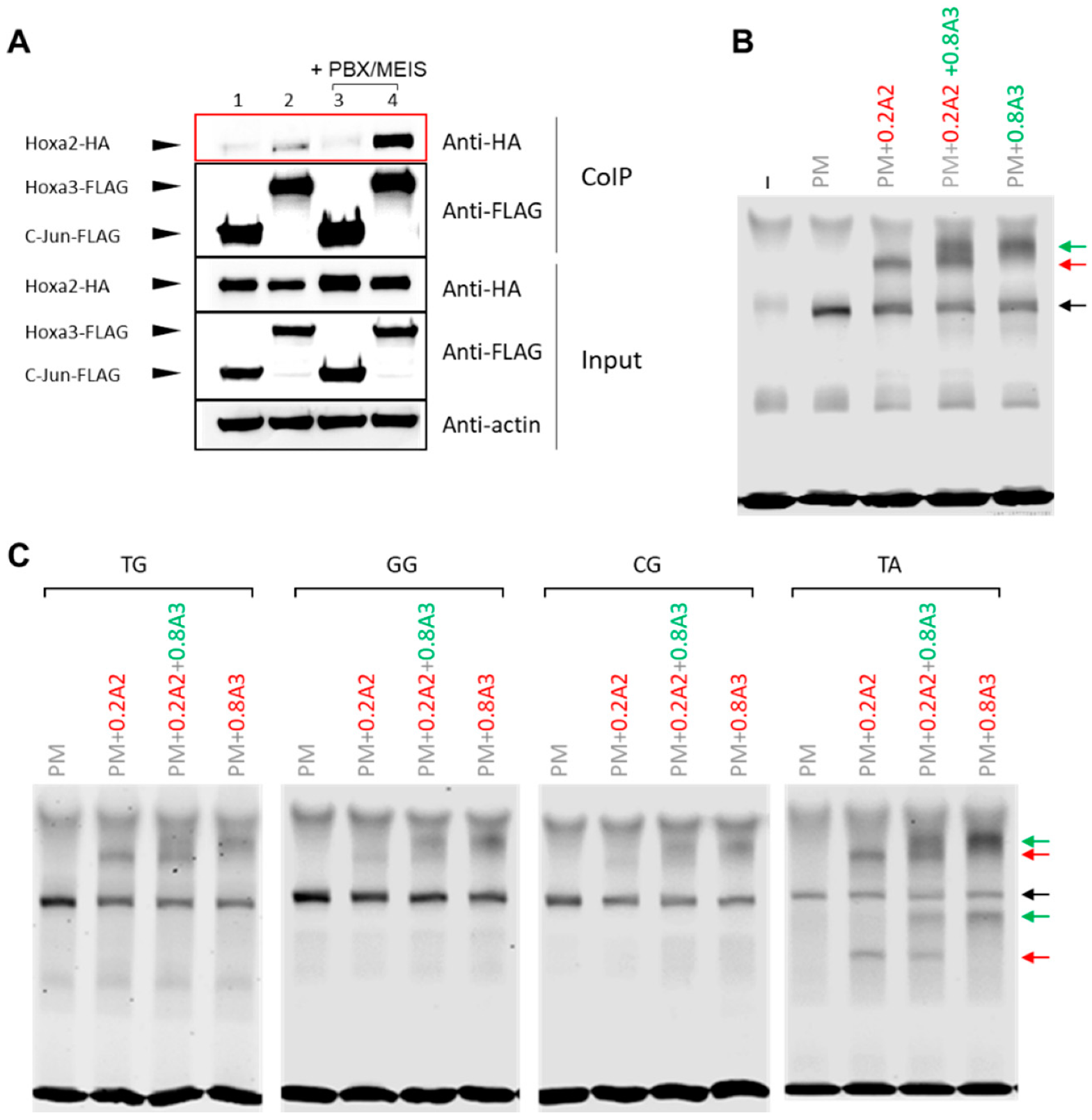
2.1. Co-Immunoprecipitation
2.2. Electrophoretic Mobility Shift Assays (EMSA)
2.3. Sequence Analysis
3. Results and Discussion
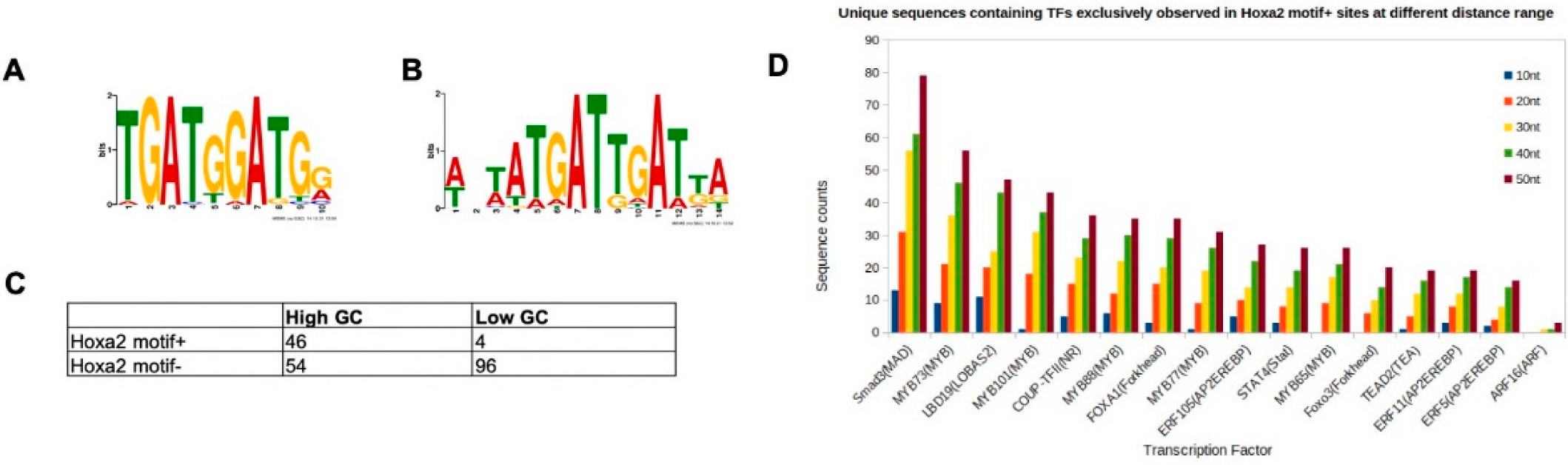
3.1. Changes to the Core Variable Sequence Affect HOXA2 Binding In Vitro
3.2. Characterisation of HOXA2-Bound TGATGGAT Motif
3.3. HOXA2 and HOXA3 Are Found in the Same Complex with TALE
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Reiter, F.; Wienerroither, S.; Stark, A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017, 43, 73–81. [Google Scholar] [CrossRef]
- Burglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobola, N.; Merabet, S. Homeodomain proteins in action: Similar DNA binding preferences, highly variable connectivity. Curr. Opin. Genet. Dev. 2017, 43, 1–8. [Google Scholar] [CrossRef]
- Noyes, M.B.; Christensen, R.G.; Wakabayashi, A.; Stormo, G.D.; Brodsky, M.H.; Wolfe, S.A. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 2008, 133, 1277–1289. [Google Scholar] [CrossRef] [Green Version]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Pearson, J.C.; Lemons, D.; McGinnis, W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005, 6, 893–904. [Google Scholar] [CrossRef]
- Rezsohazy, R.; Saurin, A.J.; Maurel-Zaffran, C.; Graba, Y. Cellular and molecular insights into Hox protein action. Development 2015, 142, 1212–1227. [Google Scholar] [CrossRef] [Green Version]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef] [Green Version]
- Gendron-Maguire, M.; Mallo, M.; Zhang, M.; Gridley, T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell 1993, 75, 1317–1331. [Google Scholar] [CrossRef]
- Rijli, F.M.; Mark, M.; Lakkaraju, S.; Dierich, A.; Dollé, P.; Chambon, P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 1993, 75, 1333–1349. [Google Scholar] [CrossRef]
- Amin, S.; Donaldson, I.J.; Zannino, D.A.; Hensman, J.; Rattray, M.; Losa, M.; Spitz, F.; Ladam, F.; Sagerstrom, C.; Bobola, N. Hoxa2 Selectively Enhances Meis Binding to Change a Branchial Arch Ground State. Dev. Cell 2015, 32, 265–277. [Google Scholar] [CrossRef] [Green Version]
- Kirilenko, P.; He, G.; Mankoo Baljinder, S.; Mallo, M.; Jones, R.; Bobola, N. Transient Activation of Meox1 Is an Early Component of the Gene Regulatory Network Downstream of Hoxa2. Mol. Cell. Biol. 2011, 31, 1301–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosin, J.M.; Li, W.; Cox, L.A.-O.; Rolfe, S.M.; Latorre, V.; Akiyama, J.A.-O.; Visel, A.A.-O.; Kuramoto, T.; Bobola, N.A.-O.; Turner, E.A.-O.; et al. A distal 594 bp ECR specifies Hmx1 expression in pinna and lateral facial morphogenesis and is regulated by the Hox-Pbx-Meis complex. Development 2016, 143, 2582–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutejova, E.; Engist, B.; Self, M.; Oliver, G.; Kirilenko, P.; Bobola, N. Six2 functions redundantly immediately downstream of Hoxa2. Development 2008, 135, 1463–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manley, N.R.; Capecchi, M.R. The role of Hoxa-3 in mouse thymus and thyroid development. Development 1995, 121, 1989–2003. [Google Scholar] [CrossRef]
- Bridoux, L.; Zarrineh, P.; Mallen, J.; Phuycharoen, M.; Latorre, V.; Ladam, F.; Losa, M.; Baker, S.M.; Sagerstrom, C.; Mace, K.A.; et al. HOX paralogs selectively convert binding of ubiquitous transcription factors into tissue-specific patterns of enhancer activation. PLoS Genet. 2020, 16, e1009162. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merabet, S.; Mann, R.S. To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins. Trends Genet. 2016, 32, 334–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selleri, L.; Zappavigna, V.; Ferretti, E. ‘Building a perfect body’: Control of vertebrate organogenesis by PBX-dependent regulatory networks. Genes Dev. 2019, 33, 258–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donaldson, I.J.; Amin, S.; Hensman, J.J.; Kutejova, E.; Rattray, M.; Lawrence, N.; Hayes, A.; Ward, C.M.; Bobola, N. Genome-wide occupancy links Hoxa2 to Wnt-β-catenin signaling in mouse embryonic development. Nucleic Acids Res. 2012, 40, 3990–4001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crocker, J.; Abe, N.; Rinaldi, L.; McGregor, A.P.; Frankel, N.; Wang, S.; Alsawadi, A.; Valenti, P.; Plaza, S.; Payre, F.; et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 2015, 160, 191–203. [Google Scholar] [CrossRef] [Green Version]
- Salomone, J.; Qin, S.; Fufa, T.D.; Cain, B.; Farrow, E.; Guan, B.; Hufnagel, R.B.; Nakafuku, M.; Lim, H.-W.; Campbell, K. Conserved Gsx2/Ind homeodomain monomer versus homodimer DNA binding defines regulatory outcomes in flies and mice. Genes Dev. 2021, 35, 157–174. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mallen, J.; Kalsan, M.; Zarrineh, P.; Bridoux, L.; Ahmad, S.; Bobola, N. Molecular Characterization of HOXA2 and HOXA3 Binding Properties. J. Dev. Biol. 2021, 9, 55. https://doi.org/10.3390/jdb9040055
Mallen J, Kalsan M, Zarrineh P, Bridoux L, Ahmad S, Bobola N. Molecular Characterization of HOXA2 and HOXA3 Binding Properties. Journal of Developmental Biology. 2021; 9(4):55. https://doi.org/10.3390/jdb9040055
Chicago/Turabian StyleMallen, Joshua, Manisha Kalsan, Peyman Zarrineh, Laure Bridoux, Shandar Ahmad, and Nicoletta Bobola. 2021. "Molecular Characterization of HOXA2 and HOXA3 Binding Properties" Journal of Developmental Biology 9, no. 4: 55. https://doi.org/10.3390/jdb9040055
APA StyleMallen, J., Kalsan, M., Zarrineh, P., Bridoux, L., Ahmad, S., & Bobola, N. (2021). Molecular Characterization of HOXA2 and HOXA3 Binding Properties. Journal of Developmental Biology, 9(4), 55. https://doi.org/10.3390/jdb9040055





