Developmental Aspects of SARS-CoV-2, Potential Role of Exosomes and Their Impact on the Human Transcriptome
Abstract
:1. Introduction
2. Emergence of SARS-CoV-2 and Relation to Other CoVs
2.1. General Description of Genes and Proteins
2.2. Replication Cycle from Entry to Egress
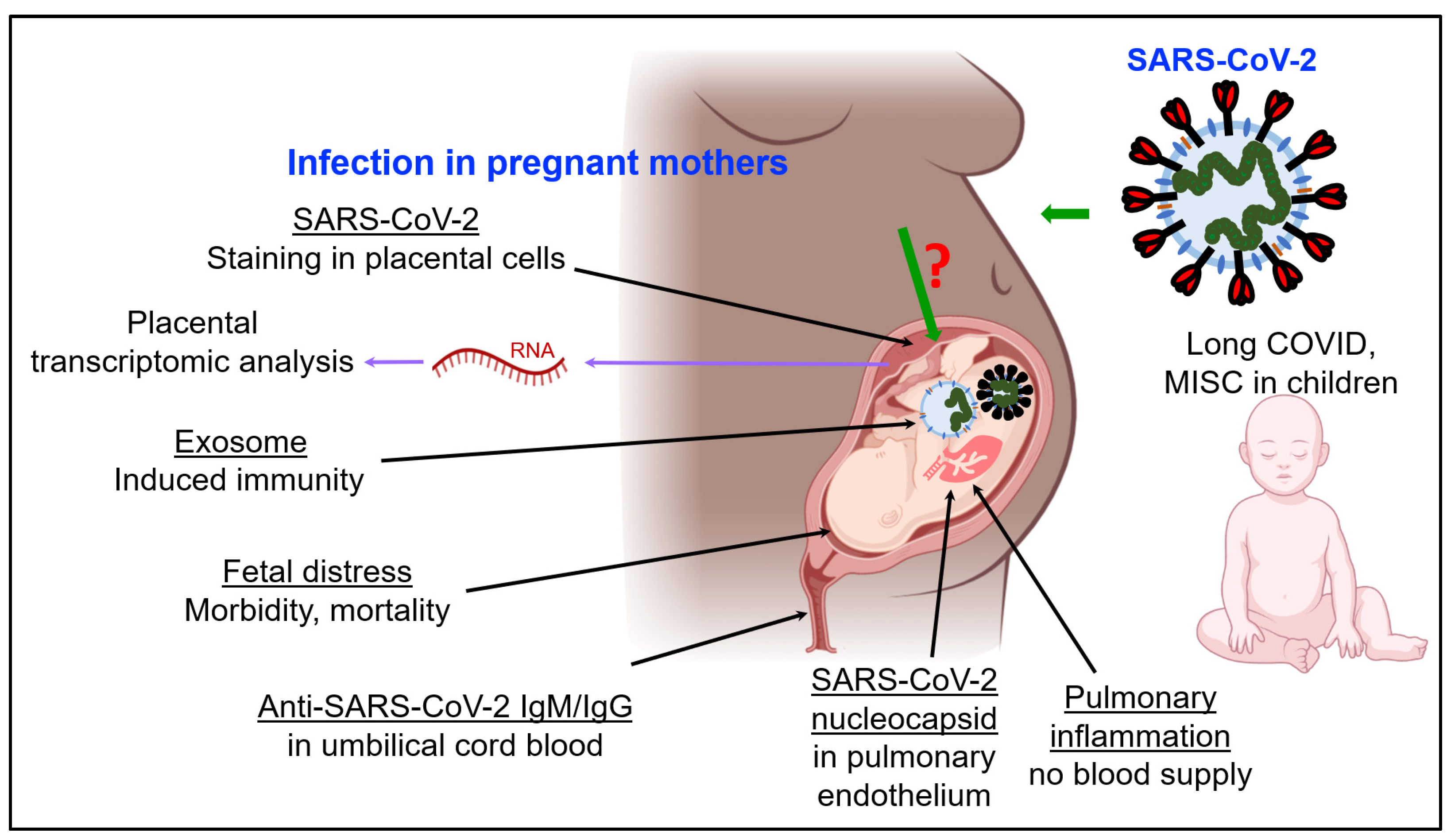
3. SARS-CoV-2 Infection and Immunity in Pregnancy and Fetal Development
3.1. Impact of COVID-19 on Fetal Development and Children
3.2. SARS-CoV-2 and Fetal Development: The Role of Exosomes
3.3. Development and Functions of Exosomes: Biogenesis and Biology
3.4. Genomic, Transcriptomic, Proteomic, and Lipidomic Landscape of Exosomes
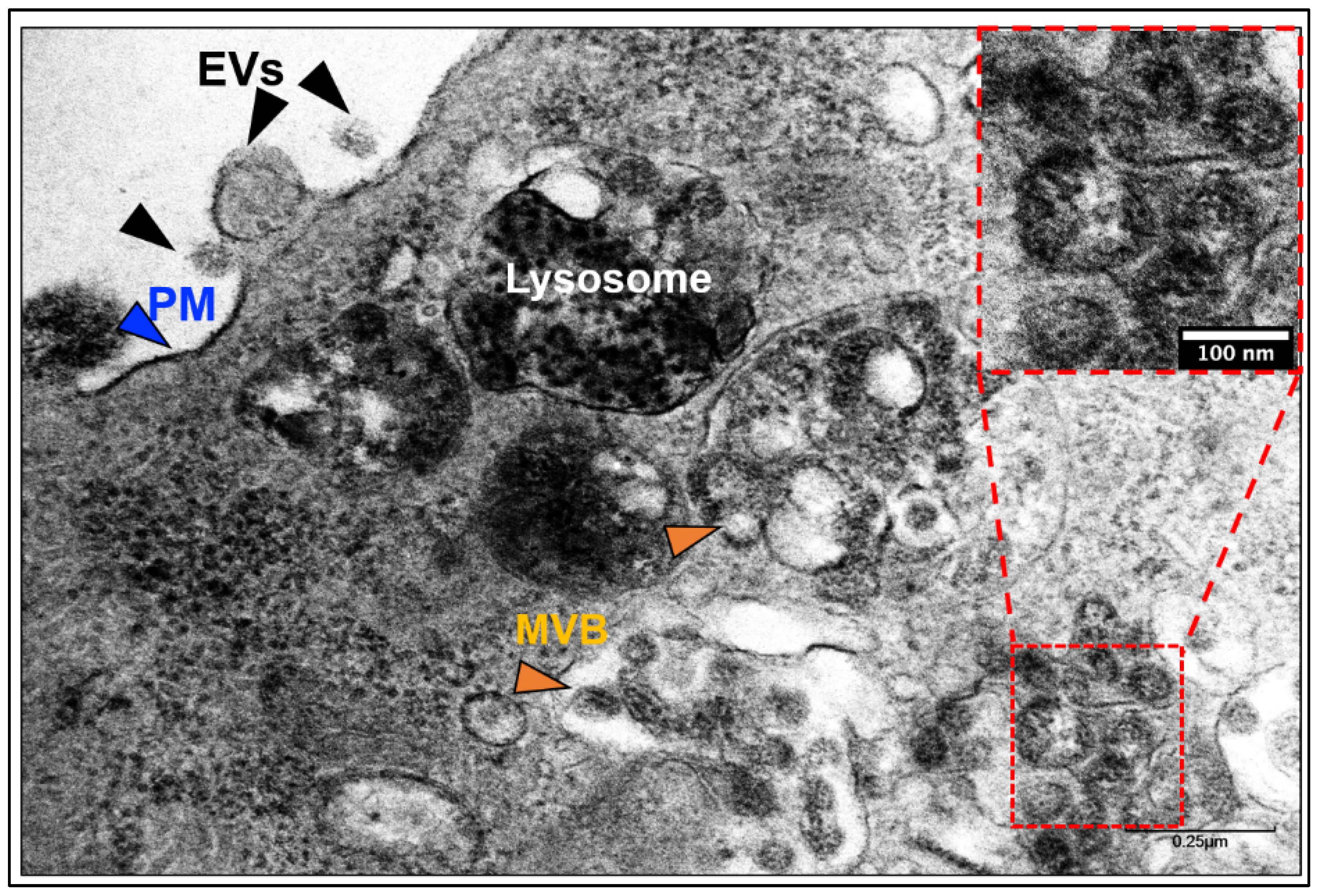
4. SARS-CoV-2 Infection and Exosomal Pathway
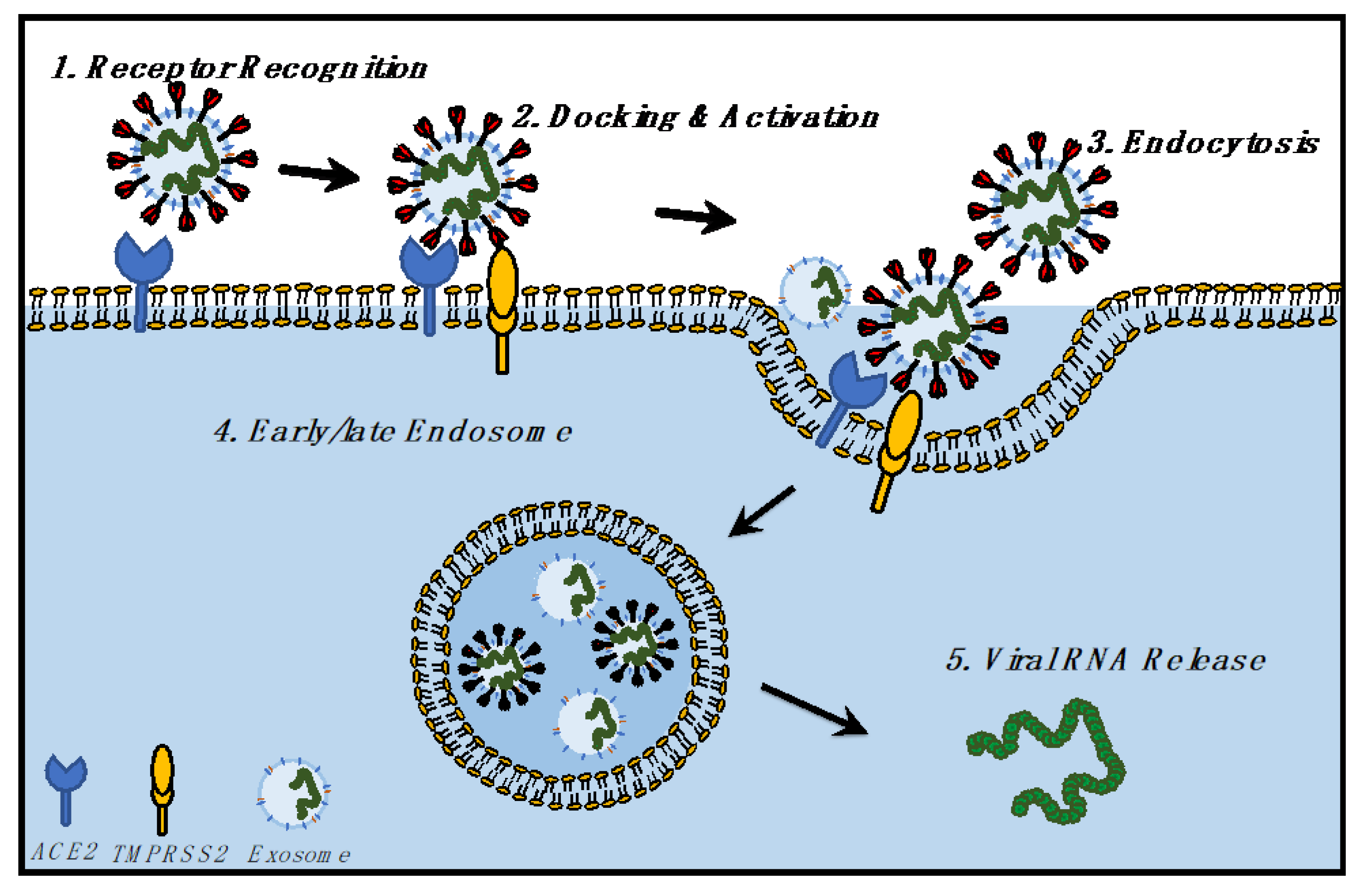
4.1. Does SARS-CoV-2 Hijack the Exosomal Pathway for Cellular Entry and Exit?
4.2. Entry of SARS-CoV-2 and Exosomes in Cells
4.3. Exit of SARS-CoV-2 and Exosomes from Cells
5. RNA-Based Analysis of COVID-19 Patients
5.1. Transcriptomic Analysis of Placental Cells in the Context of COVID-19
5.2. Transcriptome of Children Affected with COVID-19
5.3. Host Transcriptional Profiles and COVID-19 Pathogenesis
5.4. Host Transcriptomic Profiles and T Cell Behavior
5.5. Host Lung and Colon Transcriptomes Show Elevated Neutrophil Extracellular Traps and TGF-β Response, Respectively
5.6. Insights from Single-Cell Transcriptome of SARS-CoV-2 Infection in African Green Monkeys
5.7. Lessons from the SARS-CoV-2 Transcriptome
6. RNA-Based Approach to Combat COVID-19
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 26 November 2021).
- Mihalopoulos, M.; Dogra, N.; Mohamed, N.; Badani, K.; Kyprianou, N. COVID-19 and Kidney Disease: Molecular Determinants and Clinical Implications in Renal Cancer. Eur. Urol. Focus 2020, 6, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Badani, K.; Okhawere, K.; Chen, T.; Korn, T.; Razdan, S.; Meilika, K.; Wilson, M.; Tomy, T.; Ucpinar, B.; Kyprianou, N.; et al. SARS-CoV-2 RNA Detected in Abdominal Insufflation Samples During Laparoscopic Surgery. Eur. Urol. 2021. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Xie, Y.; Bowe, B. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Oh, E.S.; Vannorsdall, T.D.; Parker, A.M. Post-acute Sequelae of SARS-CoV-2 Infection and Subjective Memory Problems. JAMA Netw. Open 2021, 4, e2119335. [Google Scholar] [CrossRef]
- Alwan, N.A. The road to addressing Long Covid. Science 2021, 373, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Leon, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 long-term effects of COVID-19: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Salamanna, F.; Veronesi, F.; Martini, L.; Landini, M.P.; Fini, M. Post-COVID-19 Syndrome: The Persistent Symptoms at the Post-viral Stage of the Disease. A Systematic Review of the Current Data. Front. Med. 2021, 8, 653516. [Google Scholar] [CrossRef]
- Dolgin, E. How COVID unlocked the power of RNA vaccines. Nature 2021, 589, 189–191. [Google Scholar] [CrossRef]
- Singh, J.; Pandit, P.; McArthur, A.G.; Banerjee, A.; Mossman, K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021, 18, 166. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Tonkin-Hill, G.; Martincorena, I.; Amato, R.; Lawson, A.R.; Gerstrung, M.; Johnston, I.; Jackson, D.K.; Park, N.; Lensing, S.V.; Quail, M.A.; et al. Patterns of within-host genetic diversity in SARS-CoV-2. Elife 2021, 10, e66857. [Google Scholar] [CrossRef]
- Pegu, A.; O’Connell, S.E.; Schmidt, S.D.; O’Dell, S.; Talana, C.A.; Lai, L.; Albert, J.; Anderson, E.; Bennett, H.; Corbett, K.S. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 2021, 373, 1372–1377. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; Van Der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vale, F.F.; Vítor, J.M.; Marques, A.T.; Azevedo-Pereira, J.M.; Anes, E.; Gonçalves, J. Origin, phylogeny, variability and epitope conservation of SARS-CoV-2 worldwide. Virus Res. 2021, 304, 198526. [Google Scholar] [CrossRef]
- Vale, F.F.; Vítor, J.M.; Marques, A.T.; Azevedo-Pereira, J.M.; Anes, E.; Gonçalves, J. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Astuti, I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndr. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; De Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- Snijder, E.J.; Limpens, R.W.; de Wilde, A.H.; de Jong, A.W.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.F.G.A.; Koster, A.J.; Bárcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R. Beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e14. [Google Scholar] [CrossRef]
- Delahoy, M.J.; Ujamaa, D.; Whitaker, M.; O’Halloran, A.; Anglin, O.; Chai, S.J.; Kirley, P.D.; Alden, N.; Kawasaki, B.; Meek, J.; et al. Characteristics and Maternal and Birth Outcomes of Hospitalized Pregnant Women with Laboratory-Confirmed COVID-19—COVID-NET, 13 States, March 1–August 22, 2020. Morb. Mortal. Wkly. Rep. 2021, 70, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.B.; Gilani, Z.; Godfred-Cato, S.; Belay, E.D.; Feldstein, L.R.; Patel, M.M.; Randolph, A.G.; Newhams, M.; Thomas, D.; Magleby, R. Incidence of Multisystem Inflammatory Syndrome in Children Among US Persons Infected With SARS-CoV-2. JAMA Netw. Open 2021, 4, e2116420. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Kim, L.; Whitaker, M. Hospitalization Rates and Characteristics of Children Aged <18 Years Hospitalized with Laboratory-Confirmed COVID-19—COVID-NET, 14 States, March 1-July 25, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1081–1088. [Google Scholar]
- Havers, F.P.; Whitaker, M.; Self, J.L.; Chai, S.J.; Kirley, P.D.; Alden, N.B. Hospitalization of Adolescents Aged 12–17 Years with Laboratory-Confirmed COVID-19–COVID-NET, 14 States, March 1, 2020-April 24, 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Simpson, F.C.; Chew-Graham, C.; Lokugamage, A. Long COVID in children: The perspectives of parents and children need to be heard. Br. J. Gen. Pract. 2021, 71, 216. [Google Scholar] [CrossRef] [PubMed]
- Swann, O.V.; Holden, K.A.; Turtle, L.; Pollock, L.; Fairfield, C.J.; Drake, T.M.; Seth, S.; Egan, C.; Hardwick, H.E.; Halpin, S.; et al. Clinical characteristics of children and young people admitted to hospital with covid-19 in United Kingdom: Prospective multicentre observational cohort study. BMJ 2020, 370, m3249. [Google Scholar] [CrossRef]
- Heald-Sargent, T.; Muller, W.J.; Zheng, X.; Rippe, J.; Patel, A.B.; Kociolek, L.K. Age-Related Differences in Nasopharyngeal Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Levels in Patients with Mild to Moderate Coronavirus Disease 2019 (COVID-19). JAMA Pediatr. 2020, 174, 902–903. [Google Scholar] [CrossRef] [PubMed]
- L’Huillier, A.G.; Torriani, G.; Pigny, F.; Kaiser, L.; Eckerle, I. Culture-Competent SARS-CoV-2 in Nasopharynx of Symptomatic Neonates, Children, and Adolescents. Emerg. Infect. Dis. 2020, 26, 2494–2497. [Google Scholar] [CrossRef] [PubMed]
- Belay, E.D.; Abrams, J.; Oster, M.E.; Giovanni, J.; Pierce, T.; Meng, L.; Prezzato, E.; Balachandran, N.; Openshaw, J.J.; Rosen, H.E.; et al. Trends in Geographic and Temporal Distribution of US Children with Multisystem Inflammatory Syndrome During the COVID-19 Pandemic. JAMA Pediatr. 2021, 175, 837–845. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 698169. [Google Scholar] [CrossRef]
- Saadaoui, M.; Kumar, M.; Al Khodor, S. COVID-19 Infection during Pregnancy: Risk of Vertical Transmission, Fetal, and Neonatal Outcomes. J. Pers. Med. 2021, 11, 483. [Google Scholar] [CrossRef]
- Facchetti, F.; Bugatti, M.; Drera, E.; Tripodo, C.; Sartori, E.; Cancila, V.; Lavazza, A. SARS-CoV2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of Placenta. EBioMedicine 2020, 59, 102951. [Google Scholar] [CrossRef]
- Mullins, E.; Evans, D.; Viner, R.M.; O’Brien, P.; Morris, E. Coronavirus in pregnancy and delivery: Rapid review. Ultrasound Obstet. Gynecol. 2020, 55, 586–592. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Qian, H.; Cao, S.; Dong, B.; Yan, X.; Luo, S.; Zhou, M.; Zhou, S.; Ning, B.; Zhao, L. Is there possibility of vertical transmission of COVID-19: A systematic review. Transl. Pediatr. 2021, 10, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Tolu, L.B.; Ezeh, A.; Feyissa, G.T. Vertical transmission of Severe Acute Respiratory Syndrome Coronavirus 2: A scoping review. PLoS ONE 2021, 16, e0250196. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, C.; Biasin, M.; Cetin, I.; Vergani, P.; Mileto, D.; Spinillo, A.; Gismondo, M.R.; Perotti, F.; Callegari, C.; Mancon, S.; et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat. Commun. 2020, 11, 5128. [Google Scholar] [CrossRef] [PubMed]
- Sinaci, S.; Ocal, D.F.; Seven, B.; Anuk, A.T.; Besimoglu, B.; Keven, M.C.; Sahin, D. Vertical transmission of SARS-CoV-2: A prospective cross-sectional study from a tertiary center. J. Med. Virol. 2021, 93, 5864–5872. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Zarchi, M.; Neamatzadeh, H.; Dastgheib, S.A.; Abbasi, H.; Mirjalili, S.R.; Behforouz, A.; Ferdosian, F.; Bahrami, R. Vertical Transmission of Coronavirus Disease 19 (COVID-19) from Infected Pregnant Mothers to Neonates: A Review. Fetal Pediatr. Pathol. 2020, 39, 246–250. [Google Scholar] [CrossRef] [Green Version]
- Stonoga, E.T.; de Almeida Lanzoni, L.; Rebutini, P.Z.; de Oliveira, A.L.P.; Chiste, J.A.; Fugaça, C.A. Intrauterine Transmission of SARS-CoV-2. Emerg. Infect. Dis. 2021, 27, 638–641. [Google Scholar] [CrossRef]
- Zaigham, M.; Holmberg, A.; Karlberg, M.L.; Lindsjö, O.K.; Jokubkiene, L.; Sandblom, J. Intrauterine vertical SARS-CoV-2 infection: A case confirming transplacental transmission followed by divergence of the viral genome. BJOG 2021, 128, 1388–1394. [Google Scholar] [CrossRef]
- Schwartz, D.A.; Thomas, K.M. Characterizing COVID-19 maternal-fetal transmission and placental infection using comprehensive molecular pathology. EBioMedicine 2020, 60, 102983. [Google Scholar] [CrossRef]
- AbdelMassih, A.; Fouda, R.; Essam, R.; Negm, A.; Khalil, D.; Habib, D.; Afdal, G.; Ismail, H.-A.; Aly, H.; Genedy, I.; et al. COVID-19 during pregnancy should we really worry from vertical transmission or rather from fetal hypoxia and placental insufficiency? A systematic review. Egypt. Pediatr. Assoc. Gaz. 2021, 69, 1–13. [Google Scholar] [CrossRef]
- Mourad, M.; Jacob, T.; Sadovsky, E.; Bejerano, S.; Salazar-De Simone, G.; Bagalkot, T.R.; Zucker, J.; Yin, M.T.; Chang, J.Y.; Liu, L.; et al. Placental response to maternal SARS-CoV-2 infection. Sci. Rep. 2021, 11, 14390. [Google Scholar] [CrossRef]
- Joseph, N.T.; Dude, C.M.; Verkerke, H.P.; Les’Shon, S.I.; Dunlop, A.L.; Patel, R.M.; Easley, K.A.; Smith, A.K.; Stowell, S.R.; Jamieson, D.J.; et al. Maternal Antibody Response, Neutralizing Potency, and Placental Antibody Transfer After Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. Obstet. Gynecol. 2021, 138, 189–197. [Google Scholar] [CrossRef]
- Marinho, P.S.; Da Cunha, A.J.L.A.; Chimelli, L.; Avvad-Portari, E.; Andreiuolo, F.D.M.; de Oliveira-Szejnfeld, P.S.; Mendes, M.A.; Gomes, I.C.; Souza, L.R.Q.; Guimarães, M.Z.; et al. Case Report: SARS-CoV-2 Mother-to-Child Transmission and Fetal Death Associated With Severe Placental Thromboembolism. Front. Med. 2021, 8, 677001. [Google Scholar] [CrossRef] [PubMed]
- Arthurs, A.L.; Jankovic-Karasoulos, T.; Roberts, C.T. COVID-19 in pregnancy: What we know from the first year of the pandemic. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2021, 1867, 166248. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.P.; Khong, T.Y.; Tan, G.C. The Effects of COVID-19 on Placenta and Pregnancy: What Do We Know So Far? Diagnostics 2021, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Brion, L.P.; Chan, C.S.; Adhikari, E.H. Comment on evidence for and against vertical transmission of severe acute respiratory syndrome coronavirus 2 (coronavirus disease 2019). Am. J. Obstet. Gynecol. 2020, 223, 775. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.A.; Bugatti, M.; Santoro, A.; Facchetti, F. Molecular Pathology Demonstration of SARS-CoV-2 in Cytotrophoblast from Placental Tissue with Chronic Histiocytic Intervillositis, Trophoblast Necrosis and COVID-19. J. Dev. Biol. 2021, 9, 33. [Google Scholar] [CrossRef]
- Thomas, J.; Sun, Y.; Debelenko, L. Infrequent Placental and Fetal Involvement in SARS-CoV-2 Infection: Pathology Data from a Large Medical Center. J. Dev. Biol. 2021, 9, 45. [Google Scholar] [CrossRef]
- Bwire, G.M.; Njiro, B.J.; Mwakawanga, D.L.; Sabas, D.; Sunguya, B.F. Possible vertical transmission and antibodies against SARS-CoV-2 among infants born to mothers with COVID-19: A living systematic review. J. Med. Virol. 2021, 93, 1361–1369. [Google Scholar] [CrossRef]
- Tallarek, A.C.; Urbschat, C.; Fonseca Brito, L.; Stanelle-Bertram, S.; Krasemann, S.; Frascaroli, G.; Thiele, K.; Wieczorek, A.; Felber, N.; Lütgehetmann, M.; et al. Inefficient Placental Virus Replication and Absence of Neonatal Cell-Specific Immunity Upon Sars-CoV-2 Infection During Pregnancy. Front. Immunol. 2021, 12, 698578. [Google Scholar] [CrossRef] [PubMed]
- Tezer, H.; Demirdağ, T.B. Novel coronavirus disease (COVID-19) in children. Turk. J. Med Sci. 2020, 50, 592–603. [Google Scholar] [CrossRef]
- Gaglani, S.; Gonzalez-Kozlova, E.; Lundon, D.J.; Tewari, A.K.; Dogra, N.; Kyprianou, N. Exosomes as A Next-Generation Diagnostic and Therapeutic Tool in Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 10131. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Gonzalez-Kozlova, E.; Soleymani, T.; La Salvia, S.; Kyprianou, N.; Sahoo, S.; Tewari, A.; Cordon-Cardo, C.; Stolovitzky, G.U.S.T.A.V.O.; Dogra, N. Extracellular Vesicles Carry Distinct Proteo-Transcriptomic Signatures That are Different from Their Cancer Cell of Origin. bioRxiv 2021. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Sabapatha, A.; Gercel-Taylor, C.; Taylor, D.D. Specific Isolation of Placenta-Derived Exosomes from the Circulation of Pregnant Women and Their Immunoregulatory Consequences. Am. J. Reprod. Immunol. 2006, 56, 345–355. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Théry, C.; Duban, L.; Segura, E.; Véron, P.; Lantz, O.; Amigorena, S. Indirect activation of naïve CD4+ T cells by dendritic cell–derived exosomes. Nat. Immunol. 2002, 3, 1156–1162. [Google Scholar] [CrossRef]
- Grommé, M.; Uytdehaag, F.G.C.M.; Janssen, H.; Calafat, J.; van Binnendijk, R.S.; Kenter, M.J.H.; Tulp, A.; Verwoerd, D.; Neefjes, J. Recycling MHC class I molecules and endosomal peptide loading. Proc. Natl. Acad. Sci. USA 1999, 96, 10326–10331. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta (BBA) Biomembr. 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Gopal, S.K.; Xu, R.; Simpson, R.J.; Chen, W. Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 2015, 40, 72–81. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Gopal, S.K.; Rai, A.; Simpson, R.J. Proteomic Insights into Extracellular Vesicle Biology-Defining Exosomes and Shed Microvesicles. Expert Rev. Proteom. 2017, 14, 69–95. [Google Scholar] [CrossRef]
- Hurley, J.H. ESCRT s are everywhere. EMBO J. 2015, 34, 2398–2407. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of mRNAs and microRNAs is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron Microscopic Evidence for Externalization of the Transferrin Receptor in Vesicular Form in Sheep Reticulo-Cytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [Green Version]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef]
- Farahani, M.; Rubbi, C.; Liu, L.; Slupsky, J.R.; Kalakonda, N. CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS ONE 2015, 10, e0141429. [Google Scholar] [CrossRef] [Green Version]
- Driedonks, T.A.P.; van der Grein, S.G.; Ariyurek, Y.; Buermans, H.P.J.; Jekel, H.; Chow, F.W.N.; Wauben, M.H.M.; Buck, A.H.; Hoen, P.A.C.; Hoen, E.N.M.N. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cell. Mol. Life Sci. 2018, 75, 3857–3875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaglani, S.; Gonzalez-Kozlova, E.; Lundon, D.J.; Tewari, A.K.; Dogra, N.; Kyprianou, N. exRNA Signatures in Extracellular Vesicles and their Tumor-Lineage from Prostate Cancer. medRxiv 2020. [Google Scholar] [CrossRef]
- Bai, K.; Li, X.; Zhong, J.; Ng, E.H.Y.; Yeung, W.S.; Lee, C.-L.; Chiu, P.C.N. Placenta-Derived Exosomes as a Modulator in Maternal Immune Tolerance During Pregnancy. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Neerukonda, S.N.; Tavlarides-Hontz, P.; McCarthy, F.; Pendarvis, K.; Parcells, M.S. Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek’s Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens. Genes 2019, 10, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kharbanda, E.O.; Haapala, J.; DeSilva, M.; Vazquez-Benitez, G.; Vesco, K.K.; Naleway, A.L.; Lipkind, H.S. Spontaneous Abortion Following COVID-19 Vaccination During Pregnancy. JAMA 2021, 326, 1629. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Biava, P. Exosomes and Cell Communication: From Tumour-Derived Exosomes and Their Role in Tumour Progression to the Use of Exosomal Cargo for Cancer Treatment. Cancers 2021, 13, 822. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M. The entry of enveloped viruses into cells by endocytosis. Biochem. J. 1984, 218, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Südhof, T.C.; Rothman, J.E. Membrane Fusion: Grappling with SNARE and SM Proteins. Science 2009, 323, 474–477. [Google Scholar] [CrossRef] [Green Version]
- Qadir, F.; Aziz, M.A.; Sari, C.P.; Ma, H.; Dai, H.; Wang, X.; Raithatha, D.; Da Silva, L.G.L.; Hussain, M.; Poorkasreiy, S.P.; et al. Transcriptome reprogramming by cancer exosomes: Identification of novel molecular targets in matrix and immune modulation. Mol. Cancer 2018, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Bland, C.L.; Byrne-Hoffman, C.N.; Fernandez, A.; Rellick, S.L.; Deng, W.; Klinke, D.J. Exosomes Derived from B16F0 Melanoma Cells Alter the Transcriptome of Cytotoxic T Cells that Impacts Mitochondrial Respiration. FEBS J. 2018, 285, 1033–1050. [Google Scholar] [CrossRef] [Green Version]
- Giacomini, E.; Scotti, G.M.; Vanni, V.S.; Lazarevic, D.; Makieva, S.; Privitera, L.; Signorelli, S.; Cantone, L.; Bollati, V.; Murdica, V.; et al. Global transcriptomic changes occur in uterine fluid-derived extracellular vesicles during the endometrial window for embryo implantation. Hum. Reprod. 2021, 36, 2249–2274. [Google Scholar] [CrossRef]
- Esfandyari, S.; Elkafas, H.; Chugh, R.M.; Park, H.-S.; Navarro, A.; Al-Hendy, A. Exosomes as Biomarkers for Female Reproductive Diseases Diagnosis and Therapy. Int. J. Mol. Sci. 2021, 22, 2165. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Bagalkot, T.; Fitzgerald, W.; Sadovsky, E.; Chu, T.; Martínez-Marchal, A.; Brieño-Enríquez, M.; Su, E.J.; Margolis, L.; Sorkin, A.; et al. Term Human Placental Trophoblasts Express SARS-CoV-2 Entry Factors ACE2, TMPRSS2, and Furin. mSphere 2021, 6, e00250-21. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Luca, F.; Xu, Y.; Alazizi, A.; Leng, Y.; Hsu, C.D.; Gomez-Lopez, N. Does the Human Placenta Express the Canonical Cell Entry Mediators for SARS-CoV-2? Elife 2020, 9, e58716. [Google Scholar] [CrossRef] [PubMed]
- Daamen, A.R.; Bachali, P.; Owen, K.A.; Kingsmore, K.M.; Hubbard, E.L.; Labonte, A.C.; Robl, R.; Shrotri, S.; Grammer, A.C.; Lipsky, P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021, 11, 1–19. [Google Scholar] [CrossRef]
- Fischer, D.S.; Ansari, M.; Wagner, K.I.; Jarosch, S.; Huang, Y.; Mayr, C.H.; Strunz, M.; Lang, N.J.; D’Ippolito, E.; Hammel, M.; et al. Single-Cell RNA Sequencing Reveals ex Vivo Signatures of SARS-CoV-2-Reactive T Cells Through “Reverse Phenotyping”. Nat. Commun. 2021, 12, 4515. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R.; et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef] [PubMed]
- Speranza, E.; Williamson, B.N.; Feldmann, F.; Sturdevant, G.L.; Pérez-Pérez, L.; Meade-White, K.; Smith, B.J.; Lovaglio, J.; Martens, C.; Munster, V.J.; et al. Single-cCell RNA Sequencing Reveals SARS-CoV-2 Infection Dynamics in Lungs of African Green Monkeys. Sci. Transl. Med. 2021, 13, eabe8146. [Google Scholar] [CrossRef]
- Liu, T.; Jia, P.; Fang, B.; Zhao, Z. Differential Expression of Viral Transcripts From Single-Cell RNA Sequencing of Moderate and Severe COVID-19 Patients and Its Implications for Case Severity. Front. Microbiol. 2020, 11, 603509. [Google Scholar] [CrossRef]
- Edlow, A.G.; Li, J.Z.; Collier, A.-R.Y.; Atyeo, C.; James, K.E.; Boatin, A.A.; Gray, K.J.; Bordt, E.A.; Shook, L.L.; Yonker, L.M.; et al. Assessment of Maternal and Neonatal SARS-CoV-2 Viral Load, Transplacental Antibody Transfer, and Placental Pathology in Pregnancies During the COVID-19 Pandemic. JAMA Netw. Open 2020, 3, e2030455. [Google Scholar] [CrossRef]
- Ashary, N.; Ashary, N.; Bhide, A.; Chakraborty, P.; Colaco, S.; Mishra, A.; Chhabria, K.; Jolly, M.K.; Modi, D. Single-Cell RNA-seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors Driving Pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 2020, 8, 783. [Google Scholar] [CrossRef]
- Pierce, C.A.; Sy, S.; Galen, B.; Goldstein, D.Y.; Orner, E.; Keller, M.J.; Herold, K.C.; Herold, B.C. Natural mucosal barriers and COVID-19 in children. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Loske, J.; Röhmel, J.; Lukassen, S.; Stricker, S.; Magalhães, V.G.; Liebig, J.; Chua, R.L.; Thürmann, L.; Messingschlager, M.; Seegebarth, A.; et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol. 2021, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Yang, R.; Wen, C.; Fan, J.; Ma, J.; He, Q.; Zhao, Z.; Song, X.; Chen, H.; Shi, G.; et al. Preliminary Analyses of scRNA Sequencing and Immunohistochemistry of Children’s Lung Tissues Indicate the Expression of SARS-CoV-2 Entry-Related Genes may not be the Key Reason for the Milder Syndromes of COVID-19 in Children. Clin. Transl. Med. 2021, 11, e300. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.B.; Verma, A. Nasal ACE2 Levels and COVID-19 in Children. JAMA 2020, 323, 2386. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Brian, D.A. Subgenomic messenger RNA amplification in coronaviruses. Proc. Natl. Acad. Sci. USA 2010, 107, 12257–12262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britto, C.J.; Niu, N.; Khanal, S.; Huleihel, L.; Herazo-Maya, J.D.; Thompson, A.; Sauler, M.; Slade, M.D.; Sharma, L.; Cruz, C.S.D.; et al. BPIFA1 regulates lung neutrophil recruitment and interferon signaling during acute inflammation. Am. J. Physiol. Cell. Mol. Physiol. 2019, 316, L321–L333. [Google Scholar] [CrossRef]
- Yang, A.C.; Kern, F.; Losada, P.M.; Agam, M.R.; Maat, C.A.; Schmartz, G.P.; Fehlmann, T.; Stein, J.A.; Schaum, N.; Lee, D.P.; et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 2021, 595, 565–571. [Google Scholar] [CrossRef]
- Atlante, S.; Mongelli, A.; Barbi, V.; Martelli, F.; Farsetti, A.; Gaetano, C. The epigenetic implication in coronavirus infection and therapy. Clin. Epigenetics 2020, 12, 1–12. [Google Scholar] [CrossRef]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2020, 16, 263–270. [Google Scholar] [CrossRef]
- Sawalha, A.H.; Zhao, M.; Coit, P.; Lu, Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020, 215, 108410. [Google Scholar] [CrossRef] [PubMed]
- Castro de Moura, M.; Davalos, V.; Planas-Serra, L.; Alvarez-Errico, D.; Arribas, C.; Ruiz, M.; Aguilera-Albesa, S.; Troya, J.; Valencia-Ramos, J.; Vélez-Santamaria, V.; et al. Epigenome-Wide Association Study of COVID-19 Severity with Respiratory Failure. EBioMedicine 2021, 66, 103339. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, P.; Jayaseelan, V.P. Implication of epitranscriptomics in trained innate immunity and COVID-19. Epigenomics 2021, 13, 1077–1080. [Google Scholar] [CrossRef]
- Sen, R.; Garbati, M.R.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Jit, B.P.; Qazi, S.; Arya, R.; Srivastava, A.; Gupta, N.; Sharma, A. An immune epigenetic insight to COVID-19 infection. Epigenomics 2021, 13, 465–480. [Google Scholar] [CrossRef]
- Saksena, N.; Bonam, S.R.; Miranda-Saksena, M. Epigenetic Lens to Visualize the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Infection in COVID-19 Pandemic. Front. Genet. 2021, 12, 581726. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef]
- Danlos, F.-X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef]
- Sen, R. High-throughput approaches of diagnosis and therapies for COVID-19: Antibody panels, proteomics and metabolomics. Future Drug Discov. 2021, 3, FDD55. [Google Scholar] [CrossRef]
- Doğan, H.O.; Şenol, O.; Bolat, S.; Yıldız, N.; Büyüktuna, S.A.; Sarıismailoğlu, R.; Doğan, K.; Hasbek, M.; Hekim, S.N. Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 2020, 93, 2340–2349. [Google Scholar] [CrossRef]
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLOS Pathog. 2021, 17, e1009243. [Google Scholar] [CrossRef]
- Konigsberg, I.R.; Barnes, B.; Campbell, M.; Davidson, E.; Zhen, Y.; Pallisard, O.; Boorgula, P.; Cox, C.; Nandy, D.; Seal, S.; et al. Host Methylation Predicts SARS-CoV-2 Infection and Clinical Outcome. Commun. Med. 2021, 1, 42. [Google Scholar] [CrossRef]
- Muus, C.; Luecken, M.D.; Eraslan, G.; Sikkema, L.; Waghray, A.; Heimberg, G.; Kobayashi, Y.; Vaishnav, E.D.; Subramanian, A.; Smillie, C.; et al. Single-Cell Meta-Analysis of SARS-CoV-2 Entry Genes Across Tissues and Demographics. Nat. Med. 2021, 27, 546–559. [Google Scholar] [CrossRef]
- O’Callaghan, K.P.; Blatz, A.M.; Offit, P.A. Developing a SARS-CoV-2 Vaccine at Warp Speed. JAMA 2020, 324, 437–438. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II Study of COVID-19 RNA Vaccine BNT162b1 in Adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hui, A.; Zhang, X.; Yang, Y.; Tang, R.; Ye, H.; Ji, R.; Lin, M.; Zhu, Z.; Türeci, O.; et al. Safety and Immunogenicity of the SARS-CoV-2 BNT162b1 mRNA Vaccine in Younger and Older Chinese Adults: A Randomized, Placebo-Controlled, Double-Blind Phase 1 Study. Nat. Med. 2021, 27, 1062–1070. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Oliver, S.E.; Gargano, J.W.; Marin, M.; Wallace, M.; Curran, K.G.; Chamberland, M.; McClung, N.; Campos-Outcalt, D.; Morgan, R.L.; Mbaeyi, S.; et al. The Advisory Committee on Immunization Practices’ Interim Recommendation for Use of Moderna COVID-19 Vaccine-United States, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021, 69, 1653–1656. [Google Scholar] [CrossRef]
- Alter, G.; Yu, J.; Liu, J.; Chandrashekar, A.; Borducchi, E.N.; Tostanoski, L.H.; McMahan, K.; Jacob-Dolan, C.; Martinez, D.R.; Chang, A.; et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature 2021, 596, 268–272. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Bos, R.; Rutten, L.; van der Lubbe, J.; Bakkers, M.; Hardenberg, G.; Wegmann, F.; Zuijdgeest, D.; de Wilde, A.H.; Koornneef, A.; Verwilligen, A.; et al. Ad26 Vector-Based COVID-19 Vaccine Encoding a Prefusion-Stabilized SARS-CoV-2 Spike Immunogen Induces Potent Humoral and Cellular Immune Responses. NPJ Vaccines 2020, 5, 91. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analy-sis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and Immunogenicity of ChAdOx1 nCoV-19 Vaccine Administered in a Prime-Boost Regimen in Young and Old Adults (COV002): A Single-Blind, Randomised, Controlled, Phase 2/3 Trial. Lancet 2021, 396, 1979–1993. [Google Scholar] [CrossRef]
- Sapkal, G.N.; Yadav, P.D.; Ella, R.; Deshpande, G.R.; Sahay, R.R.; Gupta, N.; Vadrevu, K.M.; Abraham, P.; Panda, S.; Bhargava, B. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B.1.1.7 variant of SARS-CoV-2. J. Travel Med. 2021, 28. [Google Scholar] [CrossRef] [PubMed]
- Ella, R.; Reddy, S.; Jogdand, H.; Sarangi, V.; Ganneru, B.; Prasad, S.; Das, D.; Raju, D.; Praturi, U.; Sapkal, G.; et al. Safety and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine, BBV152: Interim Results from a Double-Blind, Randomised, Multicentre, Phase 2 Trial, and 3-Month Follow-Up of a Double-Blind, Randomised Phase 1 Trial. Lancet Infect. Dis. 2021, 21, 950–961. [Google Scholar] [CrossRef]
- Liu, X.; Shaw, R.H.; Stuart, A.; Greenland, M.; Aley, P.K.; Andrews, N.J.; Cameron, J.C.; Charlton, S.; Clutterbuck, E.A.; Collins, A.M.; et al. Safety and Immunogenicity of Heterologous Versus Homologous Prime-Boost Schedules with an Adenoviral Vectored and mRNA COVID-19 Vaccine (Com-COV): A Single-Blind, Randomised, Non-Inferiority Trial. Lancet 2021, 398, 856–869. [Google Scholar] [CrossRef]
- Schmidt, T.; Klemis, V.; Schub, D. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat. Med. 2021, 27, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
| Clinical Trial Identifier | Intervention | Description |
|---|---|---|
| NCT04368728 | BNT162b1 | Lipid-nanoparticle-formulated, nucleoside-modified mRNA vaccine, encodes trimerized RBD of S protein of SARS-CoV-2 |
| NCT04847050 | mRNA-1273 | Lipid nanoparticle–encapsulated mRNA-based vaccine, encodes prefusion-stabilized full length S protein of SARS-CoV-2 |
| NCT04889209 | Ad26.COV2.S | Recombinant, replication-incompetent human adenovirus type 26 vector encoding prefusion-stabilized full-length S protein of SARS-CoV-2 |
| NCT04516746 | ChAdOx1 nCoV-19 (AZD1222) | Replication-deficient simian adenovirus vector ChAdOx1 + full-length S protein SARS-CoV-2 |
| NCT04834869 | BBV152 | Whole-virion inactivated, formulated with toll-like receptor 7/8 agonist adsorbed to alum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dogra, N.; Ledesma-Feliciano, C.; Sen, R. Developmental Aspects of SARS-CoV-2, Potential Role of Exosomes and Their Impact on the Human Transcriptome. J. Dev. Biol. 2021, 9, 54. https://doi.org/10.3390/jdb9040054
Dogra N, Ledesma-Feliciano C, Sen R. Developmental Aspects of SARS-CoV-2, Potential Role of Exosomes and Their Impact on the Human Transcriptome. Journal of Developmental Biology. 2021; 9(4):54. https://doi.org/10.3390/jdb9040054
Chicago/Turabian StyleDogra, Navneet, Carmen Ledesma-Feliciano, and Rwik Sen. 2021. "Developmental Aspects of SARS-CoV-2, Potential Role of Exosomes and Their Impact on the Human Transcriptome" Journal of Developmental Biology 9, no. 4: 54. https://doi.org/10.3390/jdb9040054
APA StyleDogra, N., Ledesma-Feliciano, C., & Sen, R. (2021). Developmental Aspects of SARS-CoV-2, Potential Role of Exosomes and Their Impact on the Human Transcriptome. Journal of Developmental Biology, 9(4), 54. https://doi.org/10.3390/jdb9040054








