Abstract
Cranial neural crest (NC) cells delaminate from the neural folds in the forebrain to the hindbrain during mammalian embryogenesis and migrate into the frontonasal prominence and pharyngeal arches. These cells generate the bone and cartilage of the frontonasal skeleton, among other diverse derivatives. RNA-binding proteins (RBPs) have emerged as critical regulators of NC and craniofacial development in mammals. Conventional RBPs bind to specific sequence and/or structural motifs in a target RNA via one or more RNA-binding domains to regulate multiple aspects of RNA metabolism and ultimately affect gene expression. In this review, we discuss the roles of RBPs other than core spliceosome components during human and mouse NC and craniofacial development. Where applicable, we review data on these same RBPs from additional vertebrate species, including chicken, Xenopus and zebrafish models. Knockdown or ablation of several RBPs discussed here results in altered expression of transcripts encoding components of developmental signaling pathways, as well as reduced cell proliferation and/or increased cell death, indicating that these are common mechanisms contributing to the observed phenotypes. The study of these proteins offers a relatively untapped opportunity to provide significant insight into the mechanisms underlying gene expression regulation during craniofacial morphogenesis.
1. Introduction
During vertebrate embryogenesis, neural crest (NC) cells are induced at the neural plate border, undergo an epithelial to mesenchymal transition to delaminate from the neural folds or neural tube, migrate in stereotypical streams throughout the embryo and eventually differentiate into a wide array of derivatives. The NC can be subdivided into four axial populations: cranial, cardiac, vagal and trunk. Cranial NC cells originate from the forebrain to the hindbrain and migrate into the frontonasal prominence and pharyngeal arches. These cells generate the bone and cartilage of the frontonasal skeleton, cartilages of the jaw, middle ear, hyoid and thyroid, tendons, smooth muscle, connective tissue, melanocytes and cranial sensory ganglia of the peripheral nervous system. Cranial NC cells regulate growth and patterning of the brain, and additionally contribute to the eyes, teeth, thyroid gland, parathyroid glands and thymus [1].
RNA-binding proteins (RBPs) have emerged as critical regulators of NC and craniofacial development. Conventional RBPs bind to specific sequence and/or structural motifs in a target RNA via one or more RNA-binding domains (RBDs) to regulate multiple aspects of RNA metabolism, including transcription, splicing, modification, trafficking, translation and decay, and ultimately affect gene expression. A growing body of evidence has also identified unconventional RBPs that do not contain discernible RBDs [2,3].
In this review, we discuss the roles of RBPs other than core spliceosome components during human and/or mouse NC and craniofacial development. These include a regulator of transcription, CNBP; proteins involved in splicing such as EIF4A3, ESRP1/2, FAM50A, hnRNP H2, hnRNP K, hnRNP U, Hu proteins, PTB, RBFOX2, RBM10, RBMX and SRSF3; a regulator of nuclear export, EIF4A3; regulators of trafficking such as EIF4A3 and IMP-1; proteins involved in translation such as CNBP, DDX3X, EIF4A3, FMRP, PACT and IMP-1; and a regulator of decay, EIF4A3 (Figure 1). These proteins contain one or more RBDs, including: RNA recognition motif (ESRP1/2, hnRNP H2, Hu proteins, PTB, RBFOX2, RBM10, RBMX, SRSF3 and IMP-1), zinc-finger domain (CNBP, hnRNP H2 and RBM10), K-homology domain (hnRNP K, IMP-1 and FMRP), DEAD motif (EIF4A3 and DDX3X), ROKNT domain (hnRNP K), SAP domain (hnRNP U), Fox-1_C domain (RBFOX2), G-patch domain (RBM10), RBM1CTR domain (RBMX), Agenet domain (FMRP), FXR1P_C domain (FMRP) and double-stranded RNA-binding motif (PACT) [4]. For a review of mutations associated with craniofacial spliceosomopathies, please see [5]. Where applicable, we review data on these same RBPs from additional vertebrate species, including chicken, Xenopus and zebrafish models, which provide further insight into the mechanisms by which these proteins regulate gene expression and cell activity.
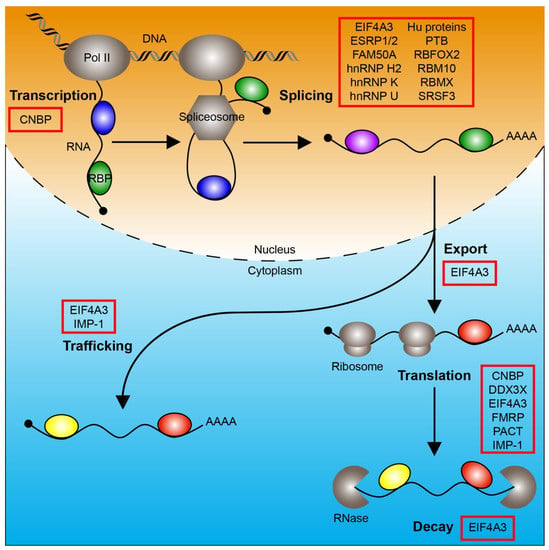
Figure 1.
RNA-binding proteins regulate RNA metabolism. The processes of transcription, splicing, nuclear export, trafficking, translation and decay are depicted. RNA-binding proteins (colored ovals) that regulate these processes and have a role in mammalian neural crest and/or craniofacial development are boxed in red. Pol II, RNA polymerase II; RBP, RNA-binding protein.
2. RBPs Involved in Transcription
CNBP
Myotonic dystrophy 2 (DM2; also known as proximal myotonic myopathy; OMIM 602668) is caused by a heterozygous expansion of 75 or more CCTG repeats in intron 1 of CNBP [6]. DM2 is characterized by muscle pain and stiffness, myotonia, progressive extremity and truncal weakness, cataracts, diabetes, cardiac arrhythmias and male hypogonadism, and may also include cognitive dysfunction, hearing loss, tremor and hypersomnia [7,8] (Table 1). Of note, in addition to its role in transcription, CNBP is also involved in translation.

Table 1.
Craniofacial defects associated with mis-expression of human RBPs.
In mice, Cnbp transcripts are expressed in the epiblast and visceral endoderm at embryonic day (E) 5.5. Approximately two days later, Cnbp transcripts and protein localize to all three germ layers in the anterior region of the embryo, including the anterior definitive endoderm underlying the future forebrain and the overlying anterior neuroectoderm. By E8.5, Cnpb expression is restricted to the headfold region. As early as E9.0, Cnbp is expressed in the forebrain, facial prominences, limb buds and somites. Expression in the craniofacial region is specifically localized to the frontonasal, maxillary and mandibular prominences and the roof of the stomodeum [9,10]. Mice that are homozygous for an allele harboring a viral insertion in intron 1 of Cnbp that abolishes transcript expression from the locus die around E10.5. These embryos display growth retardation as early as E7.5 with absence of the anterior mesendoderm and anterior neuroectoderm, and resulting truncations in the anterior neural folds at E8.5 and the forebrain at E9.5. These defects are associated with reduced proliferation in the anterior region of homozygous mutant embryos at E7.5–E8.5 and decreased expression of Myc in the anterior neuroectoderm and headfold region at E7.25 and E8.5, respectively. Further, approximately 40 percent of heterozygous neonatal pups exhibit growth retardation and craniofacial defects, including a small mandible and absence of the eyes [9]. Consistent with the phenotypes observed in human patients with DM2, mice that are heterozygous or homozygous for null alleles of Cnbp additionally exhibit postnatal muscle atrophy [11,12].
Similar to the expression patterns observed in mouse embryos, CNBP transcripts in the chick initially localize to the epiblast and hypoblast at stages XIII-XIV. At stage 4, CNBP is expressed in the neuroectoderm of the future forebrain. Expression is restricted to the anterior region of the embryo at stage 5, with strong expression in the anterior neural plate by stage 8. CNBP transcripts are detected in the telencephalon, midbrain, facial prominences, heart, wing bud and tail as early as stage 19. Akin to the findings detailed above for Cnbp homozygous mutant mouse embryos, injection of siRNA targeting CNBP into the chick forebrain at stage 9 leads to forebrain truncation [13].
In Xenopus laevis, cnbp is expressed throughout the animal pole and in the marginal zone containing the future mesoderm at early gastrula stages 10.25–10.5. During the late neural and tailbud stages 15–27, cnbp transcripts localize along the anterior to posterior dorsal axis, with specific expression in the neural tube, epidermis and somitic and lateral plate mesoderm [14]. Overexpression of cnbp mRNA leads to an expansion of the expression domain of the NC markers foxd3 and cmyc. Alternatively, overexpression of transcripts encoding dominant-negative, N-terminal truncations of Cnbp results in decreased expression of foxd3 and cmyc [15].
Similar to the findings in mice and chick, zebrafish cnbpa is expressed in the anterior region of the embryo, with increased expression at the midbrain-hindbrain border and the retina at 1 day post fertilization (dpf). At 2.5–3 dpf, cnbpa transcripts additionally localize to the craniofacial structures, pectoral fins and liver [16]. The cis elements that control zebrafish cnbpa expression during development are located in intron 1, in a region harboring conserved binding sites for NC transcription factors Pax6 and Foxd3 [17]. Knockdown of cnbpa at the one-cell stage with a translation-blocking morpholino results in growth retardation, abnormal morphology of the forebrain, midbrain and midbrain-hindbrain border, hydrocephaly, defects in craniofacial development, small eyes, fewer pigmented cells, oedema and tail curvature. The craniofacial defects include reduced first and second pharyngeal arch and neurocranial cartilage elements, disruption of pharyngeal arch 3–7 cartilages, a dysmorphic anterior ethmoid plate with large, round chondrocytes, decreased chondrocyte number in the trabeculae, malformation of the ceratohyal, reduction of the hyosymplectic and palatoquadrate and failed extension of Meckel’s cartilage at 4 dpf [16,18]. Overexpression of an N-terminal truncation of Cnbpa phenocopies knockdown with the translation-blocking morpholino [18]. These defects are associated with decreased cell proliferation and increased cell death in anterior embryonic regions at 1 dpf, particularly in the brain [16]. Further, decreased expression of the NC marker msxb is detected at 12 hours post fertilization (hpf) in morphant embryos, as well as decreased expression of the NC markers foxd3, tfap2a, crestin, sox9b and dlx2a in anterior embryonic regions at 1 dpf [16,18]. Moreover, expression of the cartilage marker col2a1 is reduced in the parachordals and ceratohyal of morphants at 2 dpf and in the ethmoid plate and trabeculae at 3 dpf [16]. Knockdown of cnbpa at the one-cell stage with a morpholino that modifies cnbpa pre-mRNA splicing generates a more severe phenotype than that observed with the translation-blocking morpholino discussed above [18]. Interestingly, morpholino-mediated knockdown of the gene underlying the craniofacial disorder Treacher Collins Syndrome, tcof1, results in decreased levels of Cnbpa protein and increased levels of reactive oxygen species at 1 dpf. The craniofacial defects in these embryos at 4 dpf can be rescued upon transgenic overexpression of cnbpa, particularly, the lengths of the head, palatoquadrate-hyosymplectic cartilages and ceratohyal cartilages, and the distance between the ceratohyal cartilages joint and lateral fins. This rescue appears to stem from a protective role of Cnbpa in preventing the upregulation of oxidative stress-responsive genes [19].
3. RBPs Involved in Splicing
3.1. EIF4A3
Robin sequence with cleft mandible and limb anomalies (also known as Richieri-Costa-Pereira syndrome; OMIM 268305) is caused by a homozygous 16-repeat expansion or compound heterozygous 16-repeat and 15-repeat expansions of an 18- or 20-nucleotide motif in the 5′ untranslated region (UTR) of EIF4A3 [20]. The syndrome is characterized by craniofacial and limb defects, laryngeal abnormalities and, commonly, learning and language disabilities. The craniofacial defects include microstomia, cleft palate, absent lower central incisors, micrognathia, abnormal fusion of the mandible and ear anomalies [21,22] (Table 1). Induced pluripotent stem cell-derived NC cells (iNCCs) generated from patients with Robin sequence with cleft mandible and limb anomalies have approximately 70% and 50% reductions in the expression of EIF4A3 mRNA and protein, respectively, and exhibit significant defects in migration. Mesenchymal stem-like cells generated from patient iNCCs have an approximately 15% reduction in EIF4A3 expression and experience impaired chondrogenic differentiation and premature osteogenic differentiation compared to control cells [23]. Of note, in addition to its role in splicing, EIF4A3 is also involved in export, trafficking, translation and decay.
Similar to patients with Robin sequence with cleft mandible and limb anomalies, animal models with reduced Eif4a3 expression display lower jaw defects. In mice, heterozygous, ubiquitous ablation of Eif4a3 using a Cmv-Cre driver results in a reduction in body size, facial malformations, delayed eye and limb development and neural tube defects at E11.5. A subset of these embryos exhibits holoprosencephaly and a mandible lacking two distinct mandibular arches, consistent with expression of EIF4A3 in the developing mandible. Heterozygous, conditional ablation of Eif4a3 in the NC lineage using the Wnt1-Cre driver leads to micrognathia at E11.5, a near complete absence of the skull, anterior facial skeletal elements and ears at E14.5, and a severely hypoplastic mandible, exencephaly, reduced eyelid closure and loss of the outer ear structures at E18.5 [23].
Similarly, morpholino-mediated knockdown of eif4a3 in zebrafish results in craniofacial defects, including underdevelopment of craniofacial cartilage and bone elements and clefting of the lower jaw, as well as a reduction in the size of the eyes, underdevelopment of the third-sixth branchial arches and brain defects at 1 dpf. These changes are accompanied by increased apoptosis in morpholino-injected embryos, particularly in the anterior-most regions of the embryo, and decreased expression of transcripts encoding the NC markers Sox9b, Foxd3, Sox10 and Tbx2 [20].
3.2. ESRP1/2
Deafness, autosomal recessive 109 (DFNB109; OMIM 618013), characterized by bilateral sensorineural hearing loss and vestibular dysplasia, is associated with compound heterozygosity for a 19-bp deletion and a missense mutation in ESRP1 (Table 1). Induced pluripotent stem cells generated from one of these patients exhibit significant changes in RNA splicing for a subset of ESRP1-dependent transcripts [24]. Alternatively, ESRP2 protein is expressed in the secondary palatal epithelium at 67–72 days of gestation in humans, with increased expression in the periderm. Relatedly, missense and nonsense mutations in ESRP2 are associated with non-syndromic cleft lip with or without cleft palate (Table 1). Expression of the nonsense mutation in human embryonic kidney (HEK) 293T cells results in a partial disruption of epithelial RNA splicing [25].
In mice, Esrp1 is expressed in the embryonic ectoderm as early as E6.5 and in the headfolds, open neural folds, definitive endoderm and foregut at E7.5. By E8.5, expression expands into the first pharyngeal arch and first pharyngeal pouch. Esrp1 transcripts are additionally detected in the pharyngeal cleft and otic vesicle at E9.5, among other sites. By E10.5, Esrp1 expression in the head region expands to Rathke’s pouch, the olfactory pit and the nasopharyngeal groove [26]. At E11.5, both Esrp1 and Esrp2 transcripts are expressed in the surface ectoderm of the medial and lateral nasal processes as well as the maxillary processes, with increased expression in the periderm [27]. By E12.5, Esrp1 transcripts are additionally present in the lens, vibrissae and pinnae, among other sites [26]. From E13.5-E15.0, Esrp1 and Esrp2 are co-expressed in the epithelium lining the secondary palatal shelves [27]. Postnatally, Esrp1 and Esrp2 expression is restricted to epithelial cells across a wide range of tissues and organs [28,29]. Global ablation of Esrp1 leads to fully penetrant, bilateral cleft lip and clefting of the primary and secondary palates, as well as defects in inner ear morphology, auditory hair cell differentiation and cell fate specification along the lateral wall of the cochlear epithelium [24,29]. Consistently, Esrp1-null embryos have reduced proliferation of both epithelial and mesenchymal cells in the medial and lateral nasal processes at E10.5, hypoplastic facial processes near mid-gestation, and persistent E-cadherin expression upon contact of the medial and lateral nasal processes at E11.5. Similarly, proliferation is reduced in the secondary palatal shelves of Esrp1-null embryos at E16.5 and palatal organ cultures established from these E13.5 embryos have persistent E-cadherin expression and medial edge epithelium presence upon contact of the shelves. These defects are accompanied by a switch in transcripts encoding ectodermal FGFR2IIIb to mesenchymal FGFR2IIIc in the facial process ectoderm at E12.0, rendering these cells incapable of responding to mesenchymal FGF10, in addition to decreased expression of transcripts encoding WNT ligands and SHH. Corresponding reductions in the expression of transcripts encoding canonical WNT targets and GLI transcription factors are detected in the facial process mesenchyme, as well as decreased WNT activity in the medial nasal and maxillary processes. Decreased expression of additional genes that have been implicated in facial clefting, such as Bmp7, Tgfb2 and Tgfb3, were also found in the Esrp1-null facial process mesenchyme, indicating that ESRP1 serves to regulate signaling crosstalk between the facial process epithelium and mesenchyme [30]. Similarly, aberrant splicing of Fgfr2 is detected in the cochlear epithelium of Esrp1-null embryos at 16.5 [24]. The same cleft lip and palate phenotypes observed in Esrp1-null embryos are recapitulated upon conditional ablation of Esrp1 in the surface ectoderm using the Crect driver, such that these E18.5 embryos exhibit hypoplasia of the premaxilla and palatine bones and dysmorphic palatal process of the maxilla bones [30]. Global, simultaneous ablation of both Esrp1 and Esrp2 additionally results in absent lungs and salivary glands and reduced kidney size at E15.5, and reduced body length, rostral shortening, absent premaxillary bone and mandibular coronoid process, hypoplasia of the palatal bone and mandible, dorsal skin epidermal hypoplasia, reduced hair follicle number, delayed hair follicle maturation and forelimb malformations at E18.5. Again, a switch in transcripts encoding ectodermal FGFR2IIIb to mesenchymal FGRF2IIIc is detected in E18.5 epidermis obtained from double knockout embryos [29].
In zebrafish, esrp1 and esrp2 transcripts co-localize in the olfactory epithelium, otic vesicle, pharynx, epidermis and notochord from 1–5 dpf, with esrp2 transcripts additionally expressed in the pronephros, hatching gland, liver and heart [31]. Both transcripts localize to the developing stomodeum and surface and oral epithelium at 2–3 dpf [27]. Combined loss-of-function of both esrp1 and esrp2 results in dysmorphology of the olfactory epithelium, clefting of the ethmoid plate, inner ear defects, abnormal basibranchial pharyngeal cartilage, altered esophagus morphology, irregular surface epithelium morphology with epithelial blebs, aberrant swim bladder epithelium and impaired fin development [27,31]. Similar to findings in mice, a switch towards expression of mesenchymal IIIc isoforms of fgfr1a, fgfr1b, fgfr2 and fgfr3 is detected in double mutant zebrafish embryos [31]. Further, aberrant, non-cartilage cells within the ethmoid plate cleft ectopically express a gene associated with facial clefting across vertebrate species, irf6 [27].
3.3. FAM50A
Intellectual developmental disorder, X-linked, syndromic, Armfield type (MRXSA; also known as Armfield X-linked mental retardation syndrome; OMIM 300261) is caused by hemizygous missense mutations in FAM50A [32]. The syndrome is characterized by intellectual disability, developmental delay, dysmorphic facial features, skeletal defects (particularly affecting the hands and feet) and ocular anomalies, and commonly includes poor or absent speech, congenital heart defects including atrial septal defect, patent ductus arteriosus, tetralogy of Fallot and right ventricle dilation, brain dysmorphologies, gastrointestinal defects, genitourinary anomalies and neurological defects such as seizures and hypotonia. The dysmorphic facial features can include macrocephaly, prominent, tall and/or broad forehead, bitemporal narrowing, faint hemangiomas between brows and at back of neck, downslanting palpebral fissures, hypotelorism, epicanthal folds, infraorbital creases, proptosis, exotropia, strabismus, keratoconus, nystagmus, depressed nasal bridge, wide nasal root, short and lightly upturned nose with underdeveloped nares, tubular nose, bulbous nose, prominent lips, bow-shaped mouth, cleft palate, hypodontia, microretrognathia, micrognathia, low-set, large and/or slightly posteriorly rotated ears and excessively folded helices [32,33] (Table 1).
In zebrafish, fam50a transcripts are ubiquitously expressed throughout gastrulation, but from 1 dpf onwards, expression is restricted to anterior structures including the eyes, mandible and brain. fam50a-null embryos exhibit defects in eye, craniofacial cartilage and brain development at 5 dpf and die approximately 1 day later. In particular, fam50-null fish display anterior-posterior shortening of the pharyngeal skeleton, delayed branchial arch patterning and a wider ceratohyal angle at 3 dpf compared to controls. These changes are accompanied by increased expression of the p53 pathway effectors tp53, mdm2 and cdkn1a at 2 dpf. The craniofacial cartilage defects of fam50a-null fish are recapitulated upon morpholino-mediated knockdown of fam50a, with morphants exhibiting increased cell cycle progression and apoptosis in the head region [32].
3.4. hnRNP H2
Mental retardation, X-linked, syndromic, Bain type (MRXSB; OMIM 300986) is associated with heterozygous mutation in HNRNPH2, the gene encoding the RBP hnRNP H2 [34]. The syndrome is characterized by intellectual disability, poor or absent speech, motor difficulties, growth retardation and musculoskeletal abnormalities, and may also include dysmorphic facial features, epilepsy, neuropsychiatric abnormalities, cortical vision impairment, acquired microcephaly and feeding difficulties [34,35]. The dysmorphic facial features can include short palpebral fissures, almond-shaped eyes, strabismus, long columella, hypoplastic alae nasi, short philtrum, full lower lip and micrognathia [34] (Table 1).
3.5. hnRNP K
Au-Kline syndrome (AUKS; OMIM 616580) is caused by heterozygous mutation in HNRNPK, the gene encoding the RBP hnRNP K, or a 264 kb deletion in chromosome 9q21.32 encompassing HNRNPK [36,37]. The syndrome is characterized by intellectual disability, developmental delay and craniofacial dysmorphologies, and may also include skeletal and connective tissue anomalies, congenital heart malformations, genitourinary anomalies, delayed psychomotor development, speech impairment, cryptorchidism, nuchal skin thickening, hand/foot abnormalities, high pain tolerance, hearing loss, vision abnormalities and agenesis of the corpus callosum [36,37]. The craniofacial dysmorphologies can include dolichocephaly, craniosynostosis, ridged metopic suture, long face, broad or sparse eyebrows, long palpebral fissures, shallow orbits, ptosis, lateral eversion of the lower eyelids, full cheeks, broad nasal bridge, bifid nasal tip, hypoplastic alae nasi, open or down-turned mouth, thin upper lip, high or cleft palate, prominent midline groove of the tongue, missing teeth, malocclusion, low-set, prominent ears, underdeveloped ear helices, preauricular pits and earlobe crease [36,37,38,39] (Table 1).
3.6. hnRNP U
Developmental and epileptic encephalopathy 54 (DEE54; OMIM 617391) is caused by heterozygous mutation in HNRNPU, the gene encoding the RBP hnRNP U [40]. DEE54 is characterized by delayed psychomotor development, seizures and severe intellectual disability, and may also include hypotonia, absent speech, delayed myelination in the brain, hyperlaxity, enlarged ventricles of the brain, microcephaly, deep-set eyes with epicanthal folds, gray sclerae, narrow palate, short second digit of the hand and autistic features [40,41,42] (Table 1).
3.7. Hu Proteins
In mice, Elavl1 encodes for the RBP HuA (also known as HuR), while Elavl4 encodes for HuD. Conditional ablation of Elavl1 in the epiblast lineage using the Sox2-Cre driver results in craniofacial and axial skeletal defects, absence of the spleen and lung dysmorphologies. Approximately 80 percent of these embryos exhibit delayed ossification in the neurocranium and upper jaw, and less than 15 percent have nasal clefting [43]. Alternatively, Elavl4-null embryos have neuronal development defects, including a transient cranial nerve phenotype characterized by impaired neurite extension of the glossopharyngeal, hypoglossal, trigeminal and acousticofacial nerves at E10.5 [44].
In chick, an anti-panELAV/HU antibody detects expression in the neural tube at the level of the pharyngeal region as early as stage 14. Immunoreactive NC-derived cells are found in the dorsal root ganglia beginning at stage 17 and in the sympathetic ganglia at stage 19 [45].
3.8. PTB
In mice, Ptbp1 encodes the RBP PTB (also known as hnRNP I). By combining null and floxed alleles of Ptbp1 with the Nestin-Cre driver, conditional ablation is achieved in the mouse central and peripheral nervous system. A subset of these Ptbp1fl/-;Nestin-Cre+/Tg postnatal day (P) 21 pups exhibit a domed cranium, in addition to brain abnormalities [46].
In Xenopus tropicalis, ptbp1 is expressed in the ectoderm and mesoderm at the gastrula stage. At neurula stages (stage 16–18), expression is maintained in the ectoderm with strong expression in the neural plate border and NC. Expression is additionally detected in the branchial arches, eyes, otic placodes and pronephros at the tailbud stage (stage 25). Finally, ptbp1 transcripts in the head region localize to the mandibular, hyoidal and branchial arches, eyes and otic vesicles at the late tailbud stage (stage 34). Expression is additionally detected in migrating trunk NC cells, the ventral aorta, lateral plate and intermediate mesoderm, dermatome and sclerotome at this stage [47].
3.9. RBFOX2
In mice, RBFOX2 is expressed in the dorsal neural tube, premigratory and migratory NC cells and somites along the rostrocaudal axis at E9.5. As development proceeds, RBFOX2 becomes restricted to the ventral neural tube. RBFOX2 is additionally detected in the dorsal root ganglia as early as E10.5 and in the trigeminal ganglion and craniofacial mesenchyme at E12.5. Conditional ablation of Rbfox2 in premigratory and migratory NC cells as well as the somites using the Pax3-Cre driver results in cranial nerve deformities at E10.5, subcutaneous edema at E17.5, and clefting of the secondary palate and shortening of the body axis at P1. The cleft palate defect stems from reduced proliferation of the palatal shelf mesenchyme at E12.5–15.5 and a subsequent failure of the palatal shelves to fuse at the midline along the anterior-posterior axis from E15.5-E18.5. The same palatal clefting phenotype is observed upon conditional ablation of Rbfox2 in the NC lineage using the Wnt1-Cre2 driver. Both Rbfox2fl/fl;Pax3-Cre+/Tg and Rbfox2fl/fl;Wnt1-Cre2+/Tg embryos at late gestation exhibit hypoplasia and reduced ossification of the majority of NC-derived bones in the frontonasal skeleton and absence of the palatal process of the palatine bone. Additionally, RBFOX2 is able to bind to several transcripts involved in NC or craniofacial development, such as Map3k7 (encoding MAP3K7/TAK1), Fn1 and Postn, in palatal mesenchyme cells. Conditional ablation of Rbfox2 using the Pax3-Cre driver leads to the differential alternative RNA splicing and subsequent downregulation of expression for a subset of these transcripts in craniofacial tissues. Transcripts encoding TGFβ pathway components are also downregulated in Rbfox2fl/fl;Pax3-Cre+/Tg craniofacial tissues, and phosphorylation of a TGFβ pathway effector, the protein serine/threonine kinase TAK1, and its targets p38 MAPK and SMAD2 are reduced in Rbfox2fl/fl;Pax3-Cre+/Tg palatal shelves. Interestingly, overexpression of TAK1 in cultured palatal mesenchyme cells restores expression of Map3k7 and Fn1 and rescues the proliferation defects observed in Rbfox2fl/fl;Pax3-Cre+/Tg embryos. Further, SMAD2 and SMAD3 are able to bind to the Rbfox2 promoter in response to TGFβ treatment of palatal mesenchyme cells to generate a positive feedback loop [48].
3.10. RBM10
TARP syndrome (TARPS; OMIM 311900) is caused by hemizygous mutation in RBM10 [49]. The syndrome is variably characterized by talipes equinovarus, atrial septal defect, Robin sequence (cleft palate, glossoptosis and micrognathia) and persistent left superior vena cava [50]. Additional fully-penetrant features include cognitive, motor and language delays, brain malformations, neurological defects, pulmonary abnormalities, failure to thrive and facial dysmorphism. Patients commonly exhibit cardiac and distal limb anomalies beyond those described in the TARP acronym, postnatal growth delay, skeletal defects and gastrointestinal abnormalities, with a subset of patients additionally displaying genital malformations, renal abnormalities, vision impairment and hearing loss [51,52]. The dysmorphic facial features can include large fontanels, round face, hypertelorism, underdevelopment of the alae nasi, prominent columella, wide mouth, downturned mouth corners, high and/or narrow palate and low-set and/or posteriorly rotated ears [52] (Table 1).
Consistent with its role in human development, Rbm10 transcripts are enriched in the first and second pharyngeal arches, limb buds and tailbud of E9.5-E10.5 mouse embryos. Expression of Rbm10 transcripts in the pharyngeal arches decreases by E11.5 [49]. Rbm10 knockout mouse embryonic stem cells as well as mouse mandibular cells isolated from E11.5 embryos exhibit decreased growth compared to wild-type cells [53].
3.11. RBMX
Mental retardation, X-linked, syndromic 11 (MRXS11; also known as Shashi X-linked mental retardation syndrome; OMIM 300238) is associated with a hemizygous mutation in the gene encoding the RBP RBMX (also known as hnRNP G) [54]. The syndrome is characterized by moderate mental retardation, craniofacial dysmorphologies, obesity and enlarged testes. The craniofacial dysmorphologies include coarse facies, prominent supraorbital ridges, narrow palpebral fissures, puffy eyelids, bulbous nose, prominent lower lip and large ears [55] (Table 1).
In Xenopus laevis, rbmx transcripts are expressed in the neural tube, NC and somites at stages 20–25. Morpholino-mediated knockdown of rbmx results in decreased expression of the NC marker slug at stages 17 and 35, as well as eye defects, jaw deformities, loss of the trigeminal ganglion and a reduction in melanocyte numbers in the trunk at tadpole stages, among other neural and axial muscle segmentation defects [56].
In zebrafish, rbmx transcripts are ubiquitously expressed during gastrulation, but become restricted to multiple sites as development progresses, including the branchial arches from 2–7 dpf. Similar to the findings in Xenopus laevis, morpholino-mediated knockdown of rbmx in zebrafish results in a reduction in the size of the head and eyes, mild cyclopia and absent jaws at 2 dpf, in addition to defects in brain development and somite patterning [57].
3.12. SRSF3
In mice, Srsf3 is broadly expressed during development, with transcripts enriched in the head region and facial processes from E8.5-E10.5. At these same timepoints, SRSF3 protein co-localizes with SOX10 in NC cells migrating away from the cranial neural folds and is expressed in the mesenchyme and overlying ectoderm of the pharyngeal arches and facial processes. Homozygous, conditional ablation of Srsf3 in the murine NC lineage using the Wnt1-Cre driver results in hypoplastic facial processes, increased distance between the nasal pits and brain malformations at E10.5. At E12.5, these conditional knockout embryos additionally exhibit facial clefting and hypoplasia of the secondary palatal shelves and tongue. A subset of embryos at these stages displays facial subepidermal blebbing, facial hemorrhaging and a wavy neural tube. By E18.5, Srsf3 conditional knockout embryos have a reduced head size, with anterior bones and cartilages that are clefted and hypoplastic and midface elements that are more severely affected and, in some cases, missing entirely. The mandible, coronoid process and Meckel’s cartilage are hypoplastic, the hyoid is not ossified or severely hypoplastic, the lesser and greater horns of the hyoid as well as the thyroid and cricoid cartilages are hypoplastic and the tracheal cartilage rings are misshapen. These defects stem from reduced proliferation of the mesenchyme underlying the cranial neural folds at E8.0 and increased cell death in the mesenchyme of pharyngeal arch 1 at E9.5. Additionally, conditional ablation of Srsf3 in the murine NC lineage leads to a significant increase in expression of multiple genes involved in NC differentiation, including Col2a1 and Zic5 [58]. PI3K is the primary effector of PDGFRα signaling during skeletal development in the mouse, and disruption of this axis leads to palatal clefting [59,60]. SRSF3 is phosphorylated at AKT consensus sites in response to PI3K-mediated PDGFRα signaling in immortalized mouse embryonic palatal mesenchyme cells, driving translocation of phosphorylated SRSF3 into the nucleus where alternative RNA splicing occurs. Relatedly, SRSF3 coordinates the alternative RNA splicing of transcripts encoding protein serine/threonine kinases in the E11.5 maxillary process mesenchyme to regulate PDGFRα-dependent intracellular signaling, specifically, phosphorylation of ERK1/2 [58].
In Xenopus laevis, srsf3 is expressed ubiquitously, with transcripts enriched in the neural tube and somites at stages 20–32. Further, overexpression of srsf3 mRNA leads to a change in the shape and location of the expression domain of the NC marker slug, among other defects in neurogenesis and mesoderm development [56].
4. RBPs Involved in Trafficking
VICKZ Proteins
In mice, Igf2bp1 encodes for the RBP IMP-1 (also known as VICKZ1 and ZBP1). At E10.5, Igf2bp1 transcripts are expressed in the forebrain, hindbrain, medial and lateral nasal processes, first and second pharyngeal arches, limb buds and tail. By E12.5, expression expands into the neural tract, eyes, tongue, heart, lung, liver and somites. At E17.5, Igf2bp1 transcripts primarily localize to the intestine, kidney and liver. Igf2bp1-null pups have a reduced body size, a short snout and, in a subset of mice, a kinked tail, wide-spread cartilage loss and dysmorphologies of the intestine, kidney and liver. Within the craniofacial region, cartilage loss was detected in the nasal bone and mandible [61]. Of note, in addition to its role in trafficking, IMP-1 is also involved in translation.
While the chick VICKZ paralogs are expressed in the neural epithelium and neural folds at the 5 somite stage, their expression is reduced in delaminating and migrating cranial NC cells at the 8 and 13 somite stages, respectively. Morpholino-mediated knockdown or expression of dominant-negative VICKZ at the 2–4 somite stage leads to enhanced cranial NC cell delamination, while this effect is reversed upon overexpression of VICKZ. Further, down-regulation of VICKZ activity decreases expression of Integrin α6 in the neural tube, while overexpression of VICKZ results in abnormal persistence of Integrin α6 expression at this site. Relatedly, immunoprecipitation of VICKZ from 3–4 day embryos leads to an enrichment in ITGA6 mRNA, suggesting that the two molecules are part of the same complex in chick embryos [62].
5. RBPs Involved in Translation
5.1. DDX3X
Intellectual developmental disorder, X-linked, syndromic, Snijders Blok type (MRXSSB; OMIM 300958) is caused by heterozygous mutations in DDX3X in females or hemizygous missense mutations in DDX3X in males [63]. In female patients, the syndrome is characterized by intellectual disability or developmental delay and can also include neurological abnormalities such as hypotonia, behavior problems, movement disorder and epilepsy, as well as microcephaly, dolichocephaly, brain dysmorphologies, joint hyperlaxity, skin pigment abnormalities, visual impairment, precocious puberty, scoliosis, hearing loss, cleft lip and/or palate and neuroblastoma. A subset of patients exhibits dysmorphic facial features, which commonly include long and/or hypotonic face, high and/or broad forehead, hypertelorism, wide nasal bridge, bulbous nose, narrow alae nasi and anteverted nostrils [63,64,65] (Table 1). Less common dysmorphic facial features include telecanthus, short palpebral fissures, epicanthal folds, smooth and/or long philtrum, thin upper vermilion, high-arched palate, micrognathia and large, protruding or low-set ears [64,66]. In male patients, the syndrome is characterized by intellectual disability and can also include neurological abnormalities such as behavior problems and movement disorder, brachycephaly, macrocephaly, facial dysmorphisms, strabismus, bifid uvula, nuchal thickening, pulmonary stenosis, cardiac anomalies, visual impairment and hearing loss [63,67,68] (Table 1).
In mice, though DDX3X protein is highly expressed throughout the embryo at E7.5, its expression becomes restricted as development proceeds, such that by E14.5, protein expression localizes to the brain, trigeminal ganglion, eyes, nasal cavity, lung, heart and liver. Conditional ablation of Ddx3x in the mouse epiblast using the Sox2-Cre driver results in male embryos exhibiting developmental delay and abnormal head fold morphology at E9.5, and neural tube closure defects, an underdeveloped brain and reduced head size at E11.5, among other allantois and myocardial trabeculae defects. These changes are accompanied by embryo-wide increases in mitosis at E9.5 and apoptosis at E8.5-E9.5. Moreover, Ddx3xfl/Y;Sox2-Cre+/Tg embryos exhibit increased DNA damage, p53 phosphorylation and expression of transcripts encoding proteins involved in cell cycle progression at E9.5 [69].
In Xenopus tropicalis, ddx3 is ubiquitously expressed at early gastrula stages in the animal half of embryos. By the end of gastrulation, expression is predominantly localized to the dorsal ectoderm. Expression of ddx3 is further restricted to the neural plate and neural plate border during neurulation, and to the head and somites at tailbud stages. Morpholino-mediated knockdown of ddx3 results in hypoplasia of the craniofacial cartilage at approximately stage 46. These defects stem from reduced expression of transcripts encoding proteins involved in neural plate border specification, including pax3 and msx1, and NC specification, including snai2 and sox9, at the end of gastrulation. Moreover, the function of Ddx3 in this context is mediated by the downstream effector Rac1, leading to activation of Akt, the subsequent phosphorylation and inhibition of Gsk3β, and ultimately the stabilization of two proteins required for NC induction, β-catenin and Snai1 [70].
Similarly, morpholino-mediated knockdown of zebrafish ddx3xb results in reduced head and brain sizes at 2 dpf [71]. DDX3X has been shown to regulate WNT/β-catenin signaling [72] and co-injection of wnt3a and ddx3xb mRNA into zebrafish embryos results in a more severe ventralization phenotype at 2 dpf than that observed upon injection of wnt3a alone. This effect is maintained upon injection of ddx3xb mRNA harboring mutations found in human male, but not female, MRXSSB patients, indicating that the de novo variants in the latter case are loss-of-function mutations with respect to regulation of the Wnt pathway [63].
5.2. FMRP
The majority of Fragile X syndrome (FXS; OMIM 300624) cases are caused by a full trinucleotide (CGG) repeat expansion of more than 200 repeats in the 5′ untranslated region of FMR1 that results in hypermethylation of FMR1 and loss of expression of the RBP FMRP [73,74,75,76]. Human FMR1 transcripts are expressed in the neural tube, central nervous system and several non-neural tissues, among them the eyes, branchial arches and cartilages, at 3–7 weeks of gestation [77]. Accordingly, FXS is characterized by mental retardation, macroorchidism and dysmorphic facial features, which can include long, narrow face, high and/or broad forehead, broad palpebral fissures, midface hypoplasia, flat nasal bridge, broad nose, upward displacement of the nasal alae, broad philtrum, large mouth, thick lips, long upper middle incisors, high-arched palate, prognathia, hypotonia affecting the lower jaw and large, low-set and/or poorly formed ears [78,79,80,81,82] (Table 1).
In mice, Fmr1 transcripts are ubiquitously expressed at E10.5, with strong expression in the neural tube. By adulthood, however, high expression is restricted to the brain, eyes, testes, ovaries, thymus, esophagus and spleen [83]. Similar to the phenotypes described for human FXS patients, Fmr1-null mice exhibit learning deficits, hyperactivity, macroorchidism and craniofacial defects including reduced skull vault height, decreased length and rear width of the inner skull, interparietal bone shortening and reduced mandible width [82,84]. Further, the NC regulator AP-2α binds the FMR1 promoter in human HeLa cells and Fmr1 expression is reduced in E18.5 head tissue of Tfap2a-null embryos [85].
In Xenopus laevis, fmr1 transcripts are expressed in the neural tube, migrating cranial NC cells and eye anlagen at stage 20. Transcripts localize to the central nervous system, NC, eyes and pharyngeal arches at early tailbud stages 25–26. fmr1 expression increases in the central nervous system and within the craniofacial region, including the eyes and otic vesicle, by the late tailbud stage, with additional expression in the notochord [86,87]. Morpholino-mediated knockdown of fmr1 in an animal-dorsal blastomere at the 8-cell stage results in smaller or absent eyes and reduced and dysmorphic cranial cartilage elements. The latter phenotypes are associated with defects in NC migration as indicated by decreased expression of foxd3 at stage 20 as well as krox20 and twist at stage 23 [87]. Consistent with findings in mice, fmr1 and tfap2a transcripts co-localize in the pharyngeal arches and craniofacial regions of Xenopus leavis embryos at stage 25. Moreover, overexpression of a dominant-negative Ap-2α and inhibition of Ap-2α through Chordin overexpression lead to significant repression of fmr1 expression in animal cap assays [85].
Morpholino-mediated knockdown of fmr1 in zebrafish leads to abnormal axon branching, reduced trigeminal neuron number and dysmorphic craniofacial cartilage. Specifically, Meckel’s cartilage is shorter and wider at 5 dpf in morphants and the angles between the ceratobranchial/ceratohyal arch cartilage and basibranchial/hyal cartilage and between Meckel’s cartilage and the rostrocaudal axis are less acute in morphants. Further, expression of a marker of migrating cranial NC cells, dlx-2a, is reduced and mis-localized in the first and second pharyngeal arches of morphants at 22 hpf [88]. Importantly, however, the phenotypes observed in morphants are not recapitulated in fmr1-null zebrafish, which are viable and fertile with no apparent defects nor altered expression of dlx-2a [89].
5.3. PACT
Dystonia 16 (DYT16; OMIM 612067) is caused by homozygous mutations in PRKRA, the gene encoding the RBP PACT in humans [90]. DYT16 is characterized by early-onset, progressive dystonia and gait abnormalities, and may also include delayed development, oromandibular dystonia, laterocollis/retrocollis, spasmodic dysphonia, bradykinesia, dysphagia, opisthotonic posturing, facial grimacing, speech and language impairment, leg pain, blepharospasm and hypomimia [90,91] (Table 1).
In mice, Prkra transcripts are expressed in mouse blastocysts, the developing ears as early as E12 and in the anterior skull base and mandible at E16 [92,93]. Consistently, Prkra-null embryos die before implantation [93]. However, mice in which a neomycin resistance cassette replaces the sequence in exon 8 encoding the PKR activation domain express no PRKRA protein (also called RAX in mice) and exhibit hypoplasia of the anterior pituitary lobe, reduced postnatal body weight, fertility defects and a number of craniofacial defects, including wide cranial sutures, hypoplastic interparietal bone, prominent bossing of the forehead, shortened nose, hypolastic nasal turbinates, hypoplastic mandibular condyle and microtia [92,94,95]. The pituitary defects stem from reduced proliferation in the anterior pituitary lobe as early as P21 [94]. The ear defects are characterized by hypoplastic pinnae and external auditory canals, malformed middle ear ossicles and reduced middle ear space and tympanic membrane, resulting in hearing impairment [92].
6. Conclusions
Multiple screens have revealed a prevalence of transcripts encoding RBPs in mouse craniofacial tissues. One such study identified an enrichment of transcripts encoding 17 proteins with RNA recognition motifs in mouse E10.5 pharyngeal arches 1 and 2 [96]. A more recent study revealed that transcripts encoding 497 RBPs are expressed in mouse craniofacial process ectoderm and/or mesenchyme from E10.5-E12.5, including Esrp1, Esrp2, Elavl4, Rbfox2, Rbm10, Rbmx, Igf2bp1 and Fmr1 [97], each of which is discussed above. However, in contrast to the well-studied roles of signaling molecules and transcription factors in regulating NC and craniofacial development, only a handful of the 1,393 RBPs identified in humans [2] and the 1,914 RBPs identified in mouse [2] have been examined in these contexts. The study of RBPs thus offers a relatively untapped opportunity to provide significant insight into the mechanisms underlying gene expression regulation during craniofacial morphogenesis.
As the majority of this review focuses on splicing regulators not associated with the spliceosome, it is useful to compare the phenotypes observed upon mis-expression of core components of the spliceosome and these additional splicing regulators. Spliceosomopathies are characterized by microcephaly, malar hypoplasia, eye anomalies, micrognathia and external ear anomalies within the craniofacial region, as well as intellectual disability, psychomotor delay, heart defects and limb abnormalities [5]. As discussed above, with the exception of malar hypoplasia, each of these phenotypes is observed upon mutation of one or more non-spliceosome-associated RBP. Given the prevalence of craniofacial dysmorphologies observed in spliceosomopathies, it has been hypothesized that NC cells have an increased need for alternative RNA splicing, perhaps owing to their multipotency, and that mutation of core components of the spliceosome leads to a common mechanism of p53 pathway disruption and NC cell apoptosis [5]. Interestingly, with the exception of EIF4A3 [98], modulation of the p53 pathway has not been reported for any of the splicing regulators discussed here, indicating that these RBPs may affect cell survival through alternative pathways.
While the target transcripts of individual RBPs do not appear to overlap in the context of craniofacial morphogenesis, even within related tissues, common cellular mechanisms are regulated by these RBPs to contribute to proper development. For example, only six transcripts are commonly differentially alternatively spliced (Exoc1, Meg3, Nfya, Smarca2, Tpm1 and Uap1) and only two transcripts are commonly differentially expressed (Lama5 and Thrb) between E12.5 Rbfox2fl/fl;Pax3-Cre+/Tg craniofacial tissue and E11.5 Srsf3fl/fl;Wnt1-Cre+/Tg maxillary process mesenchyme tissue relative to their respective controls [48,58]. Notably, however, knockdown or ablation of Esrp1/2, Rbfox2, Srsf3 and Ddx3x each results in altered expression of transcripts encoding components of developmental signaling pathways. These include the FGF, WNT, SHH, BMP and TGFβ signaling pathways in the case of ESRP1/2 [30], and the TGFβ, PDGF and Wnt signaling pathways for RBFOX2 [48], SRSF3 [58] and Ddx3x [70,72], respectively. These findings raise the intriguing possibility that RNA processing mediated by RBPs serves to regulate both intracellular and intercellular signaling during craniofacial development, often in feedback loops involving developmental signaling pathways and the phosphorylation of intracellular effectors. Further, mis-expression of several of the RBPs discussed here results in reduced cell proliferation (in the cases of CNBP, ESRP1, RBFOX2, RBM10, SRSF3 and PACT/RAX) and/or increased cell death (in the cases of Cnbp, Eif4a3, Fam50a, SRSF3 and DDX3X) across vertebrate species, indicating that these are common mechanisms underlying the observed craniofacial phenotypes.
Several outstanding questions remain regarding RBPs in NC and craniofacial development: Where and when are RBPs expressed and what upstream factors regulate this expression? How is the activity of RBPs regulated by post-translational modifications? Which proteins interact with RBPs to enhance or antagonize function, and to what extent do RBPs cross-regulate one another? Similarly, how does the network of RBPs expressed in a given cell type establish and/or reinforce a tissue-specific transcriptome? Which are the direct target transcripts of RBPs and how does mis-expression of these transcripts contribute to a disease phenotype? As highlighted above, animal models of individual RBPs provide a framework to address many of these questions. Undoubtedly, the advent of new genetic, molecular and biochemical approaches promises to reveal critical roles for additional RBPs both during development and disease.
Author Contributions
Conceptualization, T.E.F., B.J.C.D. and K.A.F.; writing—original draft preparation, K.A.F.; writing—review and editing, T.E.F., B.J.C.D. and K.A.F.; visualization, K.A.F.; funding acquisition, B.J.C.D. and K.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
B.J.C.D. was funded by National Institute of Dental and Craniofacial Research grant F31DE029364 and K.A.F. was funded by National Institute of Dental and Craniofacial Research grants R01DE027689 and K02DE028572.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank our laboratory colleagues for their helpful discussions and comments on this manuscript. We apologize to authors whose work we were unable to cite due to space limitations.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish.
References
- Mayor, R.; Theveneau, E. The neural crest. Development 2013, 140, 2247–2251. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Gebauer, F.; Schwarzl, T.; Valcárcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2020, 22, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Beauchamp, M.; Alam, S.S.; Kumar, S.; Jerome-Majewska, L.A. Spliceosomopathies and neurocristopathies: Two sides of the same coin? Dev. Dyn. 2020, 249, 924–945. [Google Scholar] [CrossRef]
- Liquori, C.L.; Ricker, K.; Moseley, M.L.; Jacobsen, J.F.; Kress, W.; Naylor, S.L.; Day, J.W.; Ranum, L.P.W. Myotonic Dystrophy Type 2 Caused by a CCTG Expansion in Intron 1 of ZNF9. Science 2001, 293, 864–867. [Google Scholar] [CrossRef]
- Thornton, C.A.; Griggs, R.C.; Moxley, R.T. Myotonic dystrophy with no trinucleotide repeat expansion. Ann. Neurol. 1994, 35, 269–272. [Google Scholar] [CrossRef]
- Udd, B.; Meola, G.; Krahe, R.; Thornton, C.; Ranum, L.; Bassez, G.; Kress, W.; Schoser, B.; Moxley, R. 140th ENMC International Workshop: Myotonic Dystrophy DM2/PROMM and other myotonic dystrophies with guidelines on management. Neuromuscul. Disord. 2006, 16, 403–413. [Google Scholar] [CrossRef]
- Chen, W.; Liang, Y.; Deng, W.; Shimizu, K.; Ashique, A.M.; Li, E.; Li, Y.-P. The zinc-finger protein CNBP is required for forebrain formation in the mouse. Development 2003, 130, 1367–1379. [Google Scholar] [CrossRef]
- Shimizu, K.; Chen, W.; Ashique, A.M.; Moroi, R.; Li, Y.-P. Molecular cloning, developmental expression, promoter analysis and functional characterization of the mouse CNBP gene. Gene 2003, 307, 51–62. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Y.; Abe, Y.; Cheney, L.; Udd, B.; Li, Y.-P. Haploinsuffciency for Znf9 in Znf9+/− Mice Is Associated with Multiorgan Abnormalities Resembling Myotonic Dystrophy. J. Mol. Biol. 2007, 368, 8–17. [Google Scholar] [CrossRef]
- Wei, C.; Stock, L.; Schneider-Gold, C.; Sommer, C.; Timchenko, N.A.; Timchenko, L. Reduction of Cellular Nucleic Acid Binding Protein Encoded by a Myotonic Dystrophy Type 2 Gene Causes Muscle Atrophy. Mol. Cell. Biol. 2018, 38. [Google Scholar] [CrossRef]
- Abe, Y.; Chen, W.; Huang, W.; Nishino, M.; Li, Y.-P. CNBP regulates forebrain formation at organogenesis stage in chick embryos. Dev. Biol. 2006, 295, 116–127. [Google Scholar] [CrossRef][Green Version]
- Flink, I.L.; Blitz, I.; Morkin, E. Characterization of cellular nucleic acid binding protein from Xenopus laevis: Expression in all three germ layers during early development. Dev. Dyn. 1998, 211, 123–130. [Google Scholar] [CrossRef]
- Armas, P.; Aguero, T.H.; Borgognone, M.; Aybar, M.J.; Calcaterra, N.B. Dissecting CNBP, a Zinc-Finger Protein Required for Neural Crest Development, in Its Structural and Functional Domains. J. Mol. Biol. 2008, 382, 1043–1056. [Google Scholar] [CrossRef]
- Weiner, A.; Allende, M.; Becker, T.; Calcaterra, N.B. CNBP mediates neural crest cell expansion by controlling cell proliferation and cell survival during rostral head development. J. Cell. Biochem. 2007, 102, 1553–1570. [Google Scholar] [CrossRef]
- Weiner, A.M.; Allende, M.L.; Calcaterra, N.B. Zebrafish cnbp intron1 plays a fundamental role in controlling spatiotemporal gene expression during embryonic development. J. Cell. Biochem. 2009, 108, 1364–1375. [Google Scholar] [CrossRef]
- Weiner, A.M.J.; Sdrigotti, M.A.; Kelsh, R.N.; Calcaterra, N.B. Deciphering the cellular and molecular roles of cellular nucleic acid binding protein during cranial neural crest development. Dev. Growth Differ. 2011, 53, 934–947. [Google Scholar] [CrossRef]
- De Peralta, M.S.P.; Mouguelar, V.S.; Sdrigotti, M.A.; Ishiy, F.A.A.; Fanganiello, R.D.; Passos-Bueno, M.R.; Coux, G.; Calcaterra, N.B. Cnbp ameliorates Treacher Collins Syndrome craniofacial anomalies through a pathway that involves redox-responsive genes. Cell Death Dis. 2016, 7, e2397. [Google Scholar] [CrossRef] [PubMed]
- Favaro, F.P.; Alvizi, L.; Zechi-Ceide, R.M.; Bertola, D.; Felix, T.M.; de Souza, J.; Raskin, S.; Twigg, S.; Weiner, A.M.; Armas, P.; et al. A Noncoding Expansion in EIF4A3 Causes Richieri-Costa-Pereira Syndrome, a Craniofacial Disorder Associated with Limb Defects. Am. J. Hum. Genet. 2014, 94, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Richieri-Costa, A.; Pereira, S.C.S. Short stature, Robin sequence, cleft mandible, pre/postaxial hand anomalies, and clubfoot: A new autosomal recessive syndrome. Am. J. Med. Genet. 1992, 42, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Favaro, F.P.; Zechi-Ceide, R.M.; Alvarez, C.W.; Maximino, L.; Antunes, L.F.B.B.; Richieri-Costa, A.; Guion-Almeida, M.L. Richieri-Costa-Pereira syndrome: A unique acrofacial dysostosis type. An overview of the Brazilian cases. Am. J. Med. Genet. Part A 2010, 155, 322–331. [Google Scholar] [CrossRef]
- Miller, E.E.; Kobayashi, G.; Musso, C.M.; Allen, M.; Ishiy, F.A.; De Caires, L.C.; Goulart, E.; Griesi-Oliveira, K.; Zechi-Ceide, R.M.; Richieri-Costa, A.; et al. EIF4A3 deficient human iPSCs and mouse models demonstrate neural crest defects that underlie Richieri-Costa-Pereira syndrome. Hum. Mol. Genet. 2017, 26, 2177–2191. [Google Scholar] [CrossRef] [PubMed]
- Rohacek, A.; Bebee, T.W.; Tilton, R.K.; Radens, C.; McDermott-Roe, C.; Peart, N.; Kaur, M.; Zaykaner, M.; Cieply, B.; Musunuru, K.; et al. ESRP1 Mutations Cause Hearing Loss due to Defects in Alternative Splicing that Disrupt Cochlear Development. Dev. Cell 2017, 43, 318–331.e5. [Google Scholar] [CrossRef]
- Cox, L.L.; Cox, T.C.; Uribe, L.M.M.; Zhu, Y.; Richter, C.T.; Nidey, N.; Standley, J.M.; Deng, M.; Blue, E.; Chong, J.; et al. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 2018, 102, 1143–1157. [Google Scholar] [CrossRef]
- Revil, T.; Jerome-Majewska, L.A. During Embryogenesis, Esrp1 Expression Is Restricted to a Subset of Epithelial Cells and Is Associated With Splicing of a Number of Developmentally Important Genes. Dev. Dyn. 2013, 242, 281–290. [Google Scholar] [CrossRef]
- Carroll, S.H.; Trevino, C.M.; Li, E.B.; Kawasaki, K.; Myers, N.; Hallett, S.A.; Alhazmi, N.; Cotney, J.; Carstens, R.P.; Liao, E.C. An Irf6-Esrp1/2 regulatory axis controls midface morphogenesis in vertebrates. Development 2020, 147. [Google Scholar] [CrossRef]
- Warzecha, C.; Sato, T.K.; Nabet, B.; Hogenesch, J.B.; Carstens, R.P. ESRP1 and ESRP2 Are Epithelial Cell-Type-Specific Regulators of FGFR2 Splicing. Mol. Cell 2009, 33, 591–601. [Google Scholar] [CrossRef]
- Bebee, T.W.; Park, J.W.; Sheridan, K.I.; Warzecha, C.; Cieply, B.W.; Rohacek, A.; Xing, Y.; Carstens, R.P. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eLife 2015, 4, e08954. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sears, M.J.; Zhang, Z.; Li, H.; Salhab, I.; Krebs, P.; Xing, Y.; Nah, H.-D.; Williams, T.; Carstens, R.P. Cleft lip and cleft palate (CL/P) in Esrp1 KO mice is associated with alterations in epithelial-mesenchymal crosstalk. Development 2020, 147. [Google Scholar] [CrossRef]
- Burguera, D.; Marquez, Y.; Racioppi, C.; Permanyer, J.; Torres-Méndez, A.; Esposito, R.; Albuixech-Crespo, B.; Fanlo, L.; D’Agostino, Y.; Gohr, A.; et al. Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat. Commun. 2017, 8, 1799. [Google Scholar] [CrossRef]
- Lee, Y.-R.; Khan, K.; Armfield-Uhas, K.; Srikanth, S.; Thompson, N.A.; Pardo, M.; Yu, L.; Norris, J.W.; Peng, Y.; Gripp, K.W.; et al. Mutations in FAM50A suggest that Armfield XLID syndrome is a spliceosomopathy. Nat. Commun. 2020, 11, 3698. [Google Scholar] [CrossRef]
- Armfield, K.; Nelson, R.; Lubs, H.A.; Häne, B.; Schroer, R.J.; Arena, F.; Schwartz, C.E.; Stevenson, R.E. X-linked mental retardation syndrome with short stature, small hands and feet, seizures, cleft palate, and glaucoma is linked to Xq28. Am. J. Med. Genet. 1999, 85, 236–242. [Google Scholar] [CrossRef]
- Bain, J.M.; Cho, M.T.; Telegrafi, A.; Wilson, A.; Brooks, S.; Botti, C.; Gowans, G.; Autullo, L.A.; Krishnamurthy, V.; Willing, M.C.; et al. Variants in HNRNPH2 on the X Chromosome Are Associated with a Neurodevelopmental Disorder in Females. Am. J. Hum. Genet. 2016, 99, 728–734. [Google Scholar] [CrossRef]
- Bain, J.M.; Thornburg, O.; Pan, C.; Rome-Martin, D.; Boyle, L.; Fan, X.; Devinsky, O.; Frye, R.; Hamp, S.; Keator, C.G.; et al. Detailed Clinical and Psychological Phenotype of the X-linked HNRNPH2-Related Neurodevelopmental Disorder. Neurol. Genet. 2021, 7, e551. [Google Scholar] [CrossRef] [PubMed]
- Au, P.Y.B.; You, J.; Caluseriu, O.; Schwartzentruber, J.; Majewski, J.; Bernier, F.P.; Marcia Marcia Ferguson Care for Rare Canada Consortium; Valle, D.; Parboosingh, J.S.; Sobreira, N.; et al. GeneMatcher Aids in the Identification of a New Malformation Syndrome with Intellectual Disability, Unique Facial Dysmorphisms, and Skeletal and Connective Tissue Abnormalities Caused by De Novo Variants in HNRNPK. Hum. Mutat. 2015, 36, 1009–1014. [Google Scholar] [CrossRef]
- Au, P.Y.B.; Care for Rare Canada Consortium; Goedhart, C.; Ferguson, M.; Breckpot, J.; Devriendt, K.; Wierenga, K.; Fanning, E.; Grange, R.K.; Graham, G.E.; et al. Phenotypic spectrum of Au-Kline syndrome: A report of six new cases and review of the literature. Eur. J. Hum. Genet. 2018, 26, 1272–1281. [Google Scholar] [CrossRef]
- Lange, L.; Pagnamenta, A.T.; Lise, S.; Clasper, S.; Stewart, H.; Akha, E.S.; Quaghebeur, G.; Knight, S.J.; Keays, D.A.; Taylor, J.C.; et al. A de novo frameshift in HNRNPK causing a Kabuki-like syndrome with nodular heterotopia. Clin. Genet. 2016, 90, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Miyake, N.; Inaba, M.; Mizuno, S.; Shiina, M.; Imagawa, E.; Miyatake, S.; Nakashima, M.; Mizuguchi, T.; Takata, A.; Ogata, K.; et al. A case of atypical Kabuki syndrome arising from a novel missense variant in HNRNPK. Clin. Genet. 2017, 92, 554–555. [Google Scholar] [CrossRef]
- Carvill, G.L.; Heavin, S.B.; Yendle, S.C.; McMahon, J.; O’Roak, B.; Cook, J.; Khan, A.; Dorschner, M.O.; Weaver, M.; Calvert, S.; et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013, 45, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, F.F.; Srour, M.; Capo-Chichi, J.-M.; Daoud, H.; Nassif, C.; Patry, L.; Massicotte, C.; Ambalavanan, A.; Spiegelman, D.; Diallo, O.; et al. De Novo Mutations in Moderate or Severe Intellectual Disability. PLoS Genet. 2014, 10, e1004772. [Google Scholar] [CrossRef]
- De Kovel, C.G.; Brilstra, E.H.; Van Kempen, M.J.; Slot, R.V.; Nijman, I.J.; Afawi, Z.; De Jonghe, P.; Djémié, T.; Guerrini, R.; Hardies, K.; et al. Targeted sequencing of 351 candidate genes for epileptic encephalopathy in a large cohort of patients. Mol. Genet. Genom. Med. 2016, 4, 568–580. [Google Scholar] [CrossRef]
- Katsanou, V.; Milatos, S.; Yiakouvaki, A.; Sgantzis, N.; Kotsoni, A.; Alexiou, M.; Harokopos, V.; Aidinis, V.; Hemberger, M.; Kontoyiannis, D.L. The RNA-Binding Protein Elavl1/HuR Is Essential for Placental Branching Morphogenesis and Embryonic Development. Mol. Cell. Biol. 2009, 29, 2762–2776. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, W.; Fujihara, H.; Mitsuhashi, T.; Yano, M.; Shibata, S.; Hayakawa, Y.; Okano, H.J.; Sakakibara, S.-I.; Takano, H.; Takano, T.; et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. USA 2005, 102, 4625–4630. [Google Scholar] [CrossRef]
- Marusich, M.F.; Furneaux, H.M.; Henion, P.D.; Weston, J.A. Hu neuronal proteins are expressed in proliferating neurogenic cells. J. Neurobiol. 1994, 25, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, T.; Tokunaga, A.; Sakamoto, R.; Sagara, H.; Noguchi, S.; Sasaoka, T.; Yoshida, N. PTB Deficiency Causes the Loss of Adherens Junctions in the Dorsal Telencephalon and Leads to Lethal Hydrocephalus. Cereb. Cortex 2012, 23, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Noiret, M.; Audic, Y.; Hardy, S. Expression analysis of the polypyrimidine tract binding protein (PTBP1) and its paralogs PTBP2 and PTBP3 during Xenopus tropicalis embryogenesis. Int. J. Dev. Biol. 2012, 56, 747–753. [Google Scholar] [CrossRef]
- Cibi, D.M.; Mia, M.M.; Shekeran, S.G.; Yun, L.S.; Sandireddy, R.; Gupta, P.; Hota, M.; Sun, L.; Ghosh, S.; Singh, M.K. Neural crest-specific deletion of Rbfox2 in mice leads to craniofacial abnormalities including cleft palate. eLife 2019, 8, e45418. [Google Scholar] [CrossRef]
- Johnston, J.J.; Teer, J.K.; Cherukuri, P.F.; Hansen, N.F.; Loftus, S.; Chong, K.; Mullikin, J.C.; Biesecker, L.G. Massively Parallel Sequencing of Exons on the X Chromosome Identifies RBM10 as the Gene that Causes a Syndromic Form of Cleft Palate. Am. J. Hum. Genet. 2010, 86, 743–748. [Google Scholar] [CrossRef]
- Gorlin, R.J.; Cervenka, J.; Anderson, R.C.; Sauk, J.J.; Bevis, W.D. Robin’s syndrome. A probably X-linked recessive subvariety exhibiting persistence of left superior vena cava and atrial septal defect. Am. J. Dis. Child. 1970, 119, 176–178. [Google Scholar] [CrossRef]
- Kaeppler, K.E.; Stetson, R.C.; Lanpher, B.C.; Collura, C.A. Infant male with TARP syndrome: Review of clinical features, prognosis, and commonalities with previously reported patients. Am. J. Med. Genet. Part A 2018, 176, 2911–2914. [Google Scholar] [CrossRef]
- Kumps, C.; D’Haenens, E.; Vergult, S.; Leus, J.; van Coster, R.; Jansen, A.; Devriendt, K.; Oostra, A.; Vanakker, O.M. Phenotypic spectrum of the RBM10 -mediated intellectual disability and congenital malformation syndrome beyond classic TARP syndrome features. Clin. Genet. 2020, 99, 449–456. [Google Scholar] [CrossRef]
- Rodor, J.; FitzPatrick, D.R.; Eyras, E.; Cáceres, J.F. The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 2016, 14, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Shashi, V.; Xie, P.; Schoch, K.; Goldstein, D.B.; Howard, T.D.; Berry, M.N.; Schwartz, C.E.; Cronin, K.; Sliwa, S.; Allen, A.; et al. The RBMX gene as a candidate for the Shashi X-linked intellectual disability syndrome. Clin. Genet. 2014, 88, 386–390. [Google Scholar] [CrossRef]
- Shashi, V.; Berry, M.N.; Shoaf, S.; Sciote, J.J.; Goldstein, D.; Hart, T.C. A Unique Form of Mental Retardation with a Distinctive Phenotype Maps to Xq26-q27. Am. J. Hum. Genet. 2000, 66, 469–479. [Google Scholar] [CrossRef][Green Version]
- Dichmann, D.S.; Fletcher, R.B.; Harland, R.M. Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev. Dyn. 2008, 237, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Tsend-Ayush, E.; O’Sullivan, L.A.; Grutzner, F.; Onnebo, S.M.; Lewis, R.S.; Delbridge, M.L.; Graves, J.; Ward, A. RBMX gene is essential for brain development in zebrafish. Dev. Dyn. 2005, 234, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Dennison, B.J.C.; Larson, E.D.; Fu, R.; Mo, J.; Fantauzzo, K.A. Srsf3 mediates alternative RNA splicing downstream of PDGFRα signaling in the facial mesenchyme. Development 2021, 148. [Google Scholar] [CrossRef] [PubMed]
- Klinghoffer, R.A.; Hamilton, T.; Hoch, R.; Soriano, P. An Allelic Series at the PDGFαR Locus Indicates Unequal Contributions of Distinct Signaling Pathways During Development. Dev. Cell 2002, 2, 103–113. [Google Scholar] [CrossRef]
- Fantauzzo, K.A.; Soriano, P. PI3K-mediated PDGFRα signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes Dev. 2014, 28, 1005–1017. [Google Scholar] [CrossRef]
- Hansen, T.V.O.; Hammer, N.A.; Nielsen, J.; Madsen, M.; Dalbaeck, C.; Wewer, U.M.; Christiansen, J.; Nielsen, F.C. Dwarfism and Impaired Gut Development in Insulin-Like Growth Factor II mRNA-Binding Protein 1-Deficient Mice. Mol. Cell. Biol. 2004, 24, 4448–4464. [Google Scholar] [CrossRef] [PubMed]
- Carmel, M.S.; Kahane, N.; Oberman, F.; Miloslavski, R.; Sela-Donenfeld, D.; Kalcheim, C.; Yisraeli, J.K. A Novel Role for VICKZ Proteins in Maintaining Epithelial Integrity during Embryogenesis. PLoS ONE 2015, 10, e0136408. [Google Scholar] [CrossRef]
- Blok, L.S.; Madsen, E.; Juusola, J.; Gilissen, C.; Baralle, D.; Reijnders, M.R.; Venselaar, H.; Helsmoortel, C.; Cho, M.T.; Hoischen, A.; et al. Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am. J. Hum. Genet. 2015, 97, 343–352. [Google Scholar] [CrossRef]
- Chanes, N.M.; Wong, J.; Lacassie, Y. Further delineation of DDX3X syndrome. Clin. Dysmorphol. 2019, 28, 149–151. [Google Scholar] [CrossRef]
- Lennox, A.L.; Hoye, M.L.; Jiang, R.; Johnson-Kerner, B.L.; Suit, L.A.; Venkataramanan, S.; Sheehan, C.; Alsina, F.C.; Fregeau, B.; Aldinger, K.; et al. Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development. Neuron 2020, 106, 404–420.e8. [Google Scholar] [CrossRef] [PubMed]
- Beal, B.; Hayes, I.; McGaughran, J.; Amor, D.J.; Miteff, C.; Jackson, V.; Van Reyk, O.; Subramanian, G.; Hildebrand, M.S.; Morgan, A.; et al. Expansion of phenotype of DDX3X syndrome: Six new cases. Clin. Dysmorphol. 2019, 28, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Kellaris, G.; Khan, K.; Baig, S.M.; Tsai, I.-C.; Zamora, F.M.; Ruggieri, P.; Natowicz, M.R.; Katsanis, N. A hypomorphic inherited pathogenic variant in DDX3X causes male intellectual disability with additional neurodevelopmental and neurodegenerative features. Hum. Genom. 2018, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Nicola, P.; Blackburn, P.R.; Rasmussen, K.J.; Bertsch, N.L.; Klee, E.W.; Hasadsri, L.; Pichurin, P.N.; Rankin, J.; Raymond, F.L.; Clayton-Smith, J.; et al. De novo DDX3X missense variants in males appear viable and contribute to syndromic intellectual disability. Am. J. Med. Genet. Part A 2019, 179, 570–578. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chan, C.-H.; Chen, C.-M.; Tsai, Y.-S.; Tsai, T.-Y.; Lee, Y.-H.W.; You, L.-R. Targeted inactivation of murine Ddx3x: Essential roles of Ddx3x in placentation and embryogenesis. Hum. Mol. Genet. 2016, 25, 2905–2922. [Google Scholar] [CrossRef][Green Version]
- Perfetto, M.; Xu, X.; Lu, C.; Shi, Y.; Yousaf, N.; Li, J.; Yien, Y.Y.; Wei, S. The RNA helicase DDX3 induces neural crest by promoting AKT activity. Development 2020, 148, dev184341. [Google Scholar] [CrossRef]
- The Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature 2014, 519, 223–228. [CrossRef]
- Cruciat, C.-M.; Dolde, C.; de Groot, R.E.A.; Ohkawara, B.; Reinhard, C.; Korswagen, H.C.; Niehrs, C. RNA Helicase DDX3 Is a Regulatory Subunit of Casein Kinase 1 in Wnt-β-Catenin Signaling. Science 2013, 339, 1436–1441. [Google Scholar] [CrossRef]
- Kremer, E.; Pritchard, M.; Lynch, M.; Yu, S.; Holman, K.; Baker, E.; Warren, S.T.; Schlessinger, D.; Sutherland, G.R.; Richards, R.I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science 1991, 252, 1711–1714. [Google Scholar] [CrossRef]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.-H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
- Pieretti, M.; Zhang, F.; Fu, Y.-H.; Warren, S.T.; Oostra, B.A.; Caskey, C.; Nelson, D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 1991, 66, 817–822. [Google Scholar] [CrossRef]
- Devys, D.; Biancalana, V.; Rousseau, F.; Boué, J.; Mandel, J.L.; Oberlé, I. Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am. J. Med. Genet. 1992, 43, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Agulhon, C.; Blanchet, P.; Kobetz, A.; Marchant, D.; Faucon, N.; Sarda, P.; Moraine, C.; Sittler, A.; Biancalana, V.; Malafosse, A.; et al. Expression of FMR1, FXR1, and FXR2 genes in human prenatal tissues. J. Neuropathol. Exp. Neurol. 1999, 58, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.P.; Bell, J. A PEDIGREE OF MENTAL DEFECT SHOWING SEX-LINKAGE. J. Neurol. Neurosurg. Psychiatry 1943, 6, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Mattei, J.F.; Mattei, M.-G.; Aumeras, C.; Auger, M.; Giraud, F. X-linked mental retardation with the fragile X. A study of 15 families. Qual. Life Res. 1981, 59, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Allen, G.A.; Singh, D.N.; Carpenter, N.J.; Hall, B.D. Preliminary communication: Photoanthropometric analysis of individuals with the fragile X syndrome. Am. J. Med. Genet. 1988, 30, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Grønskov, K.; Brøndum-Nielsen, K.; Dedic, A.; Hjalgrim, H. A nonsense mutation in FMR1 causing fragile X syndrome. Eur. J. Hum. Genet. 2011, 19, 489–491. [Google Scholar] [CrossRef] [PubMed]
- Heulens, I.; Suttie, M.; Postnov, A.; De Clerck, N.; Perrotta, C.S.; Mattina, T.; Faravelli, F.; Forzano, F.; Kooy, F.; Hammond, P. Craniofacial characteristics of fragile X syndrome in mouse and man. Eur. J. Hum. Genet. 2012, 21, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Hinds, H.L.; Ashley, C.T.; Sutcliffe, J.S.; Nelson, D.L.; Warren, S.T.; Housman, D.E.; Schalling, M. Tissue specific expression of FMR–1 provides evidence for a functional role in fragile X syndrome. Nat. Genet. 1993, 3, 36–43. [Google Scholar] [CrossRef]
- Bakker, C.E.; Verheij, C.; Willemsen, R.; van der Helm, R.; Oerlemans, F.; Vermey, M.; Bygrave, A.; Hoogeveen, A.; Oostra, B.A.; Reyniers, E.; et al. Fmr1 knockout mice: A model to study fragile X mental retardation. Cell 1994, 78, 23–33. [Google Scholar]
- Lim, J.H.; Booker, A.B.; Luo, T.; Williams, T.; Furuta, Y.; Lagutin, O.; Oliver, G.; Sargent, T.D.; Fallon, J.R. AP-2α selectively regulates fragile X mental retardation-1 gene transcription during embryonic development. Hum. Mol. Genet. 2005, 14, 2027–2034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lim, J.H.; Luo, T.; Sargent, T.D.; Fallon, J.R. Developmental expression of Xenopus Fragile X mental retardation-1 gene. Int. J. Dev. Biol. 2005, 49, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Gessert, S.; Bugner, V.; Tecza, A.; Pinker, M.; Kühl, M. FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev. Biol. 2010, 341, 222–235. [Google Scholar] [CrossRef]
- Tucker, B.; Richards, R.I.; Lardelli, M. Contribution of mGluR and Fmr1 functional pathways to neurite morphogenesis, craniofacial development and fragile X syndrome. Hum. Mol. Genet. 2006, 15, 3446–3458. [Google Scholar] [CrossRef]
- Broeder, M.J.D.; Van Der Linde, H.; Brouwer, J.R.; Oostra, B.A.; Willemsen, R.; Ketting, R.F. Generation and Characterization of Fmr1 Knockout Zebrafish. PLoS ONE 2009, 4, e7910. [Google Scholar] [CrossRef]
- Camargos, S.; Scholz, S.; Sánchez, J.S.; Paisán-Ruiz, C.; Lewis, P.; Hernandez, D.; Ding, J.; Gibbs, J.R.; Cookson, M.R.; Bras, J.; et al. DYT16, a novel young-onset dystonia-parkinsonism disorder: Identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 2008, 7, 207–215. [Google Scholar] [CrossRef]
- Zech, M.; Castrop, F.; Schormair, B.; Jochim, A.; Wieland, T.; Gross, N.; Lichtner, P.; Peters, A.; Gieger, C.; Meitinger, T.; et al. DYT16 revisited: Exome sequencing identifies PRKRA mutations in a European dystonia family. Mov. Disord. 2014, 29, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Rowe, T.M.; Rizzi, M.; Hirose, K.; Peters, G.; Sen, G.C. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc. Natl. Acad. Sci. USA 2006, 103, 5823–5828. [Google Scholar] [CrossRef]
- Bennett, R.; Blalock, W.L.; Choi, E.-J.; Lee, Y.J.; Zhang, Y.; Zhou, L.; Oh, S.P.; May, W.S. RAX is required for fly neuronal development and mouse embryogenesis. Mech. Dev. 2008, 125, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Peters, G.; Seachrist, D.D.; Keri, R.A.; Sen, G.C. The double-stranded RNA-binding protein, PACT, is required for postnatal anterior pituitary proliferation. Proc. Natl. Acad. Sci. USA 2009, 106, 10696–10701. [Google Scholar] [CrossRef]
- Dickerman, B.K.; White, C.L.; Chevalier, C.; Nalesso, V.; Charles, C.; Fouchécourt, S.; Guillou, F.; Viriot, L.; Sen, G.C.; Herault, Y. Missense Mutation in the Second RNA Binding Domain Reveals a Role for Prkra (PACT/RAX) during Skull Development. PLoS ONE 2011, 6, e28537. [Google Scholar] [CrossRef]
- Fowles, L.F.; Bennetts, J.S.; Berkman, J.L.; Williams, E.; Koopman, P.; Teasdale, R.; Wicking, C. Genomic screen for genes involved in mammalian craniofacial development. Genesis 2003, 35, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.E.; Jones, K.L.; Smith, F.J.; Williams, T.; Li, H. An Alternative Splicing Program for Mouse Craniofacial Development. Front. Physiol. 2020, 11, 1099. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; McMahon, J.J.; Tsai, Y.-H.; Wang, Z.; Silver, D.L. Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes p53-Mediated Microcephaly. PLoS Genet. 2016, 12, e1006282. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).