Exploring the Expression of Cardiac Regulators in a Vertebrate Extremophile: The Cichlid Fish Oreochromis (Alcolapia) alcalica
Abstract
1. Introduction
2. Methods
2.1. Fish maintenance and Embryo Collection
2.2. Phylogeny
2.3. RNA Extraction, Complementary DNA (cDNA) Synthesis, and Reverse-Transcription PCR (RT-PCR)
2.4. Whole-Mount In Situ Hybridisation
3. Results
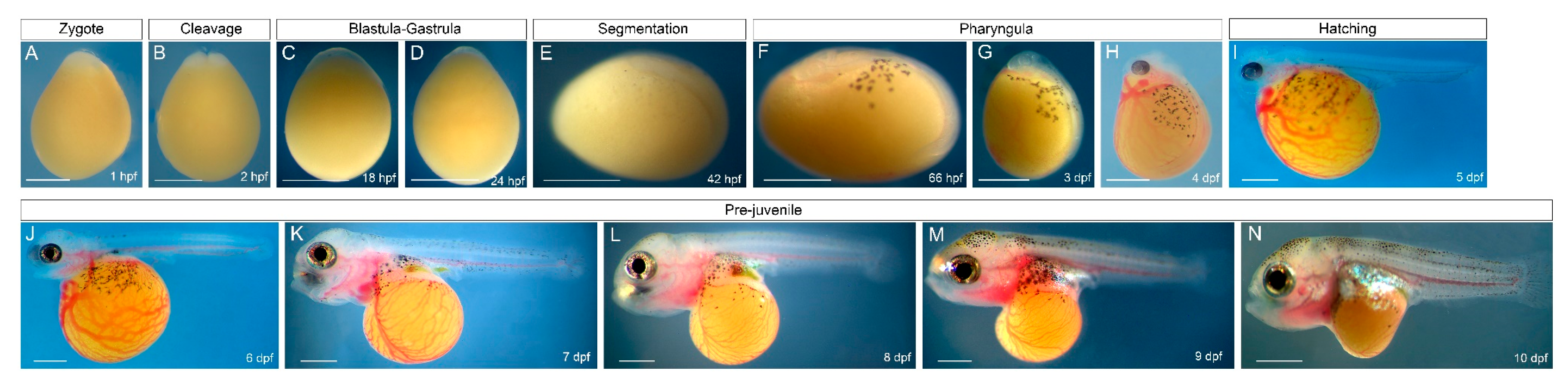
3.1. Early Development of O. alcalica
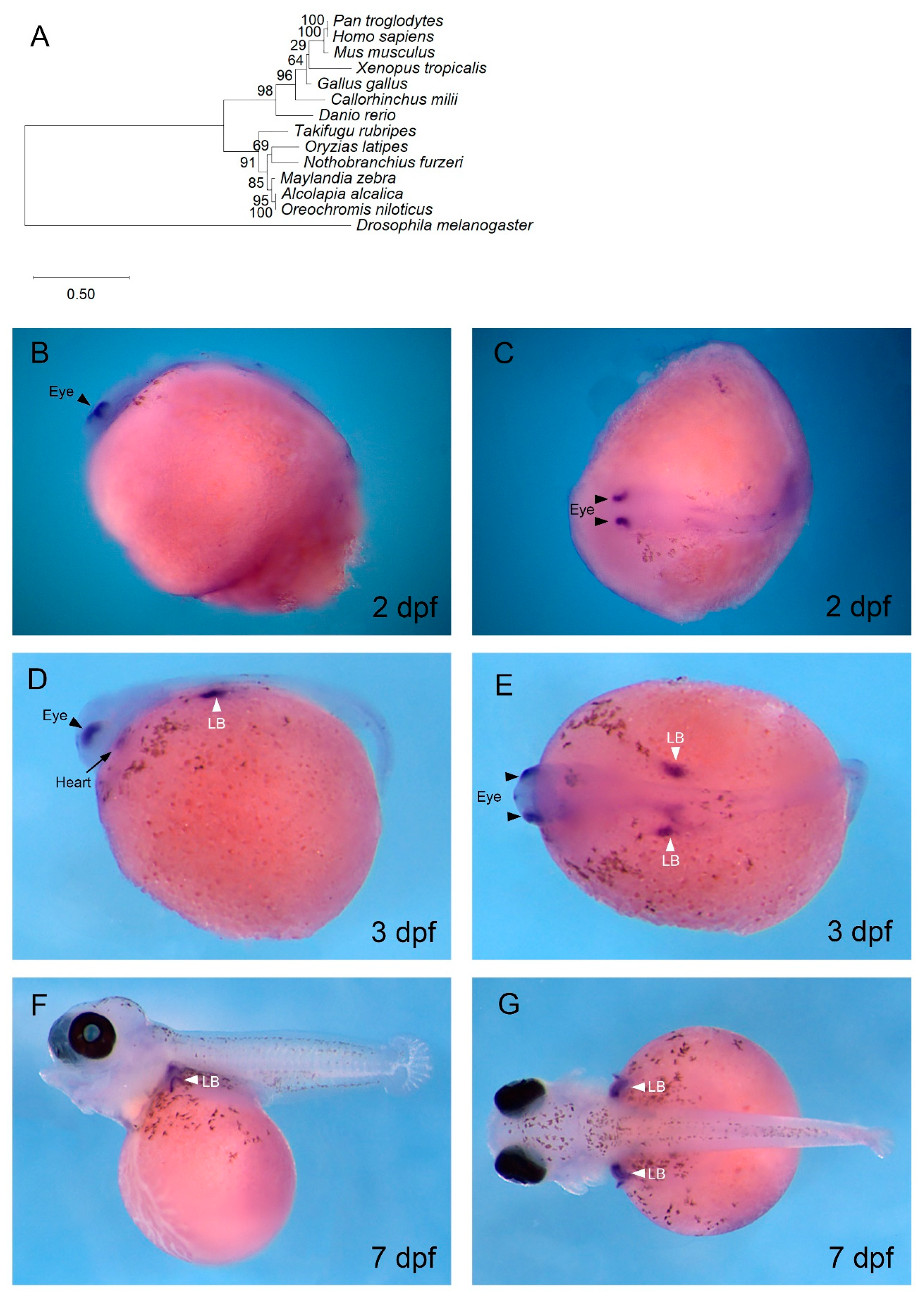
3.2. gata4 is Expressed in Cardiac and Haematopoietic Regions of O. alcalica Embryos
3.3. tbx5 is Expressed in the Developing Pectoral Fin, Eyes, and Heart of O. alcalica
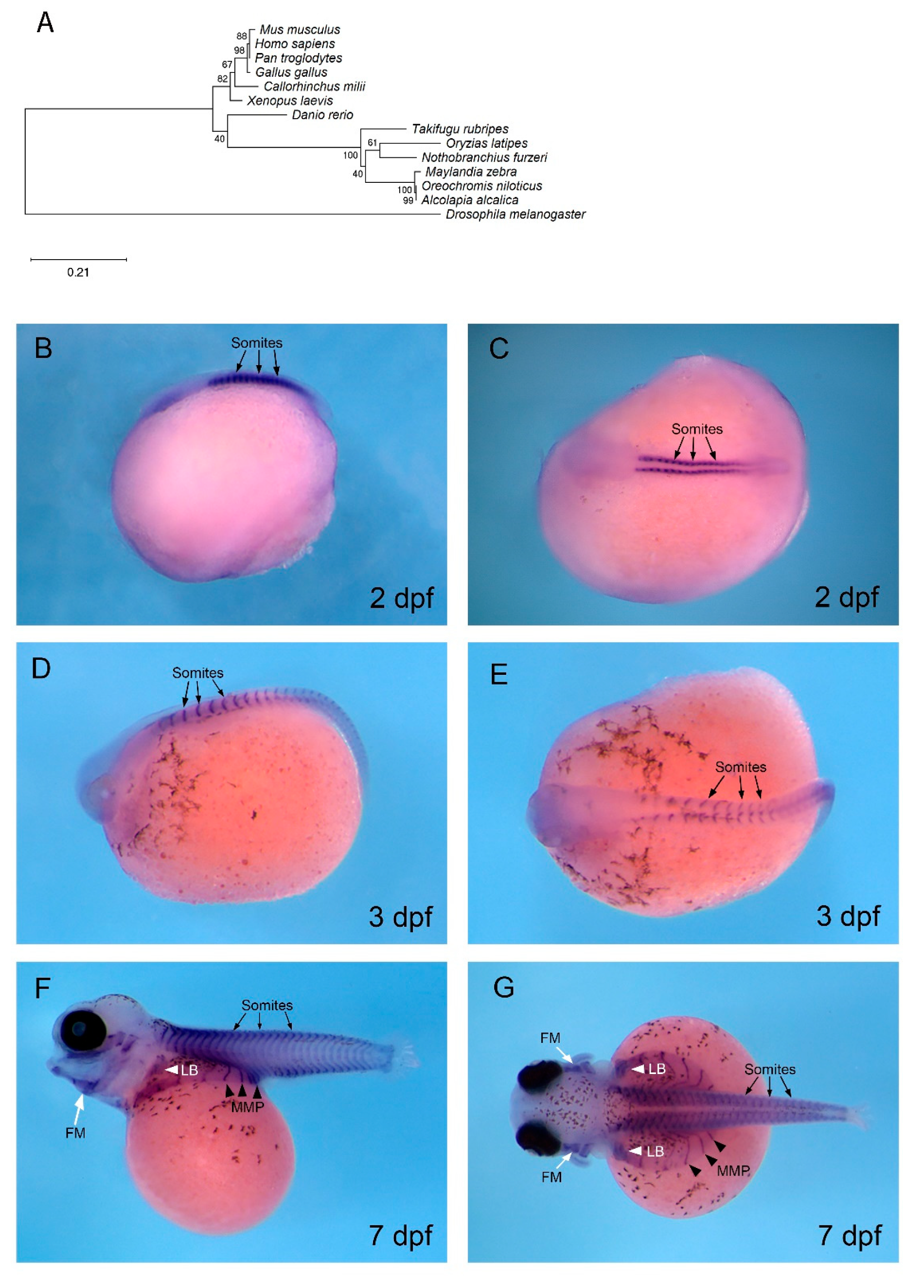
3.4. mef2c is Expressed in Developing Muscle in O. alcalica
4. Discussion
4.1. gata4
4.2. tbx5
4.3. mef2c
5. Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The Multifunctional Fish Gill: Dominant Site of Gas Exchange, Osmoregulation, Acid-Base Regulation, and Excretion of Nitrogenous Waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef]
- Hwang, P.-P.; Lee, T.-H.; Lin, L.-Y. Ion regulation in fish gills: Recent progress in the cellular and molecular mechanisms. Am. J. Physiol. Integr. Comp. Physiol. 2011, 301, R28–R47. [Google Scholar] [CrossRef]
- Helfman, G.; Collette, B.B.; Facey, D.E.; Bowen, B.W. The Diversity of Fishes: Biology, Evolution, and Ecology; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Wang, Y.; Guo, B. Adaption to extreme environments: A perspective from fish genomics. Rev. Fish. Biol. Fish. 2019, 29, 735–747. [Google Scholar] [CrossRef]
- Riesch, R.; Tobler, M.; Plath, M. Extremophile Fishes: Ecology, Evolution, and Physiology of Teleosts in Extreme Environments; Springer: New York, NY, USA, 2015. [Google Scholar]
- Garbarino, V.R.; Orr, M.E.; Rodriguez, K.A.; Buffenstein, R. Mechanisms of oxidative stress resistance in the brain: Lessons learned from hypoxia tolerant extremophilic vertebrates. Arch. Biochem. Biophys. 2015, 576, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.A. Extremophiles and their application to veterinary medicine. Environ. Technol. 2010, 31, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Jorge, C.D.; Borges, N.; Bagyan, I.; Bilstein, A.; Santos, H. Potential applications of stress solutes from extremophiles in protein folding diseases and healthcare. Extremophiles 2016, 20, 251–259. [Google Scholar] [CrossRef]
- Jeffery, W.R. Chapter 8 Evolution and Development in the Cavefish Astyanax. Curr. Top. Dev. Biol. 2009, 86, 191–221. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Iwamatsu, T. Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 2004, 121, 605–618. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D.; FishBase. World Wide Web electronic publication. Available online: https://www.scienceopen.com/document?vid=dc419213-0ca3-48cc-901c-2934ecf4441e (accessed on 2 January 2014).
- Crawford, D.L. Functional genomics does not have to be limited to a few select organisms. Genome Biol. 2001, 2, 1001. [Google Scholar]
- Kocher, T.D. Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 2004, 5, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Salzburger, W.; Meyer, A. The species flocks of East African cichlid fishes: Recent advances in molecular phylogenetics and population genetics. Naturwissenschaften 2004, 91, 277–290. [Google Scholar] [CrossRef] [PubMed]
- McGee, M.D.; Faircloth, B.C.; Borstein, S.R.; Zheng, J.; Hulsey, C.D.; Wainwright, P.C.; Alfaro, M.E. Replicated divergence in cichlid radiations mirrors a major vertebrate innovation. Proc. R. Soc. 2016, 283, 20151413. [Google Scholar] [CrossRef]
- Ford, A.G.P.; Bullen, T.R.; Pang, L.; Genner, M.J.; Bills, R.; Flouri, T.; Ngatunga, B.P.; Ruber, L.; Schliewen, U.K.; Seehausen, O.; et al. Molecular phylogeny of Oreochromis (Cichlidae: Oreochromini) reveals mito-nuclear discordance and multiple colonisation of adverse aquatic environments. Mol. Phylogenetics Evol. 2019, 136, 215–226. [Google Scholar] [CrossRef]
- Roberts, N.; Taieb, M.; Barker, P.; Damnati, B.; Icole, M.; Williamson, D. Timing of the Younger Dryas event in East Africa from lake-level changes. Nature 1993, 366, 146–148. [Google Scholar] [CrossRef]
- Tichy, H.; Seegers, L. The Oreochromis alcalicus flock (Teleostei Cichlidae) from lakes Natron and Magadi, Tanzania and Kenya: A model for the evolution of new species flocks in historical times. Ichthyol. Explor. Freshw. 1999, 10, 147–174. [Google Scholar]
- Ford, A.G.P.; Dasmahapatra, K.K.; Rüber, L.; Gharbi, K.; Cezard, T.; Day, J.J. High levels of interspecific gene flow in an endemic cichlid fish adaptive radiation from an extreme lake environment. Mol. Ecol. 2015, 24, 3421–3440. [Google Scholar] [CrossRef]
- Clarisse, L.; Van Damme, M.; Gardner, W.; Coheur, P.-F.; Clerbaux, C.; Whitburn, S.; Hadji-Lazaro, J.; Hurtmans, D. Atmospheric ammonia (NH3) emanations from Lake Natron’s saline mudflats. Sci. Rep. 2019, 9, 4441. [Google Scholar] [CrossRef]
- Randall, D.J.; Wood, C.M.; Perry, S.F.; Bergman, H.; Maloiy, G.M.O.; Mommsen, T.P.; Wright, P.A. Urea excretion as a strategy for survival in a fish living in a very alkaline environment. Nature 1989, 337, 165–166. [Google Scholar] [CrossRef]
- Narahara, A.; Bergman, H.L.; Laurent, P.; Maina, J.N.; Walsh, P.J.; Wood, C.M. Respiratory physiology of the Lake Magadi Tilapia (Oreochromis alcalicus grahami), a fish adapted to a hot, alkaline, and frequently hypoxic environment. Physiol. Zool. 1996, 69, 1114–1136. [Google Scholar] [CrossRef]
- Bergman, A.N.; Laurent, P.; Otiang’a-Owiti, G.; Bergman, H.L.; Walsh, P.J.; Wilson, P.; Wood, C.M. Physiological adaptations of the gut in the Lake Magadi tilapia, Alcolapia grahami, an alkaline- and saline-adapted teleost fish. Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2003, 136, 701–715. [Google Scholar] [CrossRef]
- Kavembe, G.D.; Franchini, P.; Irisarri, I.; Machado-Schiaffino, G.; Meyer, A. Genomics of Adaptation to Multiple Concurrent Stresses: Insights from Comparative Transcriptomics of a Cichlid Fish from One of Earth’s Most Extreme Environments, the Hypersaline Soda Lake Magadi in Kenya, East Africa. J. Mol. Evol. 2015, 81, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.M.; Brix, K.V.; De Boeck, G.; Bergman, H.L.; Bianchini, A.; Bianchini, L.F.; Maina, J.N.; Johannsson, O.E.; Kavembe, G.D.; Papah, M.B.; et al. Mammalian metabolic rates in the hottest fish on earth. Sci. Rep. 2016, 6, 26990. [Google Scholar] [CrossRef] [PubMed]
- White, L.J.; Sutton, G.; Shechonge, A.; Day, J.J.; Dasmahapatra, K.K.; Pownall, M.E. Adaptation of the carbamoyl-phosphate synthetase (CPS) enzyme in an extremophile fish. R. Soc. Open Sci. 2020, (in press).
- Bishopric, N.H. Evolution of the Heart from Bacteria to Man. Ann. New York Acad. Sci. 2005, 1047, 13–29. [Google Scholar] [CrossRef]
- Solc, D. The heart and heart conducting system in the kingdom of animals: A comparative approach to its evolution. Exp. Clin. Cardiol. 2007, 12, 113–118. [Google Scholar]
- Meijler, F.L.; Meijler, T.D. Archetype, adaptation and the mammalian heart. Neth. Hear. J. 2011, 19, 142–148. [Google Scholar] [CrossRef]
- Asnani, A.; Peterson, R.T. The zebrafish as a tool to identify novel therapies for human cardiovascular disease. Dis. Model. Mech. 2014, 7, 763–767. [Google Scholar] [CrossRef]
- Stainier, D.Y.; Lee, R.K.; Fishman, M.C. Cardiovascular development in zebrafish. I. Myocardial fate map and heart tube formation. Development 1993, 119, 31–40. [Google Scholar]
- Keegan, B.R.; Meyer, D.; Yelon, D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development 2004, 131, 3081–3091. [Google Scholar] [CrossRef]
- Yelon, D.; Horne, S.A.; Stainier, D.Y.R. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 1999, 214, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Rohr, S.; Otten, C.; Abdelilah-Seyfried, S. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ. Res. 2008, 102, 12–19. [Google Scholar] [CrossRef]
- Chen, J.N.; van Eeden, F.J.; Warren, K.S.; Chin, A.; Nusslein-Volhard, C.; Haffter, P.; Fishman, M.C. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 1997, 124, 4373–4382. [Google Scholar]
- Beis, D.; Bartman, T.; Jin, S.-W.; Scott, I.C.; D’Amico, L.A.; Ober, E.A.; Verkade, H.; Frantsve, J.; Field, H.A.; Wehman, A.; et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 2005, 132, 4193–4204. [Google Scholar] [CrossRef]
- Fossett, N.; Schulz, R.A. Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation 2001, 69, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Nemer, G.M.; Nemer, M. Regulation of heart development and function through combinatorial interactions of transcription factors. Ann. Med. 2001, 33, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Holt, M.; Oram, S. Familial heart disease with skeletal malformations. Br. Heart J. 1960, 22, 236–242. [Google Scholar] [CrossRef]
- Basson, C.T.; Cowley, G.S.; Solomon, S.D.; Weissman, B.; Poznanski, A.K.; Traill, T.A.; Seidman, J.G.; Seidman, C.E. The Clinical and Genetic Spectrum of the Holt-Oram Syndrome (Heart-Hand Syndrome). N. Engl. J. Med. 1994, 330, 885–891. [Google Scholar] [CrossRef]
- Newbury-Ecob, R.A.; Leanage, R.; Raeburn, J.A.; Young, I.D. Holt-Oram syndrome: A clinical genetic study. J. Med. Genet. 1996, 33, 300–307. [Google Scholar] [CrossRef]
- Basson, C.T.; Bachinsky, D.R.; Lin, R.C.; Levi, T.; Elkins, J.A.; Soults, J.; Grayzel, D.; Kroumpouzou, E.; Traill, T.A.; Leblanc-Straceski, J.; et al. Mutations in human cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1997, 15, 30–35. [Google Scholar] [CrossRef]
- Black, B.L.; Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Bio. 1998, 14, 167–196. [Google Scholar] [CrossRef] [PubMed]
- Lilly, B.; Zhao, B.; Ranganayakulu, G.; Paterson, B.M.; Schulz, R.A.; Olson, E.N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 1995, 267, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D.G.; Lyons, G.E.; Martin, J.F.; Olson, E.N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 1994, 120, 1251–1263. [Google Scholar] [PubMed]
- Ford, A.G.P.; Rüber, L.; Newton, J.; Dasmahapatra, K.K.; Balarin, J.D.; Bruun, K.; Day, J.J. Niche divergence facilitated by fine-scale ecological partitioning in a recent cichlid fish adaptive radiation. Evolution 2016, 70, 2718–2735. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fujimura, K.; Okada, N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Dev. Growth. Differ. 2007, 49, 301–324. [Google Scholar] [CrossRef]
- De Jong, I.M.L.; Witte, F.; Richardson, M.K. Developmental stages until hatching of the Lake Victoria cichlid Haplochromis piceatus (Teleostei: Cichlidae). J. Morphol. 2009, 270, 519–535. [Google Scholar] [CrossRef]
- Woltering, J.M.; Holzem, M.; Schneider, R.F.; Nanos, V.; Meyer, A. The skeletal ontogeny of Astatotilapia burtoni—a direct-developing model system for the evolution and development of the teleost body plan. BMC Dev. Biol. 2018, 18. [Google Scholar] [CrossRef]
- Flegler-Balon, C. Direct and Indirect Development in Fishes—Examples of Alternative Life-History Styles; Springer Science and Business Media LLC: Dordrecht, The Netherland, 1989; Volume 6, pp. 71–100. [Google Scholar]
- Balon, E.K. Alternative Ways to Become a Juvenile or a Definitive Phenotype (and on Some Persisting Linguistic Offenses). Environ. Boil. Fishes 1999, 56, 17–38. [Google Scholar] [CrossRef]
- Le Pabic, P.; Stellwag, E.J.; Scemama, J.-L. Embryonic Development and Skeletogenesis of the Pharyngeal Jaw Apparatus in the Cichlid Nile Tilapia (Oreochromis niloticus). Anat. Rec. 2009, 292, 1780–1800. [Google Scholar] [CrossRef]
- Holtzinger, A.; Evans, T. Gata4 regulates the formation of multiple organs. Development 2005, 132, 4005–4014. [Google Scholar] [CrossRef]
- Albalat, R.; Baquero, M.; Minguillon, C. Identification and characterisation of the developmental expression pattern of tbx5b, a novel tbx5 gene in zebrafish. Gene Expr. Patterns 2010, 10, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Kudoh, T.; Tsang, M.; Hukriede, N.A.; Chen, X.F.; Dedekian, M.; Clarke, C.J.; Kiang, A.; Schultz, S.; Epstein, J.A.; Toyama, R.; et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001, 11, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.; Dayhoff, M.O. Matrices for Detecting Distant Relationships. In Atlas of Protein Sequences; Dayhoff, M., Ed.; National Biomedical Research Foundation: Washington, DC, USA, 1979; pp. 353–358. [Google Scholar]
- Minko, K.; Bollerot, K.; Drevon, C.; Hallais, M.F.; Jaffredo, T. From mesoderm to blood islands: Patterns of key molecules during yolk sac erythropoiesis. Gene Expr. Patterns 2003, 3, 261–272. [Google Scholar] [CrossRef]
- Silver, L.; Palis, J. Initiation of murine embryonic erythropoiesis: A spatial analysis. Blood 1997, 89, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Chapman, D.L.; Garvey, N.; Hancock, S.; Alexiou, M.; Agulnik, S.I.; Gibson-Brown, J.J.; Cebra-Thomas, J.; Bollag, R.J.; Silver, L.M.; Papaioannou, V.E. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev. Dyn. 1996, 206, 379–390. [Google Scholar] [CrossRef]
- Gibson-Brown, J.J.; Agulnik, S.I.; Silver, L.M.; Papaioannou, V.E. Expression of T-box genes Tbx2-Tbx5 during chick organogenesis. Mech. Dev. 1998, 74, 165–169. [Google Scholar] [CrossRef]
- Gruenauer-Kloevekorn, C.; Reichel, M.B.; Duncker, G.I.W.; Froster, U.G. Molecular Genetic and Ocular Findings in Patients with Holt-Oram Syndrome. Ophthalmic Genet. 2005, 26, 1–8. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Ranganayakulu, G.; Zhao, B.; Dokidis, A.; Molkentin, J.D.; Olson, E.N.; Schulz, R.A. A Series of Mutations in the D-MEF2 Transcription Factor Reveal Multiple Functions in Larval and Adult Myogenesis in Drosophila. Dev. Biol. 1995, 171, 169–181. [Google Scholar] [CrossRef]
- Hinits, Y.; Pan, L.; Walker, C.; Dowd, J.; Moens, C.B.; Hughes, S.M. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 2012, 369, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Della Gaspera, B.; Armand, A.S.; Sequeira, I.; Lecolle, S.; Gallien, C.L.; Charbonnier, F.; Chanoine, C. The Xenopus MEF2 gene family: Evidence of a role for XMEF2C in larval tendon development. Dev. Biol. 2009, 328, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Brand-Saberi, B.; Christ, B. Evolution and development of distinct cell lineages derived from somites. Curr. Top. Dev. Biol. 2000, 48, 1–42. [Google Scholar] [CrossRef]
- Reiter, J.F.; Alexander, J.; Rodaway, A.; Yelon, D.; Patient, R.; Holder, N.; Stainier, D.Y.R. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999, 13, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Schoenebeck, J.J.; Keegan, B.R.; Yelon, D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev. Cell 2007, 13, 254–267. [Google Scholar] [CrossRef]
- Latinkic, B.V.; Kotecha, S.; Mohun, T.J. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development 2003, 130, 3865–3876. [Google Scholar] [CrossRef]
- Garg, V.; Kathiriya, I.S.; Barnes, R.; Schluterman, M.K.; King, I.N.; Butler, C.A.; Rothrock, C.R.; Eapen, R.S.; Hirayama-Yamada, K.; Joo, K.; et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003, 424, 443–447. [Google Scholar] [CrossRef]
- Rajagopal, S.K.; Ma, Q.; Obler, D.; Shen, J.; Manichaikul, A.; Tomita-Mitchell, A.; Boardman, K.; Briggs, C.; Garg, V.; Srivastava, D.; et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J. Mol. Cell. Cardiol. 2007, 43, 677–685. [Google Scholar] [CrossRef]
- Peterkin, T.; Gibson, A.; Patient, R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev. Biol. 2007, 311, 623–635. [Google Scholar] [CrossRef]
- Dobrzycki, T.; Lalwani, M.; Telfer, C.; Monteiro, R.; Patient, R. The roles and controls of GATA factors in blood and cardiac development. IUBMB Life 2019, 72, 39–44. [Google Scholar] [CrossRef]
- Ciau-Uitz, A.; Monteiro, R.; Kirmizitas, A.; Patient, R. Developmental hematopoiesis: Ontogeny, genetic programming and conservation. Exp. Hematol. 2014, 42, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.Y.; Chi, N.C.; Santoso, B.; Teng, S.; Stainier, D.Y.R.; Traver, D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010, 464, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Gore, A.V.; Pillay, L.M.; Galanternik, M.V.; Weinstein, B.M. The zebrafish: A fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e312. [Google Scholar] [CrossRef] [PubMed]
- Dzierzak, E. The emergence of definitive hematopoietic stem cells in the mammal. Curr. Opin. Hematol. 2005, 12, 197–202. [Google Scholar] [CrossRef]
- Cumano, A.; Godin, I. Ontogeny of the hematopoietic system. Annu. Rev. Immunol. 2007, 25, 745–785. [Google Scholar] [CrossRef]
- Johnson, G.R.; Moore, M.A.S. Role of stem cell migration in initiation of mouse foetal liver haemopoiesis. Nature 1975, 258, 726–728. [Google Scholar] [CrossRef]
- Houssaint, E. Differentiation of the mouse hepatic primordium. II. Extrinsic origin of the haemopoietic cell line. Cell Differ. 1981, 10, 243–252. [Google Scholar] [CrossRef]
- Keller, G.; Lacaud, G.; Robertson, S. Development of the hematopoietic system in the mouse. Exp. Hematol. 1999, 27, 777–787. [Google Scholar] [CrossRef]
- Murayama, E.; Kissa, K.; Zapata, A.; Mordelet, E.; Briolat, V.; Lin, H.-F.; Handin, R.I.; Herbomel, P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 2006, 25, 963–975. [Google Scholar] [CrossRef]
- Wolf, A.; Aggio, J.; Campbell, C.; Wright, F.; Marquez, G.; Traver, D.; Stachura, D.L. Zebrafish Caudal Haematopoietic Embryonic Stromal Tissue (CHEST) Cells Support Haematopoiesis. Sci. Rep. 2017, 7, 44644. [Google Scholar] [CrossRef]
- Ueno, H.; Weissman, I.L. The origin and fate of yolk sac hematopoiesis: Application of chimera analyses to developmental studies. Int. J. Dev. Biol. 2010, 54, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Ferkowicz, M.J.; Yoder, M.C. Blood island formation: Longstanding observations and modern interpretations. Exp. Hematol. 2005, 33, 1041–1047. [Google Scholar] [CrossRef]
- Detrich, H.W.; Kieran, M.W.; Chan, F.Y.; Barone, L.M.; Yee, K.; Rundstadler, J.A.; Pratt, S.; Ransom, D.; Zon, L.I. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 1995, 92, 10713–10717. [Google Scholar] [CrossRef] [PubMed]
- Aronson, B.E.; Stapleton, K.A.; Krasinski, S.D. Role of GATA factors in development, differentiation, and homeostasis of the small intestinal epithelium. Am. J. Physiol. Liver Physiol. Liver Physiol. 2014, 306, G474–G490. [Google Scholar] [CrossRef] [PubMed]
- Pevny, L.; Simon, M.C.; Robertson, E.; Klein, W.H.; Tsai, S.-F.; D’Agati, V.; Orkin, S.H.; Costantini, F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 1991, 349, 257–260. [Google Scholar] [CrossRef]
- Tsai, F.-Y.; Keller, G.; Kuo, F.C.; Weiss, M.; Chen, J.; Rosenblatt, M.; Alt, F.W.; Orkin, S.H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994, 371, 221–226. [Google Scholar] [CrossRef]
- Weiss, M.J.; Keller, G.; Orkin, S.H. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994, 8, 1184–1197. [Google Scholar] [CrossRef]
- Bruneau, B.G.; Nemer, G.; Schmitt, J.P.; Charron, F.; Robitaille, L.; Caron, S.; Conner, D.A.; Gessler, M.; Nemer, M.; Seidman, C.E.; et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001, 106, 709–721. [Google Scholar] [CrossRef]
- Garrity, D.M.; Childs, S.; Fishman, M.C. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development 2002, 129, 4635–4645. [Google Scholar]
- Mommersteeg, M.T.M.; Soufan, A.T.; de Lange, F.J.; van den Hoff, M.J.B.; Anderson, R.H.; Christoffels, V.M.; Moorman, A.F.M. Two distinct pools of mesenchyme contribute to the development of the atrial septum. Circ. Res. 2006, 99, 351–353. [Google Scholar] [CrossRef]
- Xie, L.; Hoffmann, A.D.; Burnicka-Turek, O.; Friedland-Little, J.M.; Zhang, K.; Moskowitz, I.P. Tbx5-Hedgehog Molecular Networks Are Essential in the Second Heart Field for Atrial Septation. Dev. Cell 2012, 23, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Gramolini, A.O.; Walsh, M.A.; Zhou, Y.-Q.; Slorach, C.; Friedberg, M.K.; Takeuchi, J.K.; Sun, H.; Henkelman, R.M.; Backx, P.H.; et al. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc. Natl. Acad. Sci. USA 2008, 105, 5519–5524. [Google Scholar] [CrossRef] [PubMed]
- Pi-Roig, A.; Martin-Blanco, E.; Minguillon, C. Distinct tissue-specific requirements for the zebrafish tbx5 genes during heart, retina and pectoral fin development. Open Biol. 2014, 4, 140014. [Google Scholar] [CrossRef] [PubMed]
- Koshiba-Takeuchi, K.; Takeuchi, J.K.; Matsumoto, K.; Momose, T.; Uno, K.; Hoepker, V.; Ogura, K.; Takahashi, N.; Nakamura, H.; Yasuda, K.; et al. Tbx5 and the retinotectum projection. Science 2000, 287, 134–137. [Google Scholar] [CrossRef]
- Anderson, E.A.T.B.; Ho, R.K. A transcriptomics analysis of the Tbx5 paralogues in zebrafish. PLoS ONE 2018, 13, e0208766. [Google Scholar] [CrossRef]
- Tamura, K.; Yonei-Tamura, S.; Belmonte, J.C.I. Differential expression of Tbx4 and Tbx5 in Zebrafish fin buds. Mech. Dev. 1999, 87, 181–184. [Google Scholar] [CrossRef]
- Begemann, G.; Ingham, P.W. Developmental regulation of Tbx5 in zebrafish embryogenesis. Mech. Dev. 2000, 90, 299–304. [Google Scholar] [CrossRef]
- Agarwal, P.; Wylie, J.N.; Galceran, J.; Arkhitko, O.; Li, C.; Deng, C.; Grosschedl, R.; Bruneau, B.G. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 2003, 130, 623–633. [Google Scholar] [CrossRef]
- Mercader, N. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev. Growth Differ. 2007, 49, 421–437. [Google Scholar] [CrossRef]
- Adachi, N.; Robinson, M.; Goolsbee, A.; Shubin, N.H. Regulatory evolution of Tbx5 and the origin of paired appendages. Proc.S Natl. Acad. Sci. USA 2016, 113, 10115–10120. [Google Scholar] [CrossRef]
- Cunningham, T.J.; Lancman, J.J.; Berenguer, M.; Dong, P.D.S.; Duester, G. Genomic Knockout of Two Presumed Forelimb Tbx5 Enhancers Reveals They Are Nonessential for Limb Development. Cell Rep. 2018, 23, 3146–3151. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Verzi, M.P.; McCulley, D.J.; De Val, S.; Dodou, E.; Black, B.L. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 2005, 287, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Vong, L.; Bi, W.; O’Connor-Halligan, K.E.; Li, C.; Cserjesi, P.; Schwarz, J.J. MEF2C is required for the normal allocation of cells between the ventricular and sinoatrial precursors of the primary heart field. Dev. Dyn. 2006, 235, 1809–1821. [Google Scholar] [CrossRef]
- Materna, S.C.; Sinha, T.; Barnes, R.M.; van Bueren, K.L.; Black, B.L. Cardiovascular development and survival require Mef2c function in the myocardial but not the endothelial lineage. Dev. Biol. 2019, 445, 170–177. [Google Scholar] [CrossRef]
- Hinits, Y.; Hughes, S.M. Mef2s are required for thick filament formation in nascent muscle fibres. Development 2007, 134, 2511–2519. [Google Scholar] [CrossRef]
- Lazic, S.; Scott, I.C. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 2011, 354, 123–133. [Google Scholar] [CrossRef]
- Ticho, B.S.; Stainier, D.Y.R.; Fishman, M.C.; Breitbart, R.E. Three zebrafish MEF2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech. Dev. 1996, 59, 205–218. [Google Scholar] [CrossRef]
- Torgersen, J.S.; Takle, H.; Andersen, Ø. Differential spatial expression of mef2 paralogs during cardiac development in Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. Part. B Biochem. Mol. Biol. 2011, 158, 181–187. [Google Scholar] [CrossRef]
- Breitbart, R.E.; Liang, C.-s.; Smoot, L.B.; Laheru, D.A.; Mahdavi, V.; Nadal-Ginard, B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development 1993, 118, 1095–1106. [Google Scholar]
- Ganassi, M.; Badodi, S.; Polacchini, A.; Baruffaldi, F.; Hughes, S.M.; Hinits, Y.; Molinari, S. Distinct functions of alternatively spliced isoforms encoded by zebrafish mef2ca and mef2cb. Biochim. Et Biophy. Acta (BBA) 2014, 1839, 559–570. [Google Scholar] [CrossRef]
- Pownall, M.E.; Gustafsson, M.K.; Emerson, C.P. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002, 18, 747–783. [Google Scholar] [CrossRef] [PubMed]
- Hinits, Y.; Osborn, D.P.S.; Hughes, S.M. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development 2009, 136, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Cole, N.J.; Currie, P.D. Insights from sharks: Evolutionary and developmental models of fin development. Dev. Dyn. 2007, 236, 2421–2431. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.K.; Moran-Rivard, L.; Velasquez, T.; Nakatsu, M.N.; Jagla, K.; Goulding, M. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 2000, 127, 413–424. [Google Scholar]
- Neyt, C.; Jagla, K.; Thisse, C.; Thisse, B.; Haines, L.; Currie, P.D. Evolutionary origins of vertebrate appendicular muscle. Nature 2000, 408, 82–86. [Google Scholar] [CrossRef]
- Cinnamon, Y.; Kahane, N.; Kalcheim, C. Characterization of the early development of specific hypaxial muscles from the ventrolateral myotome. Development 1999, 126, 4305–4315. [Google Scholar]
- Okamoto, E.; Kusakabe, R.; Kuraku, S.; Hyodo, S.; Robert-Moreno, A.; Onimaru, K.; Sharpe, J.; Kuratani, S.; Tanaka, M. Migratory appendicular muscles precursor cells in the common ancestor to all vertebrates. Nat. Ecol. Evol. 2017, 1, 1731–1736. [Google Scholar] [CrossRef]

| Gene | Forward Primer 5′–3′ | Reverse Primer 5′–3′ |
|---|---|---|
| O. alcalica gata4 | TGTCTCCGCGCTTCACCTTCTCCA | GACCGGCTCTCCCTCTGCGTTCC |
| O. alcalica mef2c | ATGGGGCGAAAGAAGAT | AGCCCACCCTGATTACTG |
| O. alcalica tbx5 | AGAGGCAGCGACGACAATGAGC | GGGGGATAGGAGGAGGGGTGATAG |
| Zebrafish gata4 | CCTACAGGCACCCCAGCAGAGCAG | CCCGCCGCCACAGAGGAGTC |
| Zebrafish mef2ca | TTGCGCGGATAATGGACGAACG | GGGGGCCGGTGGGTGACTC |
| Zebrafish tbx5b | GCTCCCCGCCACTACAAACTCAG | GATATGTCCGAAAGGGTCCAGGTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutton, G.; White, L.J.; Ford, A.G.P.; Shechonge, A.; Day, J.J.; Dasmahapatra, K.K.; Pownall, M.E. Exploring the Expression of Cardiac Regulators in a Vertebrate Extremophile: The Cichlid Fish Oreochromis (Alcolapia) alcalica. J. Dev. Biol. 2020, 8, 22. https://doi.org/10.3390/jdb8040022
Sutton G, White LJ, Ford AGP, Shechonge A, Day JJ, Dasmahapatra KK, Pownall ME. Exploring the Expression of Cardiac Regulators in a Vertebrate Extremophile: The Cichlid Fish Oreochromis (Alcolapia) alcalica. Journal of Developmental Biology. 2020; 8(4):22. https://doi.org/10.3390/jdb8040022
Chicago/Turabian StyleSutton, Gemma, Lewis J. White, Antonia G.P. Ford, Asilatu Shechonge, Julia J. Day, Kanchon K. Dasmahapatra, and Mary E. Pownall. 2020. "Exploring the Expression of Cardiac Regulators in a Vertebrate Extremophile: The Cichlid Fish Oreochromis (Alcolapia) alcalica" Journal of Developmental Biology 8, no. 4: 22. https://doi.org/10.3390/jdb8040022
APA StyleSutton, G., White, L. J., Ford, A. G. P., Shechonge, A., Day, J. J., Dasmahapatra, K. K., & Pownall, M. E. (2020). Exploring the Expression of Cardiac Regulators in a Vertebrate Extremophile: The Cichlid Fish Oreochromis (Alcolapia) alcalica. Journal of Developmental Biology, 8(4), 22. https://doi.org/10.3390/jdb8040022






