Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Rearing and Crosses
2.2. Production of BACfoxd3::GFP Transgenic Zebrafish
2.3. Forskolin and H-89 Treatments
2.4. Cell Quantification and Statistics
2.5. In Situ Hybridization
2.6. Antibody Staining and Confocal Imaging
3. Results and Discussion
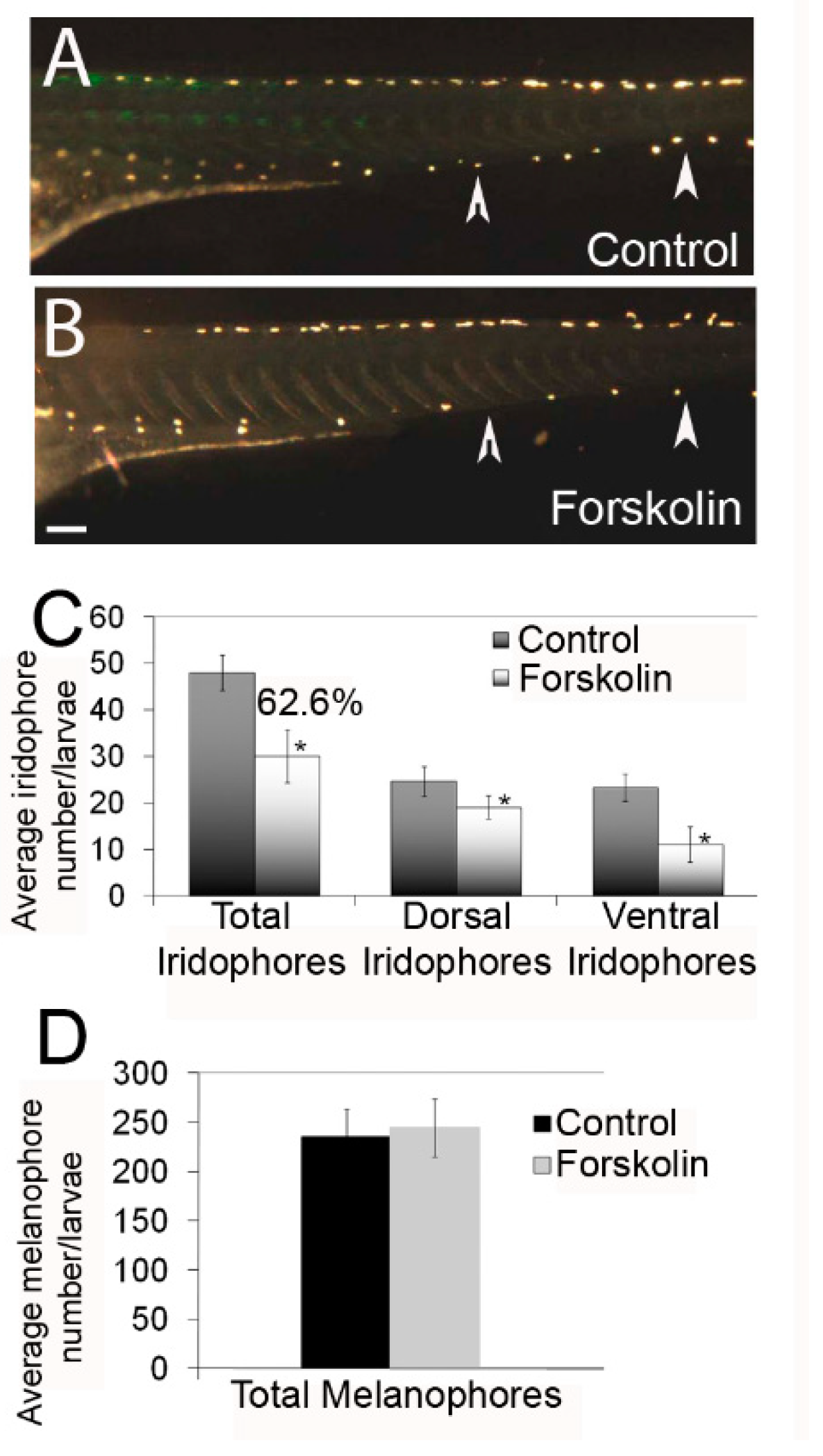
3.1. Effects of Protein Kinase A Activity Alter the Number of Differentiated Iridophores
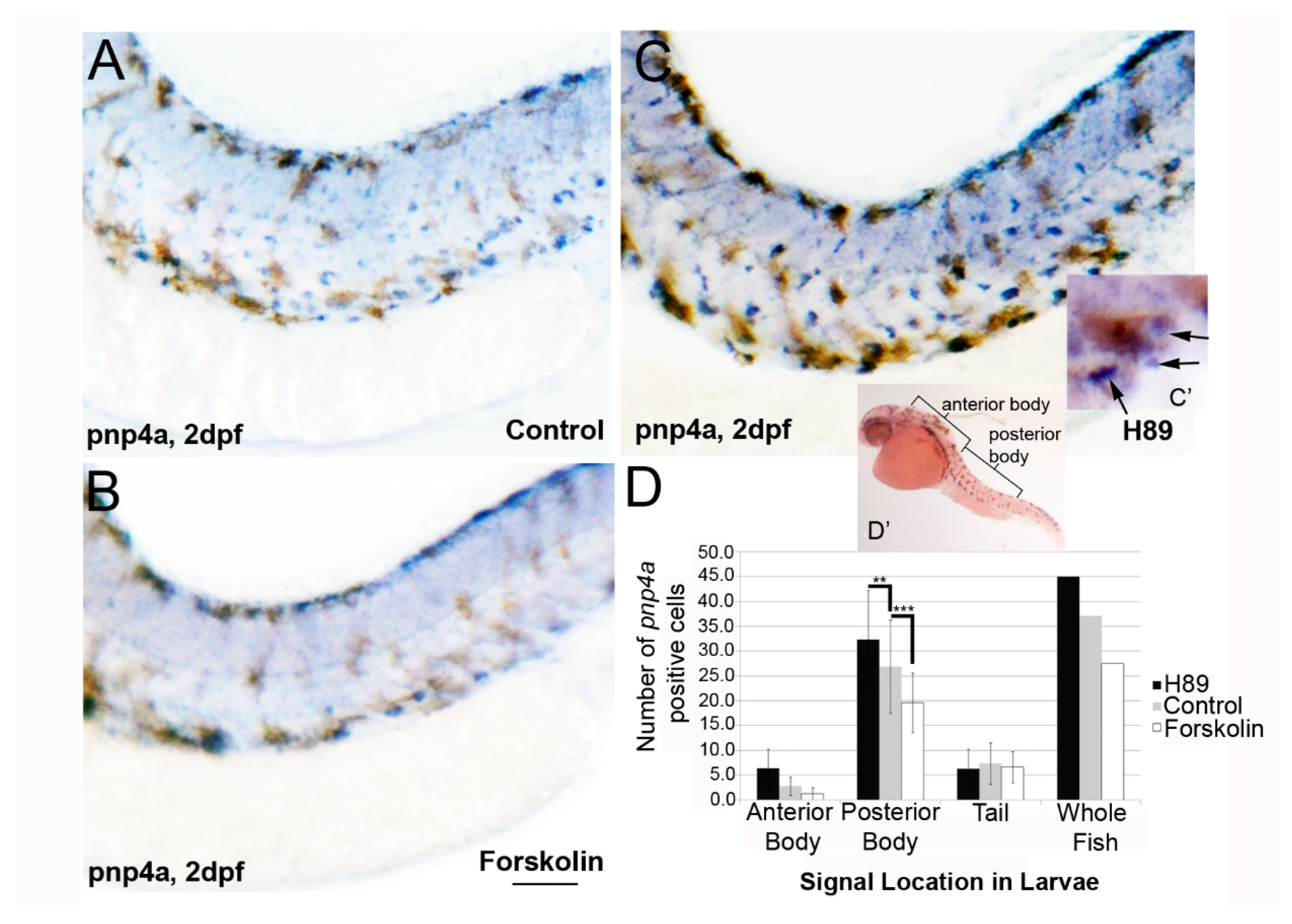
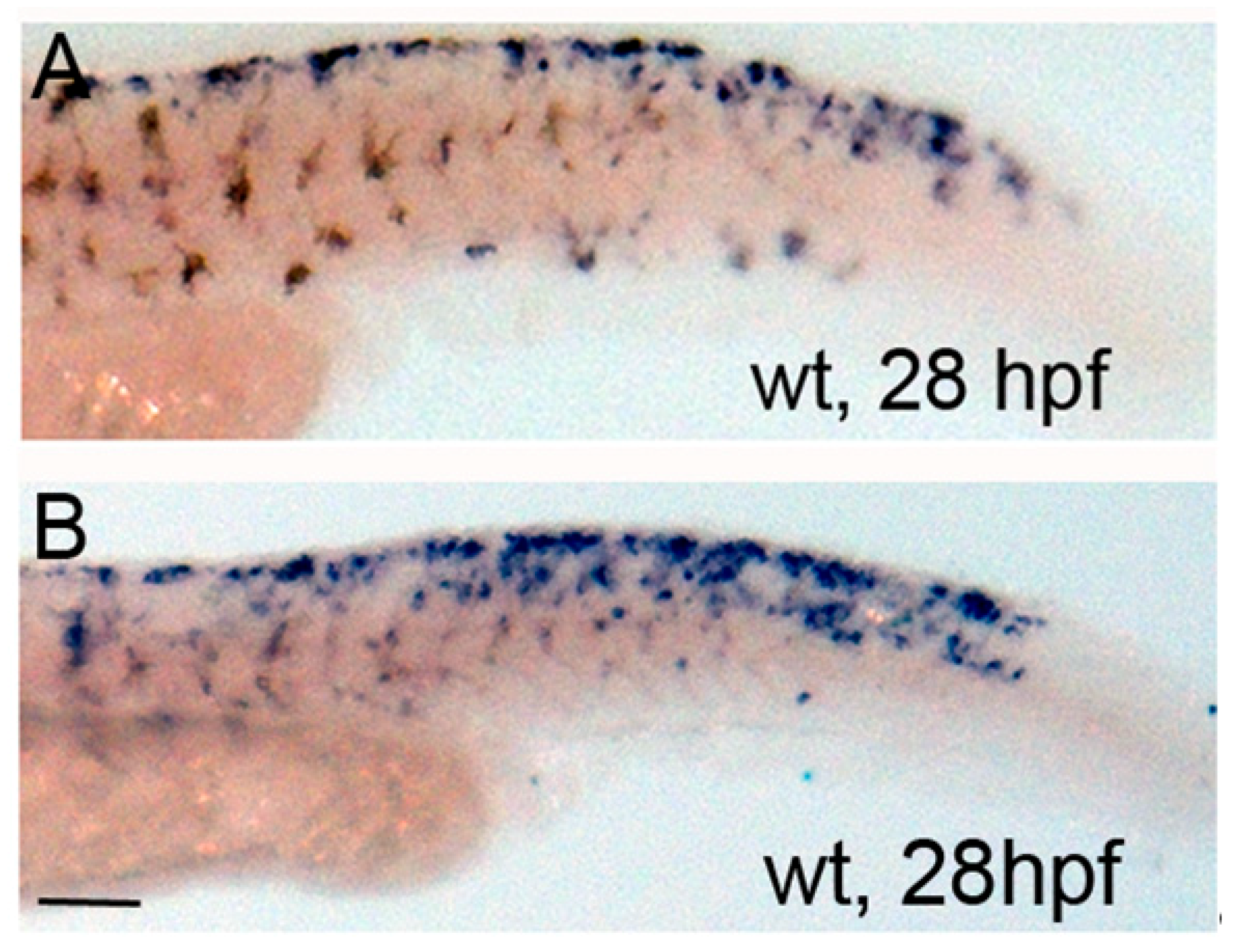
3.2. PKA Modulation Inhibits Iridophore Gene Expression
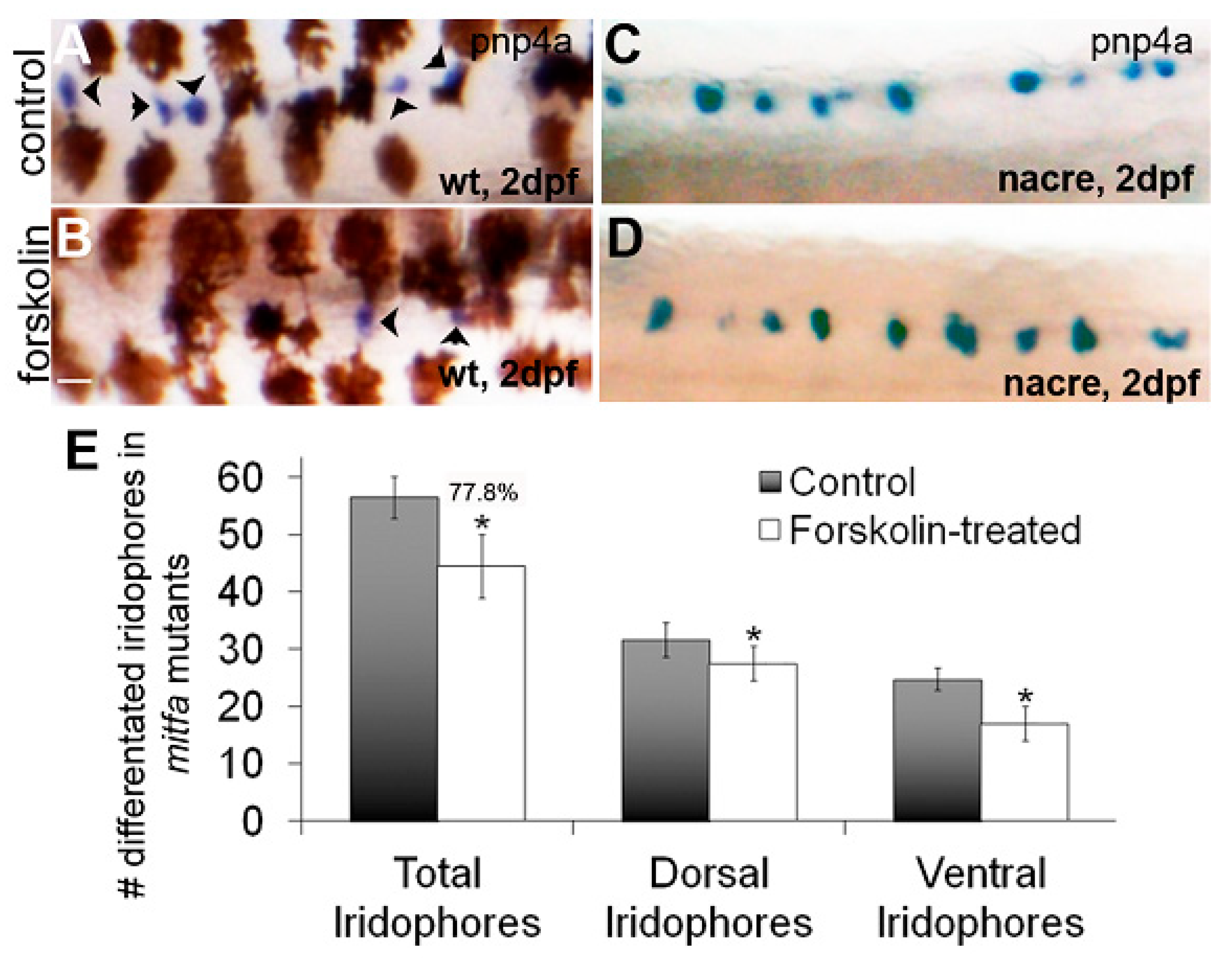
3.3. mitfa Is Partially Required for the Forskolin-Dependent Reduction in Iridophore Number
3.4. Activation of PKA Expands Neural Crest Marker foxd3 Expression
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fujii, R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000, 13, 300–319. [Google Scholar] [CrossRef] [PubMed]
- Beirl, A.J.; Linbo, T.H.; Cobb, M.J.; Cooper, C.D. oca2 regulation of chromatophore differentiation and number is cell type specific in zebrafish. Pigment Cell Melanoma Res. 2014, 27, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Clancey, L.F.; Beirl, A.J.; Linbo, T.H.; Cooper, C.D. Maintenance of Melanophore Morphology and Survival Is Cathepsin and vps11 Dependent in Zebrafish. PLoS ONE 2013, 8, e65096. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.D.; Linbo, T.H.; Raible, D.W. Kit and foxd3 genetically interact to regulate melanophore survival in zebrafish. Dev. Dyn. 2009, 238, 875–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hultman, K.A.; Bahary, N.; Zon, L.I.; Johnson, S.L. Gene Duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007, 3, e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lister, J.A.; Close, J.; Raible, D.W. Duplicate mitf genes in zebrafish: Complementary expression and conservation of melanogenic potential. Dev. Biol. 2001, 237, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.A.; Robertson, C.P.; Lepage, T.; Johnson, S.L.; Raible, D.W. Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 1999, 126, 3757–3767. [Google Scholar] [PubMed]
- Parichy, D.M.; Rawls, J.F.; Pratt, S.J.; Whitfield, T.T.; Johnson, S.L. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development 1999, 126, 3425–3436. [Google Scholar] [PubMed]
- Thomas, J.L.; Vihtelic, T.S.; denDekker, A.D.; Willer, G.; Luo, X.; Murphy, T.R.; Gregg, R.G.; Hyde, D.R.; Thummel, R. The loss of vacuolar protein sorting 11 (vps11) causes retinal pathogenesis in a vertebrate model of syndromic albinism. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3119–3128. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.; Lister, J.A.; Kunkel, G.R.; Prendergast, A.; Parichy, D.M.; Raible, D.W. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 2010, 344, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skroblin, P.; Grossmann, S.; Schafer, G.; Rosenthal, W.; Klussmann, E. Mechanisms of protein kinase A anchoring. Int. Rev. Cell Mol. Biol. 2010, 283, 235–330. [Google Scholar] [PubMed]
- Ruehr, M.L.; Russell, M.A.; Bond, M. A-kinase anchoring protein targeting of protein kinase A in the heart. J. Mol. Cell Cardiol. 2004, 37, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Sassone-Corsi, P. Cyclic AMP signalling. J. Cell Sci. 2001, 114, 1971–1972. [Google Scholar] [PubMed]
- Vandamme, J.; Castermans, D.; Thevelein, J.M. Molecular mechanisms of feedback inhibition of protein kinase A on intracellular cAMP accumulation. Cell Signal. 2012, 24, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Ge, S.; Yang, L.; Yang, M.; Ye, Z.; Yan, M.; Du, J.; Luo, Z. Ginsenosides Rb1 and Rg1 promote proliferation and expression of neurotrophic factors in primary Schwann cell cultures. Brain Res. 2010, 1357, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Sheets, L.; Ransom, D.G.; Mellgren, E.M.; Johnson, S.L.; Schnapp, B.J. Zebrafish melanophilin facilitates melanosome dispersion by regulating dynein. Curr. Biol. 2007, 17, 1721–1734. [Google Scholar] [CrossRef] [PubMed]
- Oshima, N.; Nakata, E.; Ohta, M.; Kamagata, S. Light-induced pigment aggregation in xanthophores of the medaka, Oryzias latipes. Pigment Cell Res. 1998, 11, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Fekete, D.M.; Andrisani, O.M. CtBP2 downregulation during neural crest specification induces expression of Mitf and REST, resulting in melanocyte differentiation and sympathoadrenal lineage suppression. Mol. Cell Biol. 2011, 31, 955–970. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Buchholz, F.; Muyrers, J.P.; Stewart, A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998, 20, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B.; Schilling, T.F.; Postlethwait, J.H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 1993, 119, 1203–1215. [Google Scholar] [PubMed]
- Wang, S.; Li, Z.; Shen, H.; Zhang, Z.; Yin, Y.; Wang, Q.; Zhao, X.; Ji, J. Quantitative Phosphoproteomic Study Reveals that Protein Kinase A Regulates Neural Stem Cell Differentiation Through Phosphorylation of Catenin Beta-1 and Glycogen synthase Kinase 3β. Stem Cells 2016, 34, 2090–2101. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.A.; Cooper, C.; Nguyen, K.; Modrell, M.; Grant, K.; Raible, D.W. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev. Biol. 2006, 290, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.A.; Arduini, B.L.; Berghmans, S.; George, R.E.; Kanki, J.P.; Henion, P.D.; Look, A.T. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol. 2006, 292, 174–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curran, K.; Raible, D.W.; Lister, J.A. Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev. Biol. 2009, 332, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Abel, E.V.; Aplin, A.E. FOXD3 is a mutant B-RAF-regulated inhibitor of G(1)-S progression in melanoma cells. Cancer Res. 2010, 70, 2891–2900. [Google Scholar] [CrossRef] [PubMed]
- Abel, E.V.; Basile, K.J.; Kugel, C.H., 3rd; Witkiewicz, A.K.; Le, K.; Amaravadi, R.K.; Karakousis, G.C.; Xu, X.; Xu, W.; Schuchter, L.M.; et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J. Clin. Investig. 2013, 123, 2155–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmour, D.T.; Maischein, H.M.; Nusslein-Volhard, C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron 2002, 34, 577–588. [Google Scholar] [CrossRef]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cooper, C.D.; Erickson, S.D.; Yin, S.; Moravec, T.; Peh, B.; Curran, K. Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish. J. Dev. Biol. 2018, 6, 23. https://doi.org/10.3390/jdb6040023
Cooper CD, Erickson SD, Yin S, Moravec T, Peh B, Curran K. Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish. Journal of Developmental Biology. 2018; 6(4):23. https://doi.org/10.3390/jdb6040023
Chicago/Turabian StyleCooper, Cynthia D., Steve D. Erickson, Scott Yin, Trevor Moravec, Brian Peh, and Kevin Curran. 2018. "Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish" Journal of Developmental Biology 6, no. 4: 23. https://doi.org/10.3390/jdb6040023
APA StyleCooper, C. D., Erickson, S. D., Yin, S., Moravec, T., Peh, B., & Curran, K. (2018). Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish. Journal of Developmental Biology, 6(4), 23. https://doi.org/10.3390/jdb6040023




