Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models
Abstract
1. Introduction
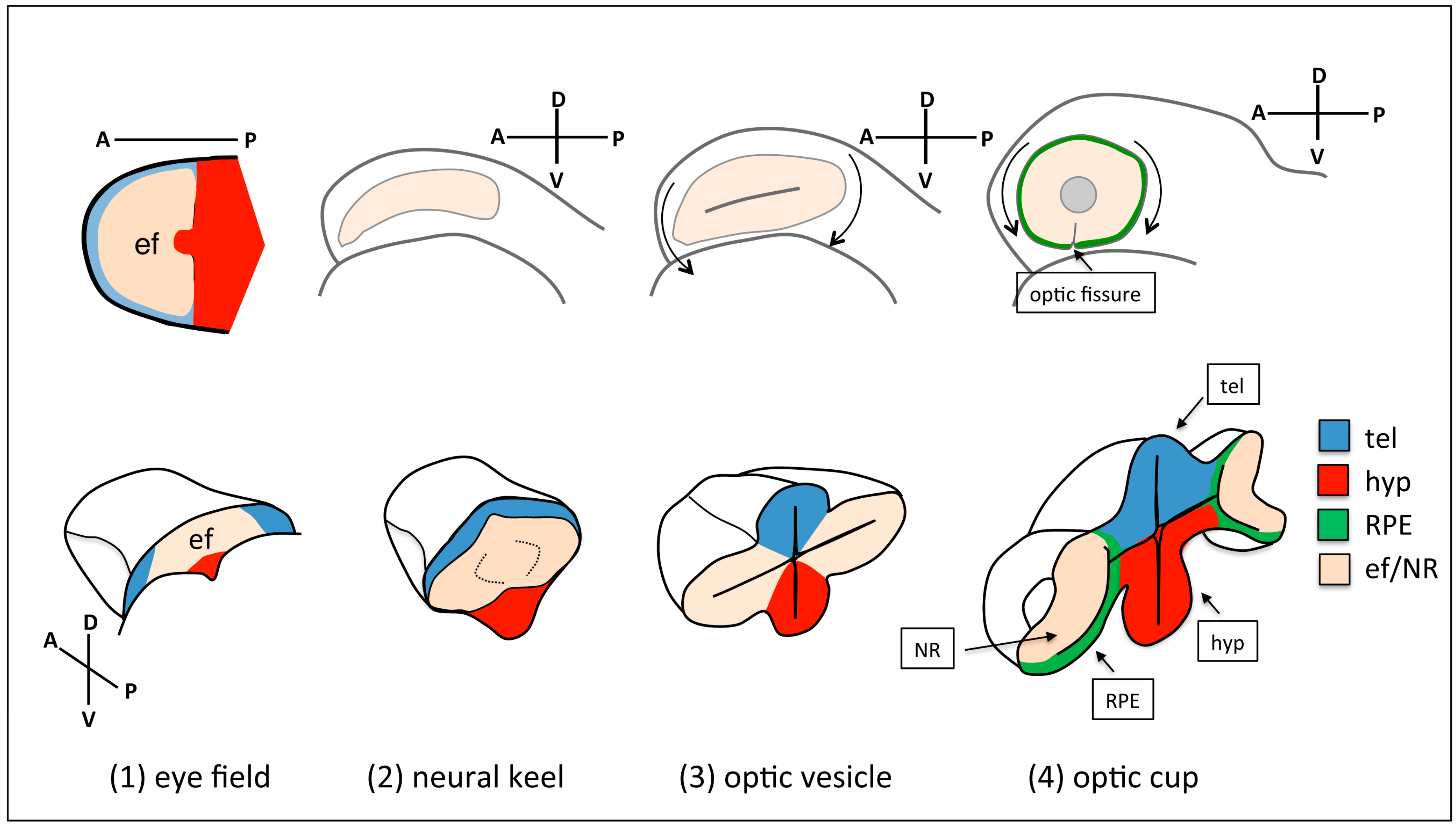
2. Overview of Eye Morphogenesis
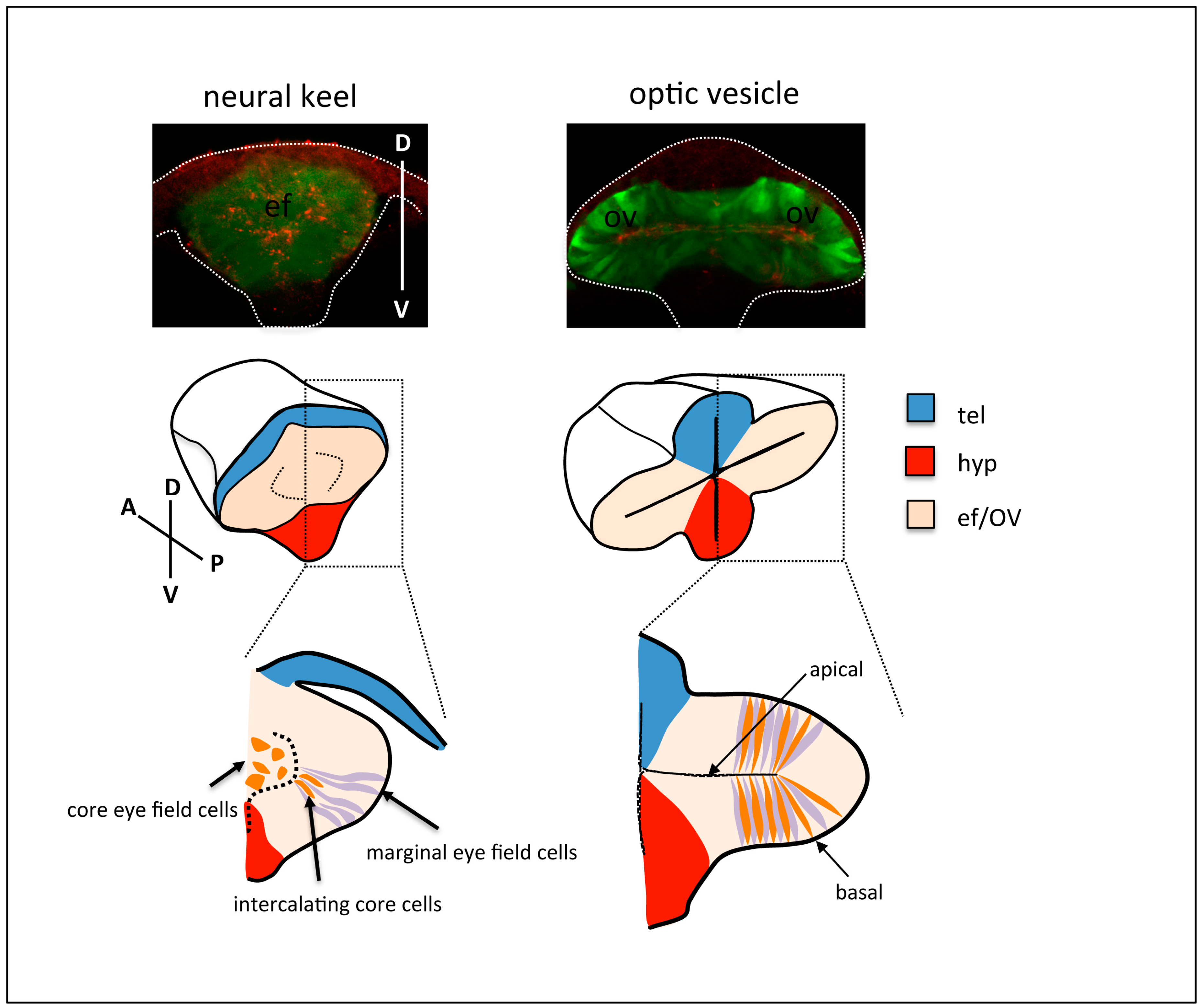
3. Visualising Global Cellular Rearrangements and Tissue Remodelling
4. Following Changes in Cell Shape and Cytoskeletal Dynamics
5. Assessing the Coordination between Fate Specification and Morphogenesis
6. Perspective
Acknowledgments
Conflicts of Interest
References
- Pantazis, P.; Supatto, W. Advances in whole-embryo imaging: A quantitative transition is underway. Nat. Rev. Mol. Cell Biol. 2014, 15, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Vacaru, A.M.; Unlu, G.; Spitzner, M.; Mione, M.; Knapik, E.W.; Sadler, K.C. In vivo cell biology in zebrafish—Providing insights into vertebrate development and disease. J. Cell Sci. 2014, 127, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.W. Growing up is stressful: Biophysical laws of morphogenesis. Curr. Opin. Genet. Dev. 2011, 21, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Colombelli, J.; Solon, J. Force communication in multicellular tissues addressed by laser nanosurgery. Cell Tissue Res. 2013, 352, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Behrndt, M.; Salbreux, G.; Campinho, P.; Hauschild, R.; Oswald, F.; Roensch, J.; Grill, S.W.; Heisenberg, C.P. Forces driving epithelial spreading in zebrafish gastrulation. Science 2012, 338, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Pestel, J.; Ramadass, R.; Gauvrit, S.; Helker, C.; Herzog, W.; Stainier, D.Y. Real-time 3d visualization of cellular rearrangements during cardiac valve formation. Development 2016, 143, 2217–2227. [Google Scholar] [CrossRef] [PubMed]
- Blaser, H.; Reichman-Fried, M.; Castanon, I.; Dumstrei, K.; Marlow, F.L.; Kawakami, K.; Solnica-Krezel, L.; Heisenberg, C.P.; Raz, E. Migration of zebrafish primordial germ cells: A role for myosin contraction and cytoplasmic flow. Dev. Cell 2006, 11, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Durdu, S.; Iskar, M.; Revenu, C.; Schieber, N.; Kunze, A.; Bork, P.; Schwab, Y.; Gilmour, D. Luminal signalling links cell communication to tissue architecture during organogenesis. Nature 2014, 515, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, S. Eye morphogenesis and patterning of the optic vesicle. Curr. Top. Dev. Biol. 2010, 93, 61–84. [Google Scholar] [PubMed]
- Fuhrmann, S.; Zou, C.; Levine, E.M. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 2014, 123, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Sowden, J.C. Genes and pathways in optic fissure closure. Semin. Cell Dev. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Livesey, F.J.; Cepko, C.L. Vertebrate neural cell-fate determination: Lessons from the retina. Nat. Rev. Neurosci. 2001, 2, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Boije, H.; MacDonald, R.B.; Harris, W.A. Reconciling competence and transcriptional hierarchies with stochasticity in retinal lineages. Curr. Opin. Neurobiol. 2014, 27, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [PubMed]
- Amram, B.; Cohen-Tayar, Y.; David, A.; Ashery-Padan, R. The retinal pigmented epithelium—From basic developmental biology research to translational approaches. Int. J. Dev. Biol. 2017, 61, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human escs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [PubMed]
- England, S.J.; Blanchard, G.B.; Mahadevan, L.; Adams, R.J. A dynamic fate map of the forebrain shows how vertebrate eyes form and explains two causes of cyclopia. Development 2006, 133, 4613–4617. [Google Scholar] [CrossRef] [PubMed]
- Rembold, M.; Loosli, F.; Adams, R.J.; Wittbrodt, J. Individual cell migration serves as the driving force for optic vesicle evagination. Science 2006, 313, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Cavodeassi, F.; Carreira-Barbosa, F.; Young, R.M.; Concha, M.L.; Allende, M.L.; Houart, C.; Tada, M.; Wilson, S.W. Early stages of zebrafish eye formation require the coordinated activity of wnt11, fz5, and the wnt/beta-catenin pathway. Neuron 2005, 47, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Loosli, F.; Staub, W.; Finger-Baier, K.C.; Ober, E.A.; Verkade, H.; Wittbrodt, J.; Baier, H. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003, 4, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.E.; Keller, P.J.; Ramialison, M.; Rembold, M.; Stelzer, E.H.; Loosli, F.; Wittbrodt, J. Nlcam modulates midline convergence during anterior neural plate morphogenesis. Dev. Biol. 2010, 339, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Kwan, K.M.; Otsuna, H.; Kidokoro, H.; Carney, K.R.; Saijoh, Y.; Chien, C.B. A complex choreography of cell movements shapes the vertebrate eye. Development 2012, 139, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Picker, A.; Cavodeassi, F.; Machate, A.; Bernauer, S.; Hans, S.; Abe, G.; Kawakami, K.; Wilson, S.W.; Brand, M. Dynamic coupling of pattern formation and morphogenesis in the developing vertebrate retina. PLoS Biol. 2009, 7, e1000214. [Google Scholar] [CrossRef] [PubMed]
- Heermann, S.; Schutz, L.; Lemke, S.; Krieglstein, K.; Wittbrodt, J. Eye morphogenesis driven by epithelial flow into the optic cup facilitated by modulation of bone morphogenetic protein. eLife 2015, 4, e05216. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.J.; Schmidt, A.D.; Wittbrodt, J.; Stelzer, E.H. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008, 322, 1065–1069. [Google Scholar] [CrossRef] [PubMed]
- Icha, J.; Schmied, C.; Sidhaye, J.; Tomancak, P.; Preibisch, S.; Norden, C. Using light sheet fluorescence microscopy to image zebrafish eye development. J. Vis. Exp. 2016, 10, e53966. [Google Scholar] [CrossRef] [PubMed]
- Ivanovitch, K.; Cavodeassi, F.; Wilson, S.W. Precocious acquisition of neuroepithelial character in the eye field underlies the onset of eye morphogenesis. Dev. Cell 2013, 27, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Cechmanek, P.B.; McFarlane, S. Retinal pigment epithelium expansion around the neural retina occurs in two separate phases with distinct mechanisms. Dev. Dyn. 2017, 246, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Sidhaye, J.; Norden, C. Concerted action of neuroepithelial basal shrinkage and active epithelial migration ensures efficient optic cup morphogenesis. eLife 2017, 6, e22689. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Morales, J.R.; Rembold, M.; Greger, K.; Simpson, J.C.; Brown, K.E.; Quiring, R.; Pepperkok, R.; Martin-Bermudo, M.D.; Himmelbauer, H.; Wittbrodt, J. Ojoplano-mediated basal constriction is essential for optic cup morphogenesis. Development 2009, 136, 2165–2175. [Google Scholar] [CrossRef] [PubMed]
- Bogdanovic, O.; Delfino-Machin, M.; Nicolas-Perez, M.; Gavilan, M.P.; Gago-Rodrigues, I.; Fernandez-Minan, A.; Lillo, C.; Rios, R.M.; Wittbrodt, J.; Martinez-Morales, J.R. Numb/numbl-opo antagonism controls retinal epithelium morphogenesis by regulating integrin endocytosis. Dev. Cell 2012, 23, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Perez, M.; Kuchling, F.; Letelier, J.; Polvillo, R.; Wittbrodt, J.; Martinez-Morales, J.R. Analysis of cellular behavior and cytoskeletal dynamics reveal a constriction mechanism driving optic cup morphogenesis. eLife 2016, 5, e15797. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Lee, C.; Williams, A.M.; Angileri, K.; Lathrop, K.L.; Gross, J.M. The hyaloid vasculature facilitates basement membrane breakdown during choroid fissure closure in the zebrafish eye. Dev. Biol. 2016, 419, 262–272. [Google Scholar] [CrossRef] [PubMed]
- McMahon, C.; Gestri, G.; Wilson, S.W.; Link, B.A. Lmx1b is essential for survival of periocular mesenchymal cells and influences fgf-mediated retinal patterning in zebrafish. Dev. Biol. 2009, 332, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Lupo, G.; Gestri, G.; O’Brien, M.; Denton, R.M.; Chandraratna, R.A.; Ley, S.V.; Harris, W.A.; Wilson, S.W. Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc. Natl. Acad. Sci. USA 2011, 108, 8698–8703. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Pillai-Kastoori, L.; Wilson, S.G.; Morris, A.C. Sox4 regulates choroid fissure closure by limiting hedgehog signaling during ocular morphogenesis. Dev. Biol. 2015, 399, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Bryan, C.D.; Chien, C.B.; Kwan, K.M. Loss of laminin alpha 1 results in multiple structural defects and divergent effects on adhesion during vertebrate optic cup morphogenesis. Dev. Biol. 2016, 416, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Heavner, W.; Pevny, L. Eye development and retinogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008391. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Bejarano, M.; Gestri, G.; Spawls, L.; Nieto-Lopez, F.; Picker, A.; Tada, M.; Brand, M.; Bovolenta, P.; Wilson, S.W.; Cavodeassi, F. Opposing shh and fgf signals initiate nasotemporal patterning of the zebrafish retina. Development 2015, 142, 3933–3942. [Google Scholar] [CrossRef] [PubMed]
- Facchinello, N.; Schiavone, M.; Vettori, A.; Argenton, F.; Tiso, N. Monitoring wnt signaling in zebrafish using fluorescent biosensors. Methods Mol. Biol. 2016, 1481, 81–94. [Google Scholar] [PubMed]
- Moro, E.; Ozhan-Kizil, G.; Mongera, A.; Beis, D.; Wierzbicki, C.; Young, R.M.; Bournele, D.; Domenichini, A.; Valdivia, L.E.; Lum, L.; et al. In vivo wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 2012, 366, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Moro, E.; Vettori, A.; Porazzi, P.; Schiavone, M.; Rampazzo, E.; Casari, A.; Ek, O.; Facchinello, N.; Astone, M.; Zancan, I.; et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol. Genet. Genomics 2013, 288, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Miesfeld, J.B.; Link, B.A. Establishment of transgenic lines to monitor and manipulate yap/taz-tead activity in zebrafish reveals both evolutionarily conserved and divergent functions of the hippo pathway. Mech. Dev. 2014, 133, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Collery, R.F.; Link, B.A. Dynamic smad-mediated bmp signaling revealed through transgenic zebrafish. Dev. Dyn. 2011, 240, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Schwend, T.; Loucks, E.J.; Ahlgren, S.C. Visualization of gli activity in craniofacial tissues of hedgehog-pathway reporter transgenic zebrafish. PLoS ONE 2010, 5, e14396. [Google Scholar] [CrossRef] [PubMed]
- Molina, G.A.; Watkins, S.C.; Tsang, M. Generation of fgf reporter transgenic zebrafish and their utility in chemical screens. BMC Dev. Biol. 2007, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Barolo, S. Transgenic wnt/tcf pathway reporters: All you need is lef? Oncogene 2006, 25, 7505–7511. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, K.K.; O’Shea, K.S. An analysis of cell shape and the neuroepithelial basal lamina during optic vesicle formation in the mouse embryo. Development 1987, 100, 185–200. [Google Scholar] [PubMed]
- Eiraku, M.; Adachi, T.; Sasai, Y. Relaxation-expansion model for self-driven retinal morphogenesis: A hypothesis from the perspective of biosystems dynamics at the multi-cellular level. Bioessays 2012, 34, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.C.; Smith, A.N.; Wagner, H.; Cohen-Tayar, Y.; Rao, S.; Wallace, V.; Ashery-Padan, R.; Lang, R.A. Wnt ligands from the embryonic surface ectoderm regulate ‘bimetallic strip’ optic cup morphogenesis in mouse. Development 2015, 142, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Beccari, L.; Marco-Ferreres, R.; Bovolenta, P. The logic of gene regulatory networks in early vertebrate forebrain patterning. Mech. Dev. 2013, 130, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Morales, J. Vertebrate eye gene regulatory networks. In Organogenetic Gene Networks; Castelli-Gair Hombria, J., Bovolenta, P., Eds.; Springer International Publishing AG: Cham, Switzerland, 2016. [Google Scholar]
- Cavodeassi, F.; Moreno-Marmol, T.; Hernandez-Bejarano, M.; Bovolenta, P. Principles of early vertebrate forebrain formation. In Organogenetic Gene Networks; Castelli-Gair Hombria, J., Bovolenta, P., Eds.; Springer International Publishing AG: Cham, Switzerland, 2016. [Google Scholar]
- Williamson, K.A.; FitzPatrick, D.R. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur. J. Med. Genet. 2014, 57, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Voronina, V.A.; Kozhemyakina, E.A.; O’Kernick, C.M.; Kahn, N.D.; Wenger, S.L.; Linberg, J.V.; Schneider, A.S.; Mathers, P.H. Mutations in the human rax homeobox gene in a patient with anophthalmia and sclerocornea. Hum. Mol. Genet. 2004, 13, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Landsend, E.S.; Utheim, O.A.; Pedersen, H.R.; Lagali, N.; Baraas, R.C.; Utheim, T.P. The genetics of congenital aniridia—A guide for the ophthalmologist. Surv. Ophthalmol. 2018, 63, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Williamson, K.A.; Rainger, J.; Floyd, J.A.; Ansari, M.; Meynert, A.; Aldridge, K.V.; Rainger, J.K.; Anderson, C.A.; Moore, A.T.; Hurles, M.E.; et al. Heterozygous loss-of-function mutations in yap1 cause both isolated and syndromic optic fissure closure defects. Am. J. Hum. Genet. 2014, 94, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Widen, S.A.; Williamson, K.A.; Ratnapriya, R.; Gerth-Kahlert, C.; Rainger, J.; Alur, R.P.; Strachan, E.; Manjunath, S.H.; Balakrishnan, A.; et al. A secreted wnt-ligand-binding domain of fzd5 generated by a frameshift mutation causes autosomal dominant coloboma. Hum. Mol. Genet. 2016, 25, 1382–1391. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavodeassi, F. Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models. J. Dev. Biol. 2018, 6, 4. https://doi.org/10.3390/jdb6010004
Cavodeassi F. Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models. Journal of Developmental Biology. 2018; 6(1):4. https://doi.org/10.3390/jdb6010004
Chicago/Turabian StyleCavodeassi, Florencia. 2018. "Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models" Journal of Developmental Biology 6, no. 1: 4. https://doi.org/10.3390/jdb6010004
APA StyleCavodeassi, F. (2018). Dynamic Tissue Rearrangements during Vertebrate Eye Morphogenesis: Insights from Fish Models. Journal of Developmental Biology, 6(1), 4. https://doi.org/10.3390/jdb6010004



