Abstract
Retinoic acid (RA), a derivative of vitamin A, is involved in signal transduction during vertebrate organogenesis. Retinoids through binding to nuclear receptors called RA receptors (RARs) and retinoid X receptors (RXRs) regulate various processes during cardiogenesis. Deregulated retinoid signaling thus has later consequences leading to cardiac malformations. In this review, we will summarize and discuss our current knowledge on the role of RA signaling during heart development, especially during patterning of the heart fields. We have also integrated recent experiments essential for our understanding of the role of RA signaling during epicardial development and myocardial growth.
1. Introduction
In vertebrates, the heart is one of the first organs to acquire its function, and continuous pump function is essential for distribution of oxygen and nutrients during fetal and post-natal life [1]. Normal cardiac morphogenesis is thus crucial for embryonic survival. Initially, at embryonic day (E) 8 in the mouse the heart tube is composed principally of contractile myocardium and endocardium essential for its pumping function. At this stage, the heart is not yet covered by the epicardium, an external tissue layer, which is only added during the establishment of cardiac chambers (Figure 1). Development of the heart with its chamber organization and three different layers is a complex morphogenetic process prone to defects due to both genetic and environmental causes (for a detailed review on heart morphogenesis see [2,3,4,5]). Congenital heart diseases (CHD) are the most common human birth defects, affecting nearly 1 in 100 live births [6]. Studying factors that control heart development can help to better understand the etiology of these defects. For a detailed summary of many other molecular pathways guiding heart formation, we suggest the following reviews [2,7,8,9,10].
Vitamin A and its metabolites, collectively called retinoids, are important in vertebrate development [11]. The heart is one of the most sensitive organs to perturbations of retinoid concentration during its development [12,13]. In the 1950s, studies using nutritional deprivation of vitamin A in a rat model demonstrated that maternal vitamin A deficiency causes severe embryonic defects including cardiac and great vessels anomalies [14,15]. Hypervitaminosis A, caused by treatment with the active (all-trans) form of retinoic acid (RA; the active derivative of vitamin A; see [16]) was one of the earliest teratogenic models of heart defects [17]. In subsequent studies, different models have been developed to address the role of RA during early development [13,18,19,20]. Among them, studies using avian embryos have proposed that the vitamin A-deficient (VAD) quail is an ideal model to study the function of vitamin A in the vertebrate embryo since it is a complete VAD model [12]. More recently, clarification of the biochemical pathways leading to retinoid synthesis and the discovery of all retinoid receptors have allowed genetic manipulation of this pathway [18,21,22]. Thus, genetic studies in mouse embryos deficient for RA-generating enzymes have been invaluable for deciphering RA function [19,23,24,25,26,27]. These studies demonstrated that RA synthesis during critical processes of heart development is controlled largely by retinaldehyde dehydrogenase 2 (RALDH2) that produces RA [19,28,29,30,31]. In 2007, Chambers et al. proposed that Cyp1B1, a P450 cytochrome, is one of the RALDH-independent components by which embryos generate RA-mediated patterning [32]. Detailed description of Cyp1B1 expression showed many overlap with expression of Raldh genes, most notably with Raldh2 in the mesoderm [32]. Interestingly, Cyp1b1 expression in the early VAD quail embryo (stage HH4-5) appears normal. Thus, we cannot exclude the possibility that low amounts of RA may be produced in early Raldh2-/- mutant embryos. Those low amounts could explain why the development of the posterior heart tube in Raldh2-/- mutant embryos is less affected than in the VAD quail (see Section 2).
Several excellent reviews have discussed the role of RA during heart formation, focusing predominantly on early events of heart development (i.e., cardiac specification and heart field patterning [13,22,33,34,35]). Here we aim to provide an update on the role of RA in several aspects of heart morphogenesis, from early events of heart formation including cardiac specification and heart field patterning to outflow tract formation and control of myocardial growth. In particular, we have summarized recent results on the requirement of RA during patterning of the heart fields in different animal models and discussed new findings on the role of RA during the outflow tract and myocardial morphogenesis. We focus on our recent work demonstrating that early expression of RA is required to regulate the anteroposterior expression of Hox genes in a cardiac progenitor cell population termed the second heart field. This population of cardiac progenitor cells contributes to major components of the heart including outflow tract, right ventricular and atrial myocardium [2]. Altering this early role of RA thus has later consequences on the production of malformations of the heart and great arteries such as those described in CHD.
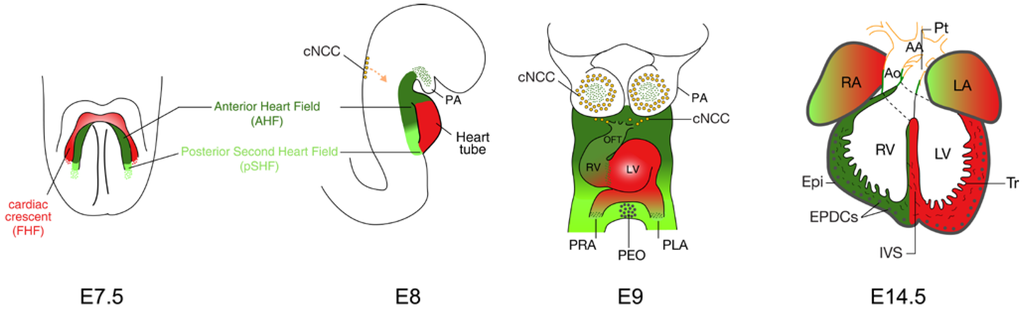
Figure 1.
Heart development and the contribution of cardiac lineages in the mouse. At E7.5, myocardial cells form the cardiac crescent (also called first heart field, FHF), with the second heart field (SHF) lying medial to it. The location and contribution of the SHF are shown in green, with the anterior heart field (AHF) subdomain in dark green and posteriorSHF (pSHF) in light green. At E8, the early cardiac tube forms through fusion of the cardiac crescent at the midline and it subsequently undergoes looping (E9). At fetal stage (E14.5), the epicardium (Epi) envelops the heart, the chambers are separated by the interventricular septum (IVS) and are connected to the pulmonary trunk (Pt) and aorta (Ao). The FHF and SHF contributions are shown in red and green respectively. The tunica media of the Ao and Pt is derived from SHF (green) and cardiac neural crest cells (cNCC; orange). AA, aortic arch; EPDCs, epicardium-derived cells; LA, left atrium; LV, left ventricle; OFT, outflow tract; PA, pharyngeal arch; PEO, pro-epicardial organ; PLA primitive left atrium; PRA, primitive right atrium; RA, right atrium; RV, right ventricle; Tr, trabeculae.
2. Retinoic Acid and Early Heart Development
Heart development begins with the specification of cardiogenic mesoderm, which in vertebrates occurs in the course of gastrulation. At this step, cardiogenic mesoderm is found in bilateral territories in lateral mesoderm [36,37,38,39,40,41,42].
In the mouse embryo, at E7.5, cardiac progenitor cells extend towards the midline to form a crescent shape in anterior lateral plate mesoderm (also referred as the first heart field (FHF)) (Figure 1). The cardiac crescent subsequently fuses at the midline to give rise to the primitive heart tube at E8.0. This tube is in connection to embryonic and extra-embryonic circulation. The myocardium (muscular layer) and endocardium (inner layer) are the main components of this simple heart. The third cellular layer, the epicardium, develops later from an extracardiac cell population called the proepicardium (or proepicardial organ). At early stages, anteroposterior polarity can be already distinguished with the future atrial and ventricular regions observed at the posterior and anterior poles of the heart tube respectively. The forming heart tube undergoes rightward looping by E8.0 in the mouse and begins to beat, as embryonic circulation initiates [1]. During looping, the heart tube elongates by addition of myocardial cells at its two poles (arterial and venous) (Figure 1). The population of cardiac progenitor cells located in the pharyngeal mesoderm that contributes to growth of the embryonic heart tube is termed the second heart field (SHF). Elegant studies in the mouse have shown that these cells contribute to the cardiac outflow tract, right ventricle and a major part of atrial myocardium, while the linear heart tube gives rise predominantly to the left ventricle (Figure 1) (see [2]). The SHF is characterized by active proliferation and expression of different markers including the transcription factors Islet 1 (Isl1) [43], Nkx2-5 [44], and T-box 1 (Tbx1) [45] and the transgenes Mlc1v-nlacZ-24 [46] and Mef2c-AHF-lacZ [47] as markers of the anterior domain of the SHF (also called the anterior heart field (AHF)) (Figure 1). Fate mapping studies using a genetic approach addressed the contribution of Isl1 expressing cells and suggested that the Isl1+ cells contribute to cardiac outflow tract, right ventricle and atrial myocardium [43]. Interestingly, a recent lineage tracing analysis has showed that Isl1 is not restricted to SHF derivatives in the heart but it is also expressed by a subset of cardiac neural crest cells [48] suggesting that results based on gene inactivation using Isl1-Cre should be interpreted with caution. Continuous recruitment of SHF cells has been demonstrated to participate to outflow tract elongation. In the absence of addition of SHF cells, heart tube elongation and looping fail, resulting in early embryonic lethality [43,44,49]. Problems in SHF deployment can in addition compromise outflow tract development resulting in a spectrum of CHD. These defects are essentially anomalies in patterning and septation of the great arteries, and ventricular and atrial septal defects (see [9,50]). Such CHDs in humans cause cyanosis due to mixed oxygenated and deoxygenated blood entering the systemic circulation.
The function of vitamin A in early heart development is a center of current interest. The VAD avian (quail) embryo model has been instrumental in demonstrating a requirement of vitamin A-derived active components on early embryogenesis and in particular on early cardiovascular development [13]. Indeed, study on VAD quail embryos revealed that while embryonic development is normal during the first 20 h of incubation (i.e., until stage HH-8 4/5 somite stage), abnormalities in heart development are detected as early as the 6–7 somite stage [51]. Later, most VAD embryos show cardia bifida while some of them form a unique heart tube with a single enlarged ventricular chamber. In all cases, the posterior region of the heart (i.e., the inflow tract) is severely affected and fails to connect to the extra-embryonic vascular system, the development of which is initiated but, as a consequence of the closed posterior heart compartment, cannot be completed resulting in total absence of omphalomesenteric veins [12,51]. In the mouse, deletion of Raldh2 causes heart defects with poor development of the atria and sinus venosus [52]. At E8.5, this defect is correlated with abnormal expression levels and spatial distribution of the T-box gene Tbx5, a marker of the prospective atrial and sinus venosus regions [53].
In the avian model, the administration of retinol or RA around HH8 is sufficient to rescue the heart phenotype of VAD quail embryos [54,55]. In 1993, Dersch and Zile have determined that the time window during which all-trans RA can rescue cardiovascular development is between 22–28 h of incubation [12]. Furthermore, Kostetskii et al. (1998) demonstrated that temporal limit of all-trans RA rescue is the 5 somite stage [56]. This study also demonstrated that the expression of RA receptors is suitable for RA action on early cardiac fields. More recently, a study using an RARα antagonist in xenopus embryos showed that the requirement for RA signaling is limited to a narrow window of time between stages 14 and 18, well before heart closure [57]. Taken together, these findings reveal an early requirement of RA for heart development and indicate that the inflow tract region (posterior compartment) of the primary heart tube is particularly sensitive to the lack of RA. Several recent studies using the VAD quail model have provided insights into the pathways operating downstream of RA. Indeed, the expression of the transcription factor GATA4 is significantly compromised in the VAD quail [58]. Gata4 is expressed most strongly at the 5 somite stage in the region of the presumptive inflow tract and posterior heart tube, and this is precisely the region that fails to develop normally in the VAD embryo. Studies showed that VAD results in reduced expression of Bmp2 throughout the sino-atrial region [59]. Furthermore, addition of exogenous Bmp2 can function additively with forced expression of Gata4 to overcome the loss of retinoid signaling, indicating the presence of an active RA-BMP-GATA network for posterior heart tube development. More recently, N-cadherin and TGFβ2 expression have been shown to be upregulated in VAD quail embryos [60,61]. Studies conducted by blocking the function of those over-expressed molecules or in the case of TGFβ2, treatment of normal embryos with the growth factor, have led to the conclusion that their deregulated activities contribute to the cardiac phenotype of the VAD quail embryo [60,61]. Interestingly, a recent transcriptomic analysis aiming to identify genes deregulated in the early Raldh2-/- mutant embryos (4 somite stage) demonstrated an up-regulation of components of the TGFβ pathway in the posterior region of the embryo including an up-regulation of TGFβ2 [62].
Other studies in mouse and avian embryos have linked RA-deficiency heart defects to anomalies of anteroposterior patterning of the early heart tube [28,63,64]. The addition of exogenous RA to chick embryo cultures between stages 5 and 8 produces various anomalies, which have initially been described as abnormal precardiac cell migration [63,65]. In fact, Yutzey et al. have shown that RA treatment produces an expansion of the atrial domain observed by the expression of the atrial-specific myosin heavy chain AMHC1 [64]. This study suggests that specification of cardiac progenitors can be altered by RA treatment. In a similar study, adding increasing doses of exogenous RA during zebrafish development also led to progressive truncation of the heart tube, the anterior region being the most sensitive to RA exposure [38]. More recent findings have confirmed that RA signaling promotes atrial cell identity within the cardiac progenitor field [28,31,35]. As mentioned earlier, the phenotype of VAD quail embryos and Raldh2-/- mutant mouse embryos illustrates the importance of RA signaling in promoting posterior fate on cardiac progenitors. Hence, pharmacological, genetic and dietary manipulations have established the crucial role of RA in early cardiac development and furthermore argued that RA functions as a potential morphogen in anteroposterior patterning of the early heart tube. Retinoids also regulate cardiac laterality, impacting on left-right positioning and looping [65,66,67,68,69]. Mouse embryos from mothers treated with an excess of RA display disrupted expression of left-right asymmetry genes and have abnormal cardiac situs [68]. Although not covered here, a detailed review of the role of RA signaling in the establishment of cardiac laterality is provided by [22,70].
Studies in zebrafish, frog and mouse have also revealed an important role of the RA signaling in restriction of the pool of cardiac progenitors [57,71,72,73]. Zebrafish embryos lacking RA signaling exhibit a surplus of cardiomyocytes [73]. Similar results are obtained on treatment with a pan-RAR antagonist, as a cell population expressing the myocardial transcription factor nkx2-5 or the cardiac myosin light chain 2 (cmlc2) was expanded. Interestingly, in the xenopus, if embryos are treated with an RA antagonist prior to gastrulation, the Nkx2-5 expression domain is expanded, as observed in zebrafish, but this initial increase in the size of the cardiac domain is not sustained [57]. More recently, investigation of RA deficient mice demonstrates that Raldh2-/- embryos display an expansion of the SHF [71,72]. Indeed, expression of genes expressed in the SHF such as Isl1, Nkx2-5, Tbx1 and Fibroblast growth factor 8 (Fgf8) are caudally expanded in Raldh2-/- embryos, indicating that RA is required to restrict the SHF (Figure 2) [71,72]. Importantly, explant culture experiments suggest that RA function is to inhibit cardiac specification rather than to limit the proliferative capacity of cardiac progenitor cells [71]. Remarkably, in the zebrafish, lack of RA signaling affects both early and late differentiating ventricular and atrial progenitor populations [74,75], while Raldh2 mutation in the mouse displays a specific effect on SHF cells [71]. This latter observation suggests that the FHF and SHF are differently affected in these species, raising the question of the timing of RA action during heart development in different species.
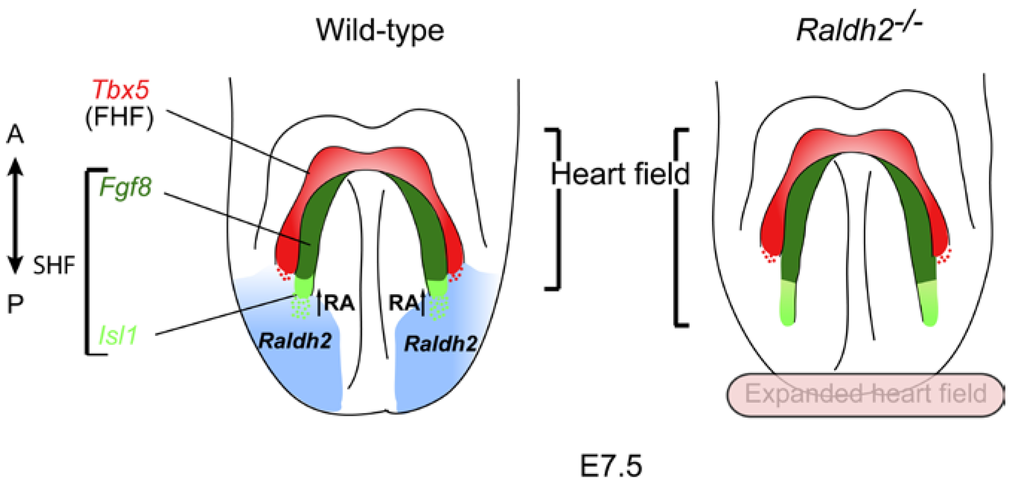
Figure 2.
Retinoic acid signaling during early cardiogenesis. Raldh2, the main enzyme responsible for Retinoic Acid (RA) synthesis during development, is expressed in the posterior lateral mesoderm. Thus, production of RA provides a signal to establish the posterior boundary of the heart field. Tbx5 is expressed in the first heart field (FHF) whereas Fgf8 and Isl1 mark the anterior and posterior domains of the second heart field (SHF). Raldh2-/- mutant embryos exhibit caudal expansion of Fgf8 and Isl1 expression within the posterior lateral mesoderm. The anterior (A)–posterior (P) axis is indicated.
2.1. How does Retinoic Acid Reduce the Number of Mesodermal Cells Adopting a Cardiac Fate?
As described above, RA deficient embryos exhibit excess of cardiac progenitor cells, however the formation of the pectoral fins is also affected in the zebrafish as well as the forelimb in the mouse [72,74]. Interestingly, Waxman et al. found that RA responsive genes are expressed in the forelimb progenitor field adjacent to the cardiac progenitor field, suggesting that action of RA on the cardiac progenitors may be indirect. Moreover, homeo box B5b (hoxb5b), a RA target gene, was indirectly required to limit expansion of cardiomyocytes [74]. However, loss of hoxb5b can only partially recapitulate the RA deficient phenotype, suggesting that other signals may also be involved downstream of RA in the coordinated development of the forelimb and heart. Consistently, RA signaling regulates anteroposterior patterning within the lateral plate mesoderm that includes the heart and forelimb fields in xenopus embryos [76]. The repressive signals from the forelimb field, which act to limit the cardiac field, are still unknown. However, these results do not exclude a direct effect of RA within the SHF.
Fgf signaling is a good RA-target candidate to be involved in coordinating forelimb and cardiac development. A recent study has demonstrated that Fgf8a, which is expressed in cardiac progenitors, is expanded posteriorly in RA deficient zebrafish embryos similar to Raldh2-/- embryos [71,72,74,77]. However, it was not clear if the ectopic Fgf8 expression in Raldh2-/- mouse embryos is a simultaneous cause of heart and forelimb defects or simply a sign of abnormal patterning. To address this question Sorrel et al. used gain- and loss-of-function approaches in zebrafish embryos. Thus, they demonstrated that proper signaling of Fgf8a downstream of RA signaling is responsible for balancing autonomous and non-autonomous interactions between the heart and forelimb fields [77]. Finally, this study proposes a feedback inhibition model in which RA coordinates heart and forelimb field development by repressing Fgf8a signaling. Repression of Fgf8 expression has also been proposed in this context in the mouse by Sirbu et al. [72] based on the existence of a RA response element (RARE) upstream of the Fgf8 promoter [78]. Fgf8 thus appears to be a critical target of RA signal in establishing the correct boundary between the heart and forelimb fields.
As mentioned above, RA deficient mouse embryos fail to undergo heart looping and have impaired atrial and sinus venosus development [19]. Investigations of Raldh2-/- mutant embryos identified that this defect is caused by a caudal expansion of Isl1+ and Fgf8+ populations (Figure 2), indicating that RA is important to establish the posterior boundary of the SHF [71,72]. In zebrafish, RA deficiency generates an excess of cardiomyocyte progenitors [73], similar to the phenotype described in mouse embryos. However, the mechanisms linking RA to transcriptional regulators of heart development remained unknown. A recent study identified Ajuba, a LIM domain protein, as a crucial regulator of SHF progenitor cell specification and expansion [75]. Interestingly, Witzel et al. used Ajuba morphant embryos to demonstrate that Ajuba specifically restricts the number of Isl1-expressing cells at the both poles of the heart. By comparing Isl1 expression to the Nkx2-5-expressing domain using the Nkx2-5:GFP transgene in Ajuba morphant embryos, the authors suggested that Ajuba restricts the SHF progenitor pool. This study also showed that Ajuba interacts with Isl1 and represses its transcriptional activity in the SHF. Furthermore, Witzel et al. show that RA regulates the Isl1-population through an Ajuba-dependent mechanism. This work demonstrated that (1) RA acts upstream of Ajuba since RA treatment produces an upregulation of Ajuba and its accumulation in the nucleus; which results to the downregulation of Isl1 expression; and (2) RA-mediated restriction of the number of cardiomyocytes in the zebrafish depends on Ajuba since treatment of Ajuba morphants with RA failed to affect the pool of cardiac progenitors expressing Isl1. How can such a mechanism define the posterior boundary of the SHF? One could postulate that the system is under a threshold mechanism. Hence, mesodermal cells exposed to a high level of RA within the lateral mesoderm would express a high amount of Ajuba in the nucleus resulting in the downregulation of Isl1 expression, thus delimiting the posterior extent of cardiac progenitor cells. To date there is no reported study on Ajuba mutant mouse embryos to confirm this hypothesis in the mouse.
2.2. When does Retinoic Acid Act to Restrict the Number of Cardiac Progenitors?
In the zebrafish, exposure to the pan-RAR antagonist BMS189453 indicates that RA-mediated restriction of the number of cardiac progenitors takes place before and during gastrulation. Interestingly, upon gastrulation, Raldh2 is expressed in involuting cells at the margin that will form mesendoderm [79]. In the mouse, during early embryogenesis Raldh2 is expressed in mesoderm adjacent to the node [30] and expands anteriorly until the 1 somite and 2 somite stages, similar to the situation in avian embryos [28]. Interestingly, a fate-mapping study established that mesodermal cells ingressing through the anterior and middle portions of the primitive streak contribute to the heart field [36]. Hence, it is likely that RA signaling acts on early migrating cardiac progenitors shortly after gastrulation. Consistently, Xavier-Neto et al. showed that a single pulse of exogenous RA given around E7 produces hearts with marked atrial dominance [31]. This observation raises the question of the spatial relationship between RA signaling and cardiac progenitors. A changing relationship between Raldh2 and cardiac progenitors was demonstrated in the mouse by Hochgreb et al. who performed double in situ hybridization for Raldh2 and Tbx5, a marker of posterior cardiac progenitors [28]. At early stages, Raldh2 and Tbx5 expression domains converge until they overlap at the late headfold stage (E7). At this stage only the posterior third of the mouse Tbx5 stripe overlaps with Raldh2 expression. Together these data indicate that Raldh2 expression is present at the right time and place to restrict the number of cardiac progenitor cells in the heart fields. This raises the question of the specificity of RA action on the SHF versus FHF. Again, RA action depends on the level of RA activity but also on the presence of effectors.
3. Retinoic Acid and Pre-Patterning of the Second Heart Field
Differences in gene expression between arterial and venous pole progenitor cells reveal that the SHF is pre-patterned (see [7]). In summary, the rostral part of the SHF, the anterior heart field (AHF), which is marked by Fgf10 expression [46] contributes to the formation of right ventricular and outflow tract myocardium [80], whereas cells in the posterior SHF [43] expressing Isl1, but not AHF markers, contribute to atrial myocardium [81]. The observation of this pre-patterning of the SHF raises the question of how it is established. As discussed above, analysis of Raldh2 mutant embryos revealed that RA signaling plays a role in establishing the boundaries of the SHF in the embryo, as indicated by the abnormal posterior expansion in expression of anterior SHF markers, including Tbx1, Fgf8 and Fgf10 [71,72]. Moreover, experiments in avian embryos showed that excess of RA results in downregulation of Tbx1 expression in pharyngeal mesoderm [82]. Antagonism between RA signaling and Tbx1, a important regulator of pharyngeal and SHF development is further revealed by the observation that Raldh2 expression domain is moved anteriorly in pharyngeal mesoderm of Tbx1 null embryos [83,84,85].
Since RA signaling affects positional identity through regulation of Hox gene expression, it has been proposed that Hox genes function in cardiac patterning. In the zebrafish, increased Hox activity mimics the effects of ectopic RA [86]. In the mouse, several novel enhancers containing RAREs, from the HoxA and HoxB complex have been identified to drive reporter gene expression in the embryonic heart [87]. Genetic lineage tracing analysis using Cre recombinase driven by the most anteriorly expressed Hox genes, including Hoxa1, Hoxb1 and Hoxa3, revealed that Hox expressing progenitor cells contribute to the both poles of the heart, in a distribution corresponding to a subset of the SHF (Figure 3). Indeed, using co-staining experiments, our lab has reported that Hoxa1, Hoxb1 and Hoxa3are expressed in distinct subdomains of the SHF that contribute to both poles of the heart tube, including atrial myocardium at the venous pole and the inferior wall of the outflow tract and sub-pulmonary myocardium at the arterial pole [88]. Interestingly, anterior Hox genes, such as Hoxa1, Hoxa3 and Hoxb1, have nested expression in the SHF, with different anterior limits of expression within the caudal AHF (Figure 3). Consistent with previous studies, reduction or excess of RA signaling perturbs the contribution of Hox expressing progenitors to the heart [88]. Diman et al. identified a novel RA-responsive Hoxa3 enhancer expressed in progenitor cells of a subset of outflow tract myocardium [89]. This may reflect the fact that only a subpopulation of the SHF may have been exposed to RA signaling at early stages. Consistently, analysis of the RA-activated cell lineage during mouse embryogenesis using a RA-activatable Cre (RARE-Cre) transgene with a conditional reporter gene, Rosa26R-lacZ (R26R-lacZ), showed RA-activation in the splanchnic mesoderm posterior to the cardiac crescent at E7.5, which may be defined as the posterior SHF [90]. Finally, combined loss of both HoxA and HoxB sister clusters results in severe defects in heart development [91], in many aspects similar in external appearance to those observed in Raldh2-/- mutants [19,52,71]. Together these observations suggest that nested Hox gene expression may play a role in pre-patterning cardiac progenitors in the SHF, downstream of RA signaling.
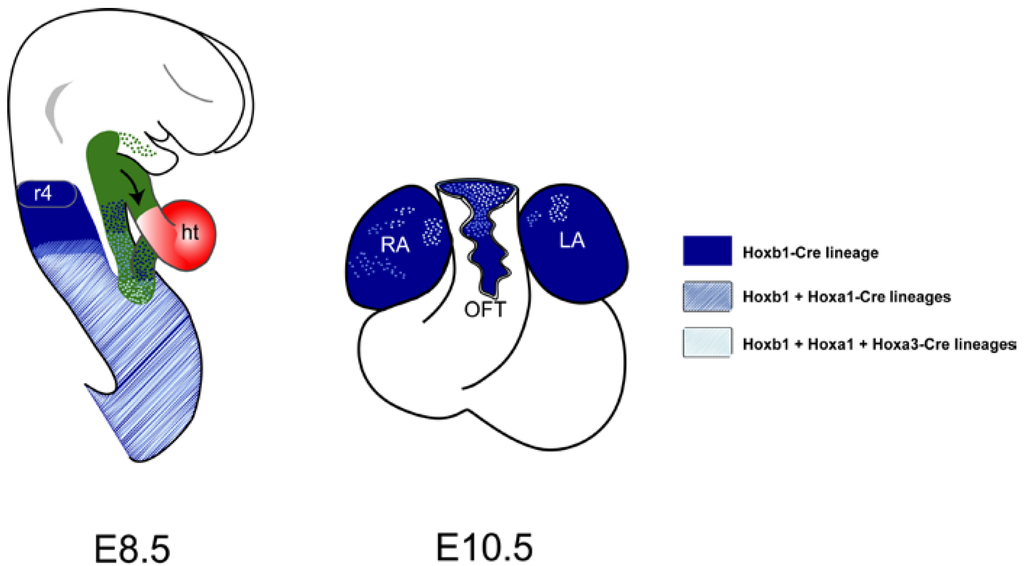
Figure 3.
Cardiac contributions of Hox-expressing cells in the second heart field (SHF). Genetic lineage analysis was performed using Hoxb1IRES-Cre, Hoxa1-EnhIII-Cre, Hoxa3IRES-Cre and R26R-lacZ lines. X-gal labeled cells are shown in blue. The location of the SHF is shown in green. At 8.5, Hoxb1-, Hoxa1- and Hoxa3-expressing cells identify distinct sub-domains along the anteroposterior axis in the SHF. At E10.5, Hoxb1-, Hoxa1- and Hoxa3-descendants contribute to both atria and the inferior wall of the outflow tract (OFT). Interestingly, descends of Hoxb1-expressing cells contribute to the proximal OFT, while Hoxa1 and Hoxa3 descendants appear more distally. ht, heart tube; LA, left atria; RA, right atria, r4, rhombomere 4.
4. Retinoic Acid and Outflow Tract Development
As mentioned above, the recruitment of cells from the SHF contributes to the elongation of the cardiac outflow tract. Later outflow tract septation is achieved by fusion and subsequent myocardialization of the ridges of the endocardial cushions and formation of the aorticopulmonary septum. Cardiac neural crest cells and SHF cells are thought to be involved in these processes and as such associated with development of the septa [92,93]. Interestingly, mouse embryos lacking Raldh2 show a failure in the deployment of SHF cells, which subsequently causes abnormal formation of the outflow tract as revealed by the absence of the y96-Myf5-nlacZ-16 transgene, a marker of the inferior wall of the outflow tract [71,88]. Studies using RARE reporter lines, demonstrated that the RARE-hsp68-lacZ and RARE-Cre;R26R-lacZ transgenes are expressed at E9.5 in the inferior wall of the outflow tract suggesting a differential contribution of RA exposed cells to the formation of this region of the heart [90,94]. Moreover, analysis of the RARE-hsp68-lacZ transgene in the Rarα1/Rarβ mutant background, in which outflow tract defects are observed, revealed the absence of X-gal+ cells in this region [94], demonstrating a role of RA signaling during outflow tract development.
4.1. Spatio-Temporal Requirement for Retinoic Acid Activity during Outflow Tract Development
RA signals are received by a heterodimer of one RAR and one RXR [21]. Lack of the RA receptors Rxrα1, Rarα1;Rxrα or Rarα1;Rarβ leads to outflow tract defects, resulting in common arterial trunk, ranging from low to high frequency respectively [94,95]. This variability of heart phenotype in retinoid receptor knockouts may be due to retinoid receptor redundancy [21]. Nevertheless, Li et al. used tamoxifen-inducible Cre line combined with a conditional Rxrα allele and a null Rarα1 allele to propose that RA signaling is required over an E9-E10.5 time-window for development of the outflow tract [94]. Anomalies of the outflow tract such as common arterial trunk can occur when the cardiac neural crest cell population is deficient or otherwise compromised [96]. However, analysis of the neural crest cell lineage demonstrated that the distribution of these cells is not compromised in RA receptor mutant embryos and that RA receptor function in the neural crest cell population is not required for normal outflow tract development [97]. These results suggest that RA signaling in an alternative tissue creates an environment in which the migrating cardiac neural crest cells are instructed to initiate outflow tract septation. Conditional loss-of-function of RA receptors within pharyngeal mesoderm demonstrated that RA signaling is required for development of the distal end of the outflow tract and consequently for outflow tract alignment and septation [94]. Interestingly, Rarα1;Rarβ mutant embryos failed to activate a Mef2c enhancer in SHF cells at E9.5, suggesting a failure of renewal of distal outflow tract progenitors in the SHF. Mef2c appears to be a target of Isl1, as indicated by an enhancer element within the Mef2c gene that is regulated by Isl1 and GATA4 [47]. Consistently, expression of GATA4, is specifically compromised in the posterior SHF of Rarα1;Rarβ mutant embryos as it was also observed in VAD quail embryos [58]. Li et al. demonstrated that the septation defect in RA receptor mutant embryos was caused by retention of proximal outflow tract tissue in the distal region of the heart tube and that anomaly is associated with elevated levels of TGFβ2 in this region [94]. Previous works have shown that TGFβ2 expression is negatively and indirectly regulated by RA signaling [58,98]. Thus, reducing TGFβ2 levels rescued the septation but not the alignment defects in half of the Rarα1; Rarβ mutant embryos. This result suggests that different downstream pathways of RA are required for outflow tract septation and alignment of the great arteries. In conclusion, RA signaling thus plays sequential roles during the development of the SHF, (1) in delimiting the posterior boundary of the SHF and (2) later in the specification of SHF cells that give rise to the distal region of the outflow tract.
5. Role of Retinoic Acid in Epicardium during Myocardial Growth
The epicardium, the outer layer of the developing heart, is required for normal growth and maturation of the adjacent compact myocardial layer (Figure 4) (for recent reviews see [99,100,101]). Indeed, in avian embryos, removal of the epicardium leads to an arrest in cardiomyocyte proliferation [102]. Several lines of evidence support a role of the epicardium in the transduction of retinoid signaling during cardiac growth at fetal stages. First, expression of Raldh2 and activity of the RARE-hsp68-lacZ reporter transgene are detected in the epicardial and subepicardial layers from E11.5 onwards [29,103]. Analysis of the RA-activated cell lineage (RARE-Cre;R26RlacZ) confirmed observations obtained with the RARE-hsp68-lacZ transgene [90]. Indeed, X-gal+ cells are observed in the epicardium but also in some sub-epicardial myocardial cells. This suggests that RA originating in the epicardium may induce myocardial compact zone growth. Secondly, interfering with RA signaling by genetic alteration of RA receptors (Rarα;Rarγ or Rxrα) or using blockers of those receptors causes embryonic lethality around E15.5 associated with severe hypoplasia of the ventricular myocardium especially in its compact zone, similar to phenotype of other mutants affecting epicardial function [104,105,106]. These data support a model in which RA induces growth through cross-talk between two adjacent regions of the heart, the epicardium and myocardium [107].
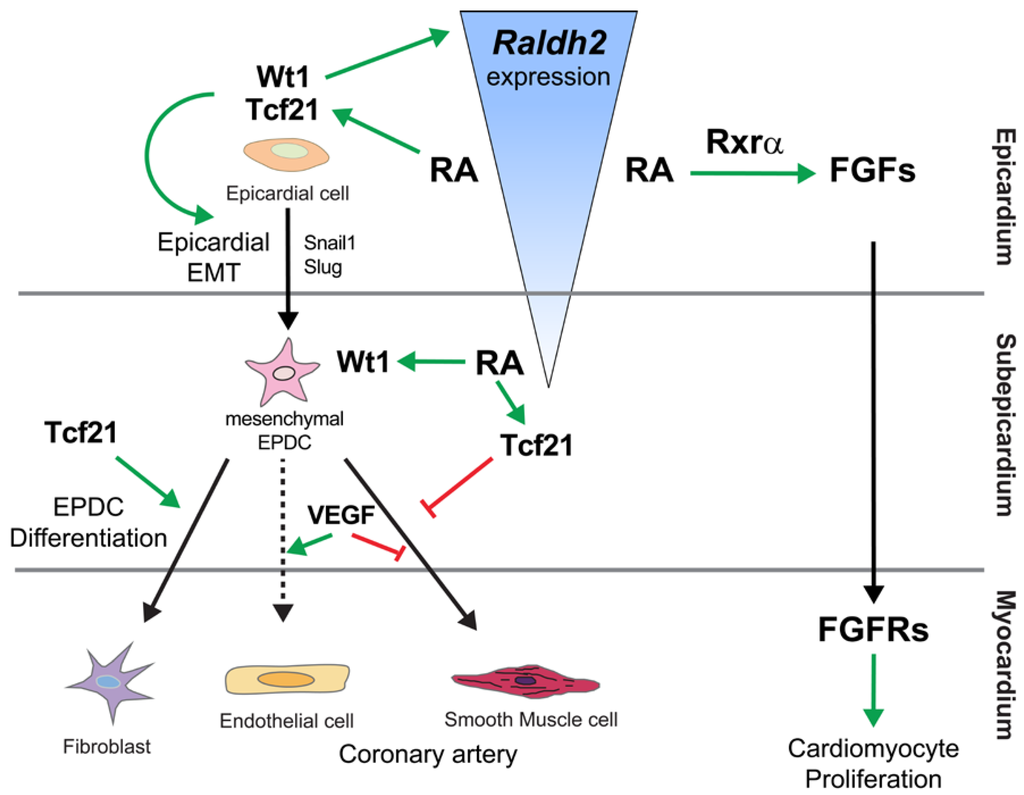
Figure 4.
Summary of the role of retinoic acid signaling in the regulation of coronary formation and myocardial growth. Retinoic acid (RA) signaling activates the expression of several Fgfs including Fgf2, Fgf9 and probably Fgf16 and Fgf20. A major role of RA signaling in epicardial cells is to induce the expression of the transcription factors Wt1 and Tcf21 (Pod1) [108]. These transcription factors control epithelial-to-mesenchymal transition (EMT) of epicardial cells through the activation Snail and Slug. Mesenchymal epicardium-derived cells (EPDCs) give rise to interstitial fibroblasts, coronary smooth muscle cells and coronary endothelial cells in the heart. However, different opinions exist on the origin of the coronary endothelial cell from EPDCs since a recent study demonstrated that some murine coronary endothelial cells arise from both the sinus venosus and the endocardium but not from proepicardial cells [100,109,110]. Raldh2 is highly expressed in epicardial cells, and progressively lost in EPDCs when these cells differentiate. Thus, RA signaling, together with VEGF, acts on the differentiation of several EPDC lineages including fibroblasts, coronary endothelial and smooth muscle cells.
5.1. Epicardial Retinoic Acid Induces Myocardial Growth
As indicated above, RA signaling is important for the interaction between epicardial and myocardial layers to support myocardial proliferation. In the mouse, Rxrα mutant embryos fail to expand their ventricular myocardial compact zone [104]. Furthermore, specific epicardial-cell deletion of Rxrα using the Gata5-Cre line indicated that reception of RA signal in the epicardium is crucial for myocardial growth [106]. Recent studies using food-based maternal RA supplementation (E7.5 to E9.5) to rescue early lethality of Raldh2-/- embryos, showed that at E12.5 RA rescued-Raldh2-/- hearts exhibited a thin underdeveloped ventricular compact zone, similar to the phenotype observed in Rxrα mutant embryos [103,111]. Moreover, in null or RA rescued-Raldh2 mutant embryos, normal expression of Tbx18, a marker of the proepicardial organ and the epicardium is observed, demonstrating that formation of this tissue occurs normally in the absence of Raldh2 function [111]. These data suggest that RA activity is required after the epicardium develops to induce myocardial growth.
Several approaches have been used to identify the pathways by which retinoids regulate cardiomyocyte proliferation. Epicardial cell cultures demonstrated that treatment with RA leads to the expression of mitogens that stimulate cardiomyocyte proliferation [112]. Using RAR antagonism and an anti-erythropoietin (Epo) receptor antibody on heart slice cultures, Stuckmann et al. demonstrated that RA and Epo pathways act in parallel to induce cell proliferation in ventricular myocardium [102]. Interestingly, this study showed that RA and Epo do not directly induce proliferation of cardiomyocytes but rather induce the secretion of a soluble cardiac mitogen from the epicardium. In 2005, Lavine et al. demonstrated that RA but not Epo treatment in both organ culture and cultured primary epicardial cells upregulated Fgf9 [113]. Furthermore, this study showed that Fgf9, and probably Fgf16 and 20, constitutes an epicardial-derived signal that regulates myocardial proliferation. Indeed, Fgf9 deficient mice and mice carrying a myocardial-specific deletion of Fgfr1/Fgfr2 display ventricular hypoplasia caused by decreased cardiomyocyte proliferation [113]. Consistent with these findings, fetal RA rescued-Raldh2 mutant embryos also have a reduction in cardiac Fgf2 and Fgf9 mRNA and the intracellular Fgf target phosphorylated Erk1/2 [103]. Finally, RA signaling events outside the heart may also be responsible for induction of an epicardial mitogen. Indeed, Brade et al. found that hepatic expression of Epo, is dependent on both Raldh2 and Rxrα [111]. Altogether these studies support a model in which a network of RA and Epo signals in the epicardium induces expression of Fgf ligands that directly regulate cardiomyocyte proliferation (Figure 4).
5.2. Retinoic Acid is Necessary for Epithelial-to-Mesenchymal Transition into Epicardium-Derived Cells
A subset of the epicardial cells undergoes epithelial-to-mesenchymal transformation (EMT), supplying mesenchymal cells to the outer surface of the heart (Figure 4). These epicardium-derived cells (EPDCs) migrate into the myocardium and differentiate into interstitial fibroblasts and coronary vascular smooth muscle cells [100]. Epicardial and subepicardial EPDCs express various transcription factors, including Tcf21 (Pod1/Capsulin/Epicardin, bHLH protein), Wilms’ tumor 1 (Wt1, zinc finger C2H2 protein), Nfatc1 (nuclear factor of activated T-cells 1), Snai1 (zinc finger C2H2 protein) and Tbx18. Wt1 deficient mice have epicardial defects associated with reduced numbers of EPDC and subsequent coronary defects [114]. Wt1 is crucial for epicardial adhesion and EMT through direct activation of the transcription factors Snail1 and Slug [115]. In addition, Wt1-/- mouse embryos show decreased expression of Raldh2 [116], and epicardial EMT is partially rescued by RA supplementation in Wt1 deficient mouse [117,118]. These data indicate that perturbation of RA signaling contributes to defective EMT in Wt1 null hearts. The Wt1 and RA signaling pathways are also cross-inductive as epicardial RA has been shown to induce Wt1 expression [118,119].
Lineage tracing experiments have suggested that epicardium contributes to the formation of coronary endothelial and smooth muscle cells [107]. To date, the contribution of EPDCs to coronary system in the mouse continues to be debated (see [101]). Interestingly, differentiation of coronary smooth muscle cells is delayed compared with endothelial cells, and coronary smooth muscle cells mature only after endothelial tube formation. Differentiation of EPDCs into coronary smooth muscle cells corresponds to the time when Raldh2 expression is progressively lost in EPDCs [107]. Braitsch et al. showed that Tcf21 and Raldh2 are co-expressed in approximately 80% EPDCs [118]. They propose an interesting model for the regulation of EPDC differentiation in which RA signaling, downstream of Raldh2 expression, induces Tcf21 to inhibit smooth muscle cell differentiation in subepicardial EPDCs [108]. Upon invasion of the myocardium, EPDC expression of Raldh2 is downregulated, concomitantly with decreased Tcf21 expression in some but not all EPDCs. Thus, downregulation of Tcf21 expression allows EPDC differentiation into smooth muscle cells. In contrast, Tcf21 expression persists in EPDCs that differentiate into fibroblasts, suggesting a bimodal action of this factor. In addition, Azumbaja et al. showed that whereas an early role of RA and VEGF signaling is necessary to prevent differentiation of EPDCs into coronary smooth muscle cells, the latter action of myocardially secreted VEGF is to orient uncommitted EPDCs towards a coronary endothelial phenotype [120]. Thus, these two signals act until a primary coronary endothelial plexus is formed by vasculogenesis. However, it remains to be established whether RA and VEGF signaling inhibit coronary smooth muscle cells additively or synergistically, and whether these signals are in the same pathway. This latter point is supported by a recent study revealing a reduction of VEGFR2 expression in Raldh2 knockout embryos [103]. Finally, all these studies provide evidence that RA is a crucial signal involved in the spatiotemporal control of coronary smooth muscle cell differentiation (Figure 4).
6. Conclusions
Since the first experimental data demonstrating the relationship between maternal vitamin A deficiency and congenital heart defects, there have been major advances in our understanding of the role of RA signaling during heart development. In particular, the action of RA signaling on cardiac progenitor cells has gained renewed interpretation with the identification of the SHF. Although a number of studies have shown that RA signaling is essential to restrict the pool of cardiac progenitor cells, it is not clear whether RA acts equivalently on the FHF and SHF. A recent study has shown that the expression of anterior Hox genes in the SHF and the contribution of these progenitor cells to the heart are sensitive to RA dosage. While nested domain of Hox genes expression in the SHF appears important for the pre-patterning of the heart, much remains to be learned in terms of transcriptional targets of RA signaling during this process. Although cardiac defects in Hox deficient embryos have yet to be studied in detail, it will be important to compare these phenotypes to those of RA deficient mice. During later development, RA signaling regulates myocardial growth through the activation of mitogenic factors and transcription factors in the epicardium. During regeneration of the injured zebrafish heart, Raldh2 expression in the epicardium is increased, suggesting that RA signaling contributes to cardiac reprograming [121]. As a note, a recent study in the mouse has shown that the RA signaling pathway is activated in postischemic hearts and may play a role in the regulation of damage and repair during remodeling [122]. It will therefore be important to resolve postnatal RA actions in order to provide clinical benefits in treating damaged cardiac muscle.
Acknowledgements
We thank Robert Kelly critical reading of the manuscript. N.E.R. received a Ph.D. fellowship from the “Ministère de l’Enseignement Supérieur et de la Recherche”. Grant support was provided by the Association Française contre les Myopathies (NMH-Decrypt Project), the Agence Nationale pour la Recherche (ANR-13-BSV2-0003-01), and the Institut National de la Santé et de la Recherche Médicale to S.Z.
Author Contribution
S.Z wrote the paper with contributions from N.E.R and N.B. S.Z. and N.E.R. made the artwork. N.E.R. and N.B. made thoughtful suggestions in the preparation of the manuscript.
Conflict of Interest
The authors declare no conflict of interest.
References
- Kirby, M.L. Cardiac Development; University Press: Oxford, UK, 2007. [Google Scholar]
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835. [Google Scholar] [CrossRef]
- Perez-Pomares, J.M.; Gonzalez-Rosa, J.M.; Munoz-Chapuli, R. Building the vertebrate heart—An evolutionary approach to cardiac development. Int. J. Dev. Biol. 2009, 53, 1427–1443. [Google Scholar] [CrossRef]
- Brand, T. Heart development: Molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 2003, 258, 1–19. [Google Scholar] [CrossRef]
- Xu, H.; Baldini, A. Genetic pathways to mammalian heart development: Recent progress from manipulation of the mouse genome. Semin. Cell. Dev. Biol. 2007, 18, 77–83. [Google Scholar] [CrossRef]
- Hoffman, J.I.; Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002, 39, 1890–1900. [Google Scholar] [CrossRef]
- Vincent, S.D.; Buckingham, M.E. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 2010, 90, 1–41. [Google Scholar] [CrossRef]
- Srivastava, D. Making or breaking the heart: From lineage determination to morphogenesis. Cell 2006, 126, 1037–1048. [Google Scholar] [CrossRef]
- Zaffran, S.; Kelly, R.G. New developments in the second heart field. Differentiation 2012, 84, 17–24. [Google Scholar] [CrossRef]
- Rochais, F.; Mesbah, K.; Kelly, R.G. Signaling pathways controlling second heart field development. Circul. Res. 2009, 104, 933–942. [Google Scholar] [CrossRef]
- Rhinn, M.; Dolle, P. Retinoic acid signalling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef]
- Dersch, H.; Zile, M.H. Induction of normal cardiovascular development in the vitamin A-deprived quail embryo by natural retinoids. Dev. Biol. 1993, 160, 424–433. [Google Scholar] [CrossRef]
- Zile, M.H. Vitamin A-not for your eyes only: Requirement for heart formation begins early in embryogenesis. Nutrients 2010, 2, 532–550. [Google Scholar] [CrossRef]
- Wilson, J.G.; Warkany, J. Cardiac and aortic arch anomalies in the offspring of vitamin A deficient rats correlated with similar human anomalies. Pediatrics 1950, 5, 708–725. [Google Scholar]
- Wilson, J.G.; Warkany, J. Congenital anomalies of heart and great vessels in offspring of vitamin A-deficient rats. Am. J. Dis. Child. 1950, 79, 963. [Google Scholar]
- Maden, M. Role and distribution of retinoic acid during cns development. Int. Rev. Cytol. 2001, 209, 1–77. [Google Scholar] [CrossRef]
- Lammer, E.J.; Chen, D.T.; Hoar, R.M.; Agnish, N.D.; Benke, P.J.; Braun, J.T.; Curry, C.J.; Fernhoff, P.M.; Grix, A.W., Jr.; Lott, I.T.; et al. Retinoic acid embryopathy. N. Engl. J. Med. 1985, 313, 837–841. [Google Scholar] [CrossRef]
- Duester, G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008, 134, 921–931. [Google Scholar] [CrossRef]
- Niederreither, K.; Subbarayan, V.; Dolle, P.; Chambon, P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999, 21, 444–448. [Google Scholar] [CrossRef]
- Thompson, J.N. The role of vitamin A in reproduction. In The Fat Soluble Vitamins; University of Wisconcin Press: Madison, WI, USA, 1969; pp. 267–281. [Google Scholar]
- Mark, M.; Ghyselinck, N.B.; Chambon, P. Function of retinoic acid receptors during embryonic development. Nucl. Recept. Sign. 2009, 7. [Google Scholar] [CrossRef]
- Niederreither, K.; Dolle, P. Retinoids and heart development. In Heart Development and Regeneration; Rosenthal, N., Harvey, R.P., Eds.; Academic Press: San Diego, CA, USA, 2010; Volume 1. [Google Scholar]
- Chambon, P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996, 10, 940–954. [Google Scholar]
- Molotkov, A.; Duester, G. Genetic evidence that retinaldehyde dehydrogenase raldh1 (aldh1a1) functions downstream of alcohol dehydrogenase adh1 in metabolism of retinol to retinoic acid. J. Biol. Chem. 2003, 278, 36085–36090. [Google Scholar] [CrossRef]
- Molotkova, N.; Molotkov, A.; Duester, G. Role of retinoic acid during forebrain development begins late when raldh3 generates retinoic acid in the ventral subventricular zone. Dev. Biol 2007, 303, 601–610. [Google Scholar] [CrossRef]
- Zhao, X.; Sirbu, I.O.; Mic, F.A.; Molotkova, N.; Molotkov, A.; Kumar, S.; Duester, G. Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr. Biol. 2009, 19, 1050–1057. [Google Scholar]
- Maclean, G.; Dolle, P.; Petkovich, M. Genetic disruption of cyp26b1 severely affects development of neural crest derived head structures, but does not compromise hindbrain patterning. Dev. Dyn. 2009, 238, 732–745. [Google Scholar] [CrossRef]
- Hochgreb, T.; Linhares, V.L.; Menezes, D.C.; Sampaio, A.C.; Yan, C.Y.; Cardoso, W.V.; Rosenthal, N.; Xavier-Neto, J. A caudorostral wave of raldh2 conveys anteroposterior information to the cardiac field. Development 2003, 130, 5363–5374. [Google Scholar] [CrossRef]
- Moss, J.B.; Xavier-Neto, J.; Shapiro, M.D.; Nayeem, S.M.; McCaffery, P.; Drager, U.C.; Rosenthal, N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev. Biol. 1998, 199, 55–71. [Google Scholar] [CrossRef]
- Niederreither, K.; McCaffery, P.; Drager, U.C.; Chambon, P.; Dolle, P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (raldh-2) gene during mouse development. Mech. Dev. 1997, 62, 67–78. [Google Scholar] [CrossRef]
- Xavier-Neto, J.; Neville, C.M.; Shapiro, M.D.; Houghton, L.; Wang, G.F.; Nikovits, W., Jr.; Stockdale, F.E.; Rosenthal, N. A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 1999, 126, 2677–2687. [Google Scholar]
- Chambers, D.; Wilson, L.; Maden, M.; Lumsden, A. Raldh-independent generation of retinoic acid during vertebrate embryogenesis by cyp1b1. Development 2007, 134, 1369–1383. [Google Scholar] [CrossRef]
- Duester, G. Retinoid signaling in control of progenitor cell differentiation during mouse development. Semin. Cell. Dev. Biol. 2013, 24, 694–700. [Google Scholar] [CrossRef]
- Hoover, L.L.; Burton, E.G.; Brooks, B.A.; Kubalak, S.W. The expanding role for retinoid signaling in heart development. Sci. World J. 2008, 8, 194–211. [Google Scholar] [CrossRef]
- Xavier-Neto, J.; Rosenthal, N.; Silva, F.A.; Matos, T.G.; Hochgreb, T.; Linhares, V.L. Retinoid signaling and cardiac anteroposterior segmentation. Genesis 2001, 31, 97–104. [Google Scholar] [CrossRef]
- Tam, P.P.; Parameswaran, M.; Kinder, S.J.; Weinberger, R.P. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: The role of ingression and tissue movement during gastrulation. Development 1997, 124, 1631–1642. [Google Scholar]
- Garcia-Martinez, V.; Schoenwolf, G.C. Primitive-streak origin of the cardiovascular system in avian embryos. Dev. Biol. 1993, 159, 706–719. [Google Scholar] [CrossRef]
- Stainier, D.Y.; Fishman, M.C. Patterning the zebrafish heart tube: Acquisition of anteroposterior polarity. Dev. Biol. 1992, 153, 91–101. [Google Scholar] [CrossRef]
- Jacobson, A.G.; Sater, A.K. Features of embryonic induction. Development 1988, 104, 341–359. [Google Scholar]
- Tonissen, K.F.; Drysdale, T.A.; Lints, T.J.; Harvey, R.P.; Krieg, P.A. Xnkx-2.5, a xenopus gene related to nkx-2.5 and tinman: Evidence for a conserved role in cardiac development. Dev. Biol. 1994, 162, 325–328. [Google Scholar] [CrossRef]
- Evans, S.M.; Yan, W.; Murillo, M.P.; Ponce, J.; Papalopulu, N. Tinman, a drosophila homeobox gene required for heart and visceral mesoderm specification, may be represented by a family of genes in vertebrates: Xnkx-2.3, a second vertebrate homologue of tinman. Development 1995, 121, 3889–3899. [Google Scholar]
- Raffin, M.; Leong, L.M.; Rones, M.S.; Sparrow, D.; Mohun, T.; Mercola, M. Subdivision of the cardiac nkx2.5 expression domain into myogenic and nonmyogenic compartments. Dev. Biol. 2000, 218, 326–340. [Google Scholar] [CrossRef]
- Cai, C.L.; Liang, X.; Shi, Y.; Chu, P.H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Prall, O.W.; Menon, M.K.; Solloway, M.J.; Watanabe, Y.; Zaffran, S.; Bajolle, F.; Biben, C.; McBride, J.J.; Robertson, B.R.; Chaulet, H.; et al. An nkx2–5/bmp2/smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 2007, 128, 947–959. [Google Scholar] [CrossRef]
- Baldini, A. Dissecting contiguous gene defects: Tbx1. Curr. Opin. Genet. Dev. 2005, 15, 279–284. [Google Scholar] [CrossRef]
- Kelly, R.G.; Brown, N.A.; Buckingham, M.E. The arterial pole of the mouse heart forms from fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell. 2001, 1, 435–440. [Google Scholar] [CrossRef]
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.M.; Black, B.L. Mef2c is a direct transcriptional target of isl1 and gata factors in the anterior heart field during mouse embryonic development. Development 2004, 131, 3931–3942. [Google Scholar] [CrossRef]
- Engleka, K.A.; Manderfield, L.J.; Brust, R.D.; Li, L.; Cohen, A.; Dymecki, S.M.; Epstein, J.A. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circul. Res. 2012, 110, 922–926. [Google Scholar] [CrossRef]
- Park, E.J.; Ogden, L.A.; Talbot, A.; Evans, S.; Cai, C.L.; Black, B.L.; Frank, D.U.; Moon, A.M. Required, tissue-specific roles for fgf8 in outflow tract formation and remodeling. Development 2006, 133, 2419–2433. [Google Scholar] [CrossRef]
- Dyer, L.A.; Kirby, M.L. The role of secondary heart field in cardiac development. Dev. Biol. 2009, 336, 137–144. [Google Scholar] [CrossRef]
- Heine, U.I.; Roberts, A.B.; Munoz, E.F.; Roche, N.S.; Sporn, M.B. Effects of retinoid deficiency on the development of the heart and vascular system of the quail embryo. Virchows Archiv. B 1985, 50, 135–152. [Google Scholar]
- Niederreither, K.; Vermot, J.; Messaddeq, N.; Schuhbaur, B.; Chambon, P.; Dolle, P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development 2001, 128, 1019–1031. [Google Scholar]
- Bruneau, B.G.; Logan, M.; Davis, N.; Levi, T.; Tabin, C.J.; Seidman, J.G.; Seidman, C.E. Chamber-specific cardiac expression of tbx5 and heart defects in holt-oram syndrome. Dev. Biol. 1999, 211, 100–108. [Google Scholar] [CrossRef]
- Sporn, M.B.; Roberts, A.B. Suppression of carcinogenesis by retinoids: Interactions with peptide growth factors and their receptors as a key mechanism. Princess Takamatsu Symp. 1985, 16, 149–158. [Google Scholar]
- Thompson, J.N.; Howell, J.M.; Pitt, G.A.; McLaughlin, C.I. The biological activity of retinoic acid in the domestic fowl and the effects of vitamin A deficiency on the chick embryo. Brit. J. Nutr. 1969, 23, 471–490. [Google Scholar] [CrossRef]
- Kostetskii, I.; Yuan, S.Y.; Kostetskaia, E.; Linask, K.K.; Blanchet, S.; Seleiro, E.; Michaille, J.J.; Brickell, P.; Zile, M. Initial retinoid requirement for early avian development coincides with retinoid receptor coexpression in the precardiac fields and induction of normal cardiovascular development. Dev. Dyn. 1998, 213, 188–198. [Google Scholar] [CrossRef]
- Collop, A.H.; Broomfield, J.A.; Chandraratna, R.A.; Yong, Z.; Deimling, S.J.; Kolker, S.J.; Weeks, D.L.; Drysdale, T.A. Retinoic acid signaling is essential for formation of the heart tube in xenopus. Dev. Biol. 2006, 291, 96–109. [Google Scholar] [CrossRef]
- Kostetskii, I.; Jiang, Y.; Kostetskaia, E.; Yuan, S.; Evans, T.; Zile, M. Retinoid signaling required for normal heart development regulates gata-4 in a pathway distinct from cardiomyocyte differentiation. Dev. Biol. 1999, 206, 206–218. [Google Scholar] [CrossRef]
- Ghatpande, S.; Brand, T.; Zile, M.; Evans, T. Bmp2 and gata4 function additively to rescue heart tube development in the absence of retinoids. Dev. Dyn. 2006, 235, 2030–2039. [Google Scholar] [CrossRef]
- Ghatpande, S.K.; Zhou, H.R.; Cakstina, I.; Carlson, C.; Rondini, E.A.; Romeih, M.; Zile, M.H. Transforming growth factor beta2 is negatively regulated by endogenous retinoic acid during early heart morphogenesis. Dev. Growth Differ. 2010, 52, 433–455. [Google Scholar] [CrossRef]
- Romeih, M.; Cakstina, I.; Zile, M.H. Retinoic acid is a negative physiological regulator of n-cadherin during early avian heart morphogenesis. Dev. Growth Differ. 2009, 51, 753–767. [Google Scholar] [CrossRef]
- Paschaki, M.; Schneider, C.; Rhinn, M.; Thibault-Carpentier, C.; Dembele, D.; Niederreither, K.; Dolle, P. Transcriptomic analysis of murine embryos lacking endogenous retinoic acid signaling. PLoS ONE 2013, 8, e62274. [Google Scholar] [CrossRef]
- Osmond, M.K.; Butler, A.J.; Voon, F.C.; Bellairs, R. The effects of retinoic acid on heart formation in the early chick embryo. Development 1991, 113, 1405–1417. [Google Scholar]
- Yutzey, K.E.; Rhee, J.T.; Bader, D. Expression of the atrial-specific myosin heavy chain amhc1 and the establishment of anteroposterior polarity in the developing chicken heart. Development 1994, 120, 871–883. [Google Scholar]
- Dickman, E.D.; Smith, S.M. Selective regionalisation of cardiomyocyte gene expression and cardiac morphogenesis. Dev. Dyn. 1996, 206, 39–48. [Google Scholar] [CrossRef]
- Chazaud, C.; Chambon, P.; Dolle, P. Retinoic acid is required in the mouse embryo for left-right asymmetry determination and heart morphogenesis. Development 1999, 126, 2589–2596. [Google Scholar]
- Tsukui, T.; Capdevila, J.; Tamura, K.; Ruiz-Lozano, P.; Rodriguez-Esteban, C.; Yonei-Tamura, S.; Magallon, J.; Chandraratna, R.A.; Chien, K.; Blumberg, B.; et al. Multiple left-right asymmetry defects in shh(-/-) mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of lefty-1. Proc. Nat. Acad. Sci. USA 1999, 96, 11376–11381. [Google Scholar] [CrossRef]
- Wasiak, S.; Lohnes, D. Retinoic acid affects left-right patterning. Dev. Biol. 1999, 215, 332–342. [Google Scholar] [CrossRef]
- Zile, M.H.; Kostetskii, I.; Yuan, S.; Kostetskaia, E.; St Amand, T.R.; Chen, Y.; Jiang, W. Retinoid signaling is required to complete the vertebrate cardiac left/right asymmetry pathway. Dev. Biol. 2000, 223, 323–338. [Google Scholar] [CrossRef]
- Duester, G. Retinoic acid regulation of the somitogenesis clock. Birth Defects Res. Part C Embryo Today Rev. 2007, 81, 84–92. [Google Scholar] [CrossRef]
- Ryckebusch, L.; Wang, Z.; Bertrand, N.; Lin, S.C.; Chi, X.; Schwartz, R.; Zaffran, S.; Niederreither, K. Retinoic acid deficiency alters second heart field formation. Proc. Nat. Acad. Sci. USA 2008, 105, 2913–2918. [Google Scholar] [CrossRef]
- Sirbu, I.O.; Zhao, X.; Duester, G. Retinoic acid controls heart anteroposterior patterning by down-regulating isl1 through the fgf8 pathway. Dev. Dyn. 2008, 237, 1627–1635. [Google Scholar] [CrossRef]
- Keegan, B.R.; Feldman, J.L.; Begemann, G.; Ingham, P.W.; Yelon, D. Retinoic acid signaling restricts the cardiac progenitor pool. Science 2005, 307, 247–249. [Google Scholar] [CrossRef]
- Waxman, J.S.; Keegan, B.R.; Roberts, R.W.; Poss, K.D.; Yelon, D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev. Cell. 2008, 15, 923–934. [Google Scholar] [CrossRef]
- Witzel, H.R.; Jungblut, B.; Choe, C.P.; Crump, J.G.; Braun, T.; Dobreva, G. The lim protein ajuba restricts the second heart field progenitor pool by regulating isl1 activity. Dev. Cell. 2012, 23, 58–70. [Google Scholar] [CrossRef]
- Deimling, S.J.; Drysdale, T.A. Retinoic acid regulates anterior-posterior patterning within the lateral plate mesoderm of xenopus. Mech. Dev. 2009, 126, 913–923. [Google Scholar] [CrossRef]
- Sorrell, M.R.; Waxman, J.S. Restraint of fgf8 signaling by retinoic acid signaling is required for proper heart and forelimb formation. Dev. Biol. 2011, 358, 44–55. [Google Scholar] [CrossRef]
- Brondani, V.; Klimkait, T.; Egly, J.M.; Hamy, F. Promoter of fgf8 reveals a unique regulation by unliganded raralpha. J. Mol. Biol. 2002, 319, 715–728. [Google Scholar] [CrossRef]
- Begemann, G.; Schilling, T.F.; Rauch, G.J.; Geisler, R.; Ingham, P.W. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development 2001, 128, 3081–3094. [Google Scholar]
- Zaffran, S.; Kelly, R.G.; Meilhac, S.M.; Buckingham, M.E.; Brown, N.A. Right ventricular myocardium derives from the anterior heart field. Circul. Res. 2004, 95, 261–268. [Google Scholar] [CrossRef]
- Galli, D.; Dominguez, J.N.; Zaffran, S.; Munk, A.; Brown, N.A.; Buckingham, M.E. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as pitx2c is expressed. Development 2008, 135, 1157–1167. [Google Scholar] [CrossRef]
- Roberts, C.; Ivins, S.M.; James, C.T.; Scambler, P.J. Retinoic acid down-regulates tbx1 expression in vivo and in vitro. Dev. Dyn. 2005, 232, 928–938. [Google Scholar] [CrossRef]
- Ivins, S.; Lammerts van Beuren, K.; Roberts, C.; James, C.; Lindsay, E.; Baldini, A.; Ataliotis, P.; Scambler, P.J. Microarray analysis detects differentially expressed genes in the pharyngeal region of mice lacking tbx1. Dev. Biol. 2005, 285, 554–569. [Google Scholar] [CrossRef]
- Guris, D.L.; Duester, G.; Papaioannou, V.E.; Imamoto, A. Dose-dependent interaction of tbx1 and crkl and locally aberrant ra signaling in a model of del22q11 syndrome. Dev. Cell. 2006, 10, 81–92. [Google Scholar] [CrossRef]
- Ryckebusch, L.; Bertrand, N.; Mesbah, K.; Bajolle, F.; Niederreither, K.; Kelly, R.G.; Zaffran, S. Decreased levels of embryonic retinoic acid synthesis accelerate recovery from arterial growth delay in a mouse model of digeorge syndrome. Circul. Res. 2010, 106, 686–694. [Google Scholar] [CrossRef]
- Waxman, J.S.; Yelon, D. Increased hox activity mimics the teratogenic effects of excess retinoic acid signaling. Dev. Dyn. 2009, 238, 1207–1213. [Google Scholar] [CrossRef]
- Nolte, C.; Jinks, T.; Wang, X.; Martinez Pastor, M.T.; Krumlauf, R. Shadow enhancers flanking the hoxb cluster direct dynamic hox expression in early heart and endoderm development. Dev. Biol. 2013, 383, 158–173. [Google Scholar] [CrossRef]
- Bertrand, N.; Roux, M.; Ryckebusch, L.; Niederreither, K.; Dolle, P.; Moon, A.; Capecchi, M.; Zaffran, S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev. Biol. 2011, 353, 266–274. [Google Scholar] [CrossRef]
- Diman, N.Y.; Remacle, S.; Bertrand, N.; Picard, J.J.; Zaffran, S.; Rezsohazy, R. A retinoic acid responsive hoxa3 transgene expressed in embryonic pharyngeal endoderm, cardiac neural crest and a subdomain of the second heart field. PloS One 2011, 6, e27624. [Google Scholar]
- Dolle, P.; Fraulob, V.; Gallego-Llamas, J.; Vermot, J.; Niederreither, K. Fate of retinoic acid-activated embryonic cell lineages. Dev. Biol. 2010, 239, 3260–3274. [Google Scholar]
- Soshnikova, N.; Dewaele, R.; Janvier, P.; Krumlauf, R.; Duboule, D. Duplications of hox gene clusters and the emergence of vertebrates. Dev. Biol. 2013, 378, 194–199. [Google Scholar] [CrossRef]
- Ward, C.; Stadt, H.; Hutson, M.; Kirby, M.L. Ablation of the secondary heart field leads to tetralogy of fallot and pulmonary atresia. Dev. Biol. 2005, 284, 72–83. [Google Scholar] [CrossRef]
- Waldo, K.; Miyagawa-Tomita, S.; Kumiski, D.; Kirby, M.L. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: Aortic sac to ventricular septal closure. Dev. Biol. 1998, 196, 129–144. [Google Scholar] [CrossRef]
- Li, P.; Pashmforoush, M.; Sucov, H.M. Retinoic acid regulates differentiation of the secondary heart field and tgfbeta-mediated outflow tract septation. Dev. Cell. 2010, 18, 480–485. [Google Scholar] [CrossRef]
- Lee, R.Y.; Luo, J.; Evans, R.M.; Giguere, V.; Sucov, H.M. Compartment-selective sensitivity of cardiovascular morphogenesis to combinations of retinoic acid receptor gene mutations. Circul. Res. 1997, 80, 757–764. [Google Scholar]
- Kirby, M.L.; Gale, T.F.; Stewart, D.E. Neural crest cells contribute to normal aorticopulmonary septation. Science 1983, 220, 1059–1061. [Google Scholar]
- Jiang, X.; Choudhary, B.; Merki, E.; Chien, K.R.; Maxson, R.E.; Sucov, H.M. Normal fate and altered function of the cardiac neural crest cell lineage in retinoic acid receptor mutant embryos. Mech. Dev. 2002, 117, 115–122. [Google Scholar] [CrossRef]
- Kubalak, S.W.; Hutson, D.R.; Scott, K.K.; Shannon, R.A. Elevated transforming growth factor beta2 enhances apoptosis and contributes to abnormal outflow tract and aortic sac development in retinoic x receptor alpha knockout embryos. Development 2002, 129, 733–746. [Google Scholar]
- Braitsch, C.M.; Yutzey, K.E. Transcriptional control of cell lineage development in epicardium-derived cells. J. Dev. Biol. 2013, 1, 92–111. [Google Scholar] [CrossRef]
- Gittenberger-de Groot, A.C.; Winter, E.M.; Poelmann, R.E. Epicardium-derived cells (EPDCs) in development, cardiac disease and repair of ischemia. J. Cell. Mol. Med. 2010, 14, 1056–1060. [Google Scholar]
- Pires-Gomes, A.A.; Perez-Pomares, J.M. The epicardium and coronary artery formation. J. Dev. Biol. 2013, 1, 186–202. [Google Scholar] [CrossRef]
- Stuckmann, I.; Evans, S.; Lassar, A.B. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev. Biol. 2003, 255, 334–349. [Google Scholar] [CrossRef]
- Lin, S.C.; Dolle, P.; Ryckebusch, L.; Noseda, M.; Zaffran, S.; Schneider, M.D.; Niederreither, K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc. Nat. Acad. Sci. USA 2010, 107, 9234–9239. [Google Scholar]
- Sucov, H.M.; Dyson, E.; Gumeringer, C.L.; Price, J.; Chien, K.R.; Evans, R.M. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994, 8, 1007–1018. [Google Scholar] [CrossRef]
- Gruber, P.J.; Kubalak, S.W.; Pexieder, T.; Sucov, H.M.; Evans, R.M.; Chien, K.R. RXR alpha deficiency confers genetic susceptibility for aortic sac, conotruncal, atrioventricular cushion, and ventricular muscle defects in mice. J. Clin. Invest. 1996, 98, 1332–1343. [Google Scholar] [CrossRef]
- Merki, E.; Zamora, M.; Raya, A.; Kawakami, Y.; Wang, J.; Zhang, X.; Burch, J.; Kubalak, S.W.; Kaliman, P.; Izpisua Belmonte, J.C.; et al. Epicardial retinoid x receptor alpha is required for myocardial growth and coronary artery formation . Proc. Nat. Acad. Sci. USA 2005, 102, 18455–18460. [Google Scholar] [CrossRef]
- Perez-Pomares, J.M.; Phelps, A.; Sedmerova, M.; Carmona, R.; Gonzalez-Iriarte, M.; Munoz-Chapuli, R.; Wessels, A. Experimental studies on the spatiotemporal expression of wt1 and raldh2 in the embryonic avian heart: A model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs). Dev. Biol. 2002, 247, 307–326. [Google Scholar] [CrossRef]
- Acharya, A.; Baek, S.T.; Huang, G.; Eskiocak, B.; Goetsch, S.; Sung, C.Y.; Banfi, S.; Sauer, M.F.; Olsen, G.S.; Duffield, J.S.; et al. The bhlh transcription factor tcf21 is required for lineage-specific emt of cardiac fibroblast progenitors. Development 2012, 139, 2139–2149. [Google Scholar] [CrossRef]
- Red-Horse, K.; Ueno, H.; Weissman, I.L.; Krasnow, M.A. Coronary arteries form by developmental reprogramming of venous cells. Nature 2010, 464, 549–553. [Google Scholar] [CrossRef]
- Vrancken Peeters, M.P.; Gittenberger-de Groot, A.C.; Mentink, M.M.; Hungerford, J.E.; Little, C.D.; Poelmann, R.E. The development of the coronary vessels and their differentiation into arteries and veins in the embryonic quail heart. Dev. Dyn. 1997, 208, 338–348. [Google Scholar] [CrossRef]
- Brade, T.; Kumar, S.; Cunningham, T.J.; Chatzi, C.; Zhao, X.; Cavallero, S.; Li, P.; Sucov, H.M.; Ruiz-Lozano, P.; Duester, G. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial igf2. Development 2011, 138, 139–148. [Google Scholar] [CrossRef]
- Chen, T.; Chang, T.C.; Kang, J.O.; Choudhary, B.; Makita, T.; Tran, C.M.; Burch, J.B.; Eid, H.; Sucov, H.M. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev. Biol. 2002, 250, 198–207. [Google Scholar]
- Lavine, K.J.; Yu, K.; White, A.C.; Zhang, X.; Smith, C.; Partanen, J.; Ornitz, D.M. Endocardial and epicardial derived fgf signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell. 2005, 8, 85–95. [Google Scholar] [CrossRef]
- Moore, A.W.; McInnes, L.; Kreidberg, J.; Hastie, N.D.; Schedl, A. Yac complementation shows a requirement for wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999, 126, 1845–1857. [Google Scholar]
- Zhou, B.; von Gise, A.; Ma, Q.; Hu, Y.W.; Pu, W.T. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev. Biol. 2010, 338, 251–261. [Google Scholar] [CrossRef]
- Guadix, J.A.; Ruiz-Villalba, A.; Lettice, L.; Velecela, V.; Munoz-Chapuli, R.; Hastie, N.D.; Perez-Pomares, J.M.; Martinez-Estrada, O.M. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of raldh2. Development 2011, 138, 1093–1097. [Google Scholar] [CrossRef]
- Norden, J.; Grieskamp, T.; Lausch, E.; van Wijk, B.; van den Hoff, M.J.; Englert, C.; Petry, M.; Mommersteeg, M.T.; Christoffels, V.M.; Niederreither, K.; et al. Wt1 and retinoic acid signaling in the subcoelomic mesenchyme control the development of the pleuropericardial membranes and the sinus horns. Circul. Res. 2010, 106, 1212–1220. [Google Scholar] [CrossRef]
- Braitsch, C.M.; Combs, M.D.; Quaggin, S.E.; Yutzey, K.E. Pod1/tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev. Biol. 2012, 368, 345–357. [Google Scholar] [CrossRef]
- Martinez-Estrada, O.M.; Lettice, L.A.; Essafi, A.; Guadix, J.A.; Slight, J.; Velecela, V.; Hall, E.; Reichmann, J.; Devenney, P.S.; Hohenstein, P.; et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of snail and e-cadherin. Nat. Genet. 2010, 42, 89–93. [Google Scholar] [CrossRef]
- Azambuja, A.P.; Portillo-Sanchez, V.; Rodrigues, M.V.; Omae, S.V.; Schechtman, D.; Strauss, B.E.; Costanzi-Strauss, E.; Krieger, J.E.; Perez-Pomares, J.M.; Xavier-Neto, J. Retinoic acid and vegf delay smooth muscle relative to endothelial differentiation to coordinate inner and outer coronary vessel wall morphogenesis. Circul. Res. 2010, 107, 204–216. [Google Scholar] [CrossRef]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011, 20, 397–404. [Google Scholar] [CrossRef]
- Bilbija, D.; Haugen, F.; Sagave, J.; Baysa, A.; Bastani, N.; Levy, F.O.; Sirsjo, A.; Blomhoff, R.; Valen, G. Retinoic acid signalling is activated in the postischemic heart and may influence remodelling. PLoS ONE 2012, 7, e44740. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).