A Residual N-Terminal Peptide Enhances Signaling of Depalmitoylated Hedgehog to the Patched Receptor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Lines
2.2. Generation of Recombinant Proteins
2.3. Protein Expression
2.4. Chromatography
2.5. Verification of Mouse Mesenchymal Stromal Cells
2.6. Shh Reporter Assays
2.7. qRT-PCR
2.8. Confocal Microscopy of Drosophila Eye Discs
2.9. Bioanalytical and Statistical Analysis
2.10. Molecular Modeling
3. Results
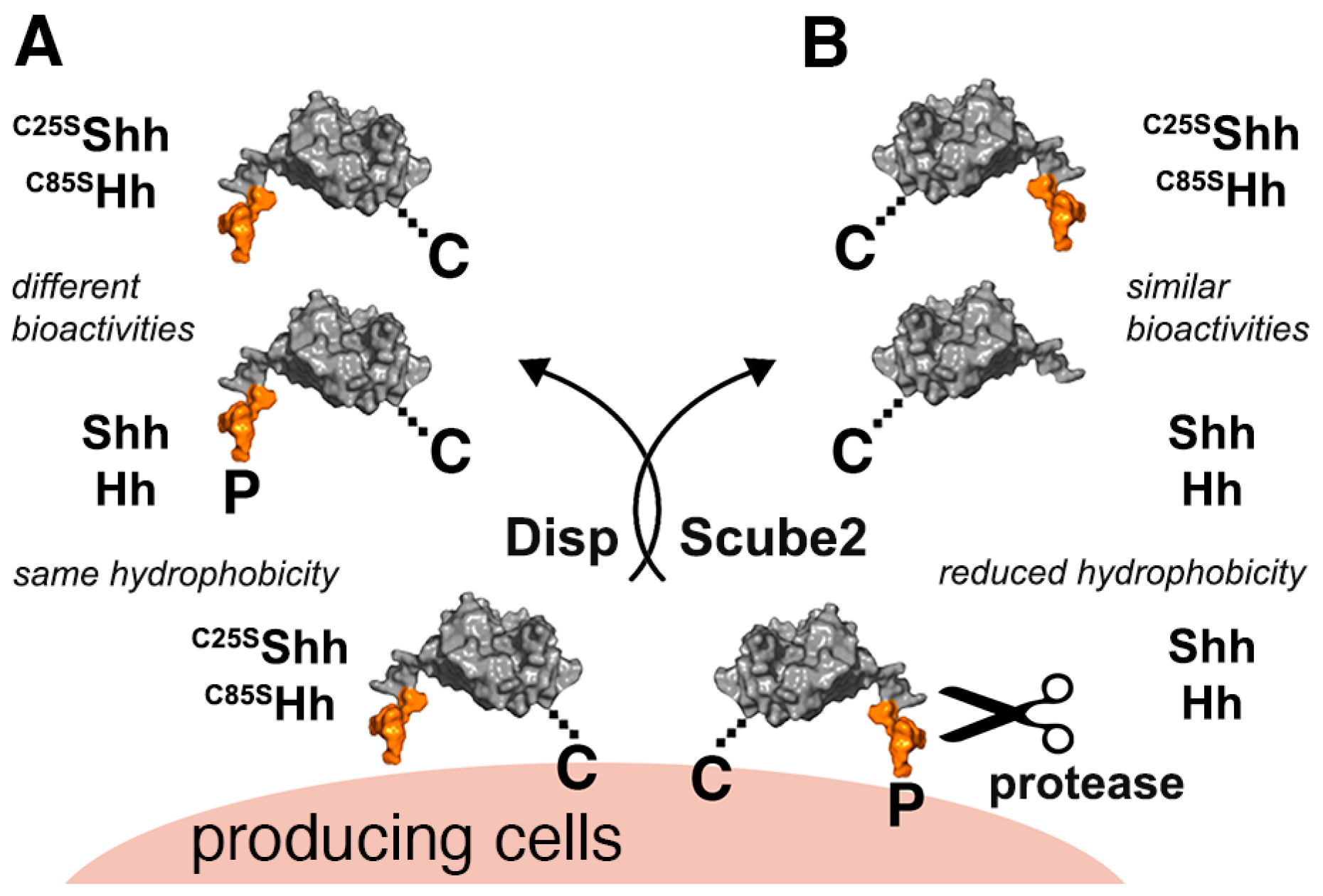
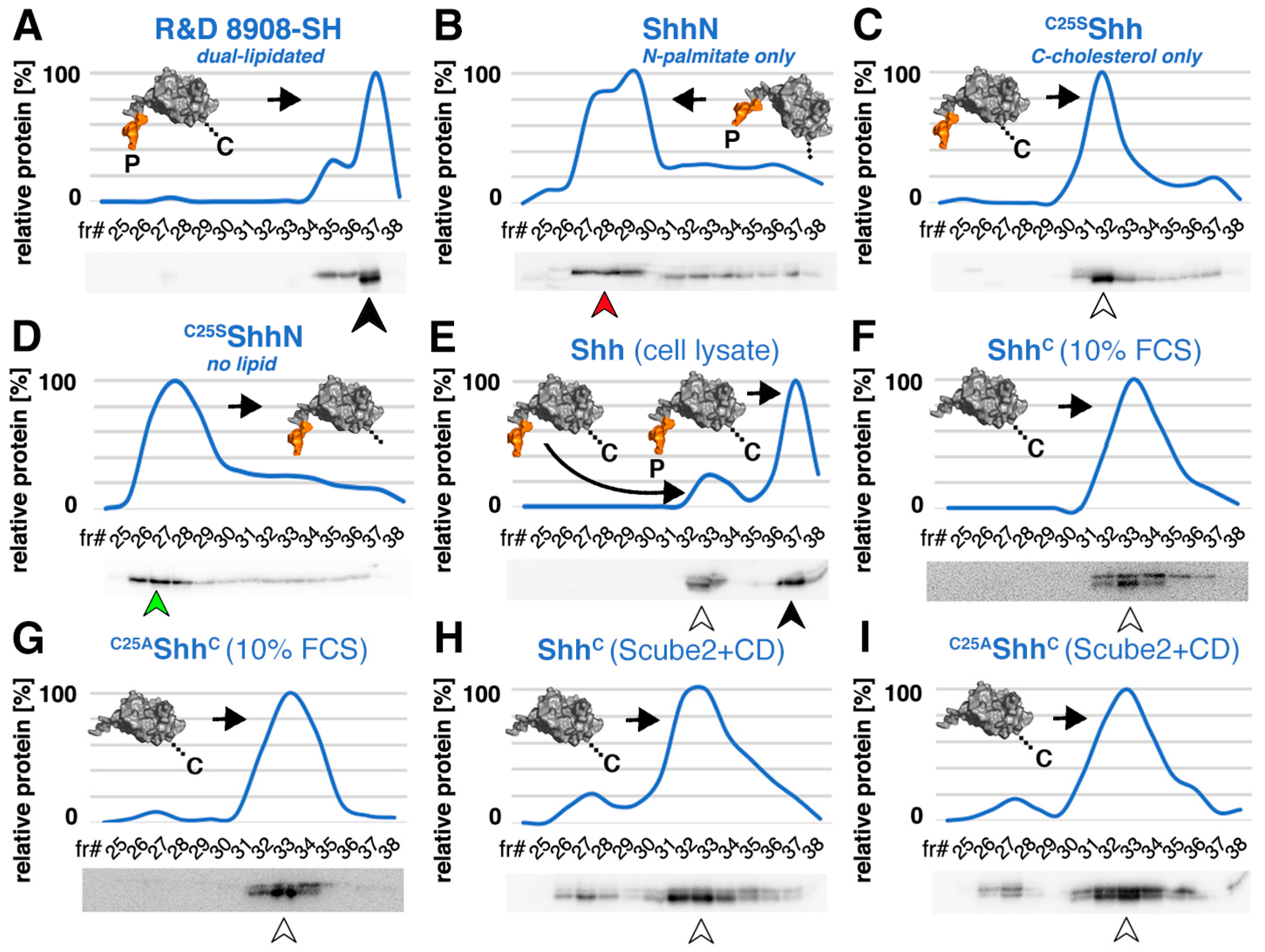
3.1. Serum Promotes the Release of Selectively Depalmitoylated ShhC from the Dual-Lipidated Cellular Precursor
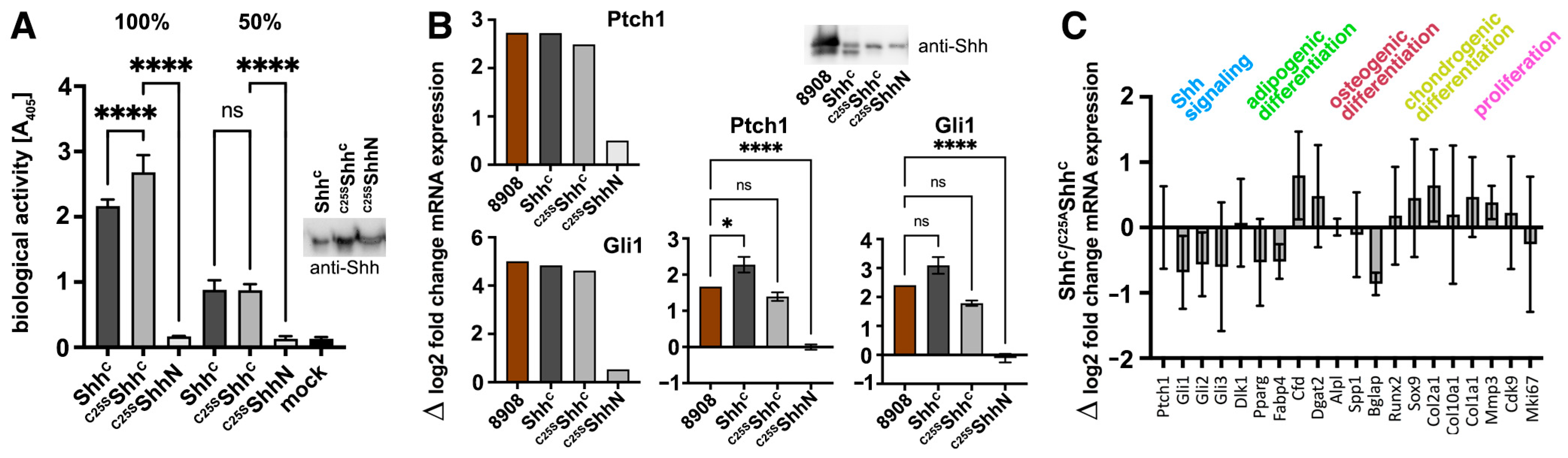
3.2. ShhC Induces In Vitro Differentiation of C3H10T1/2 and NIH3T3 Cells
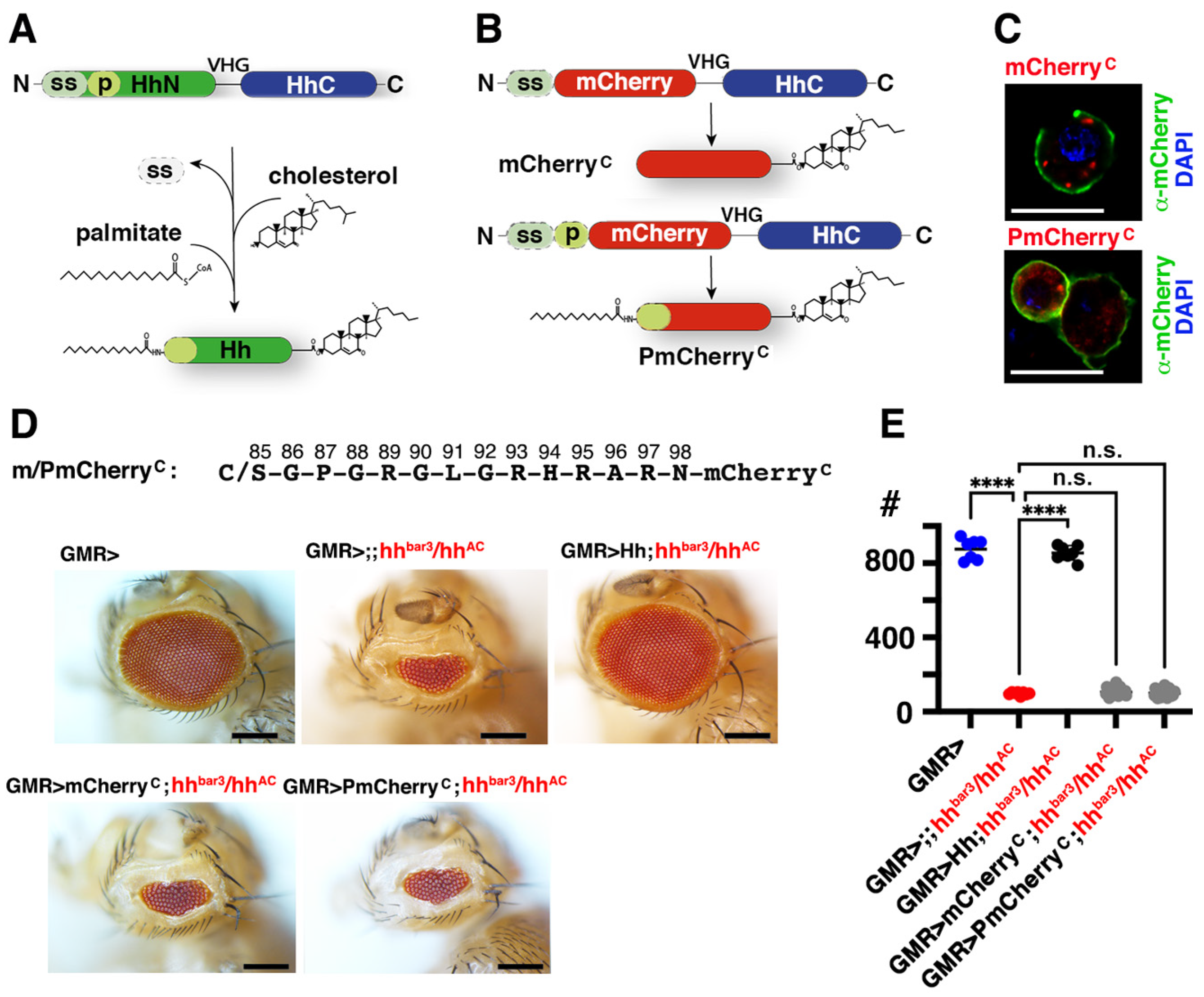
3.3. N-Terminal Amino Acids Contribute to ShhC Biofunction In Vitro
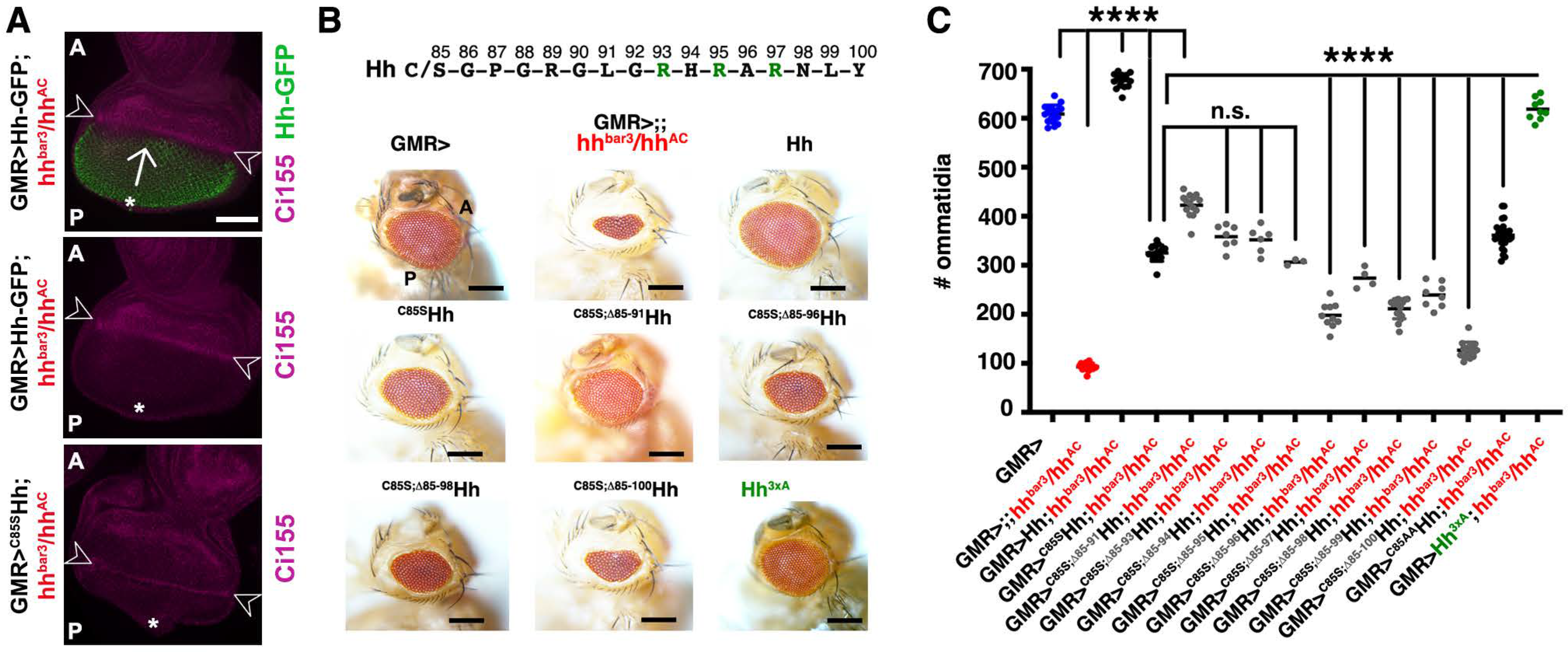
3.4. The N-Terminal Hh Peptide Contributes to Morphogenetic Furrow Progression in the Drosophila Eye Disc
3.5. Isolated Palmitoylated or Unpalmitoylated N-Terminal Peptides Are Not Active In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Luca, A.; Cerrato, V.; Fuca, E.; Parmigiani, E.; Buffo, A.; Leto, K. Sonic hedgehog patterning during cerebellar development. Cell Mol. Life Sci. 2016, 73, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kalderon, D. Coupling of Hedgehog and Hippo pathways promotes stem cell maintenance by stimulating proliferation. J. Cell Biol. 2014, 205, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 2008, 15, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Frank, D.B.; Kadzik, R.S.; Morley, M.P.; Rathi, K.S.; Wang, T.; Zhou, S.; Cheng, L.; Lu, M.M.; Morrisey, E.E. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 2015, 526, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, M. Hedgehog and wing development in Drosophila: A morphogen at work? Bioessays 2000, 22, 460–468. [Google Scholar] [CrossRef]
- Groves, I.; Placzek, M.; Fletcher, A.G. Of mitogens and morphogens: Modelling Sonic Hedgehog mechanisms in vertebrate development. Philos. Trans. R. Soc. Lond B. Biol. Sci. 2020, 375, 20190660. [Google Scholar] [CrossRef] [PubMed]
- Yauch, R.L.; Gould, S.E.; Scales, S.J.; Tang, T.; Tian, H.; Ahn, C.P.; Marshall, D.; Fu, L.; Januario, T.; Kallop, D.; et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008, 455, 406–410. [Google Scholar] [CrossRef]
- Porter, J.A.; Young, K.E.; Beachy, P.A. Cholesterol modification of hedgehog signaling proteins in animal development. Science 1996, 274, 255–259. [Google Scholar] [CrossRef]
- Chamoun, Z.; Mann, R.K.; Nellen, D.; von Kessler, D.P.; Bellotto, M.; Beachy, P.A.; Basler, K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 2001, 293, 2080–2084. [Google Scholar] [CrossRef]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef]
- Coupland, C.E.; Andrei, S.A.; Ansell, T.B.; Carrique, L.; Kumar, P.; Sefer, L.; Schwab, R.A.; Byrne, E.F.X.; Pardon, E.; Steyaert, J.; et al. Structure, mechanism, and inhibition of Hedgehog acyltransferase. Mol. Cell 2021, 81, 5025–5038.e5010. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.; Nellen, D.; Bellotto, M.; Hafen, E.; Senti, K.A.; Dickson, B.J.; Basler, K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 1999, 99, 803–815. [Google Scholar] [CrossRef]
- Ma, Y.; Erkner, A.; Gong, R.; Yao, S.; Taipale, J.; Basler, K.; Beachy, P.A. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 2002, 111, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Hollway, G.E.; Maule, J.; Gautier, P.; Evans, T.M.; Keenan, D.G.; Lohs, C.; Fischer, D.; Wicking, C.; Currie, P.D. Scube2 mediates Hedgehog signalling in the zebrafish embryo. Dev. Biol. 2006, 294, 104–118. [Google Scholar] [CrossRef]
- Johnson, J.L.; Hall, T.E.; Dyson, J.M.; Sonntag, C.; Ayers, K.; Berger, S.; Gautier, P.; Mitchell, C.; Hollway, G.E.; Currie, P.D. Scube activity is necessary for Hedgehog signal transduction in vivo. Dev. Biol. 2012, 368, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Dawber, R.J.; Hebbes, S.; Herpers, B.; Docquier, F.; van den Heuvel, M. Differential range and activity of various forms of the Hedgehog protein. BMC Dev. Biol. 2005, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.D.; Kraus, P.; Gaiano, N.; Nery, S.; Kohtz, J.; Fishell, G.; Loomis, C.A.; Treisman, J.E. An acylatable residue of Hedgehog is differentially required in Drosophila and mouse limb development. Dev. Biol. 2001, 233, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Li, Y.J.; Kawakami, T.; Xu, S.M.; Chuang, P.T. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004, 18, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Goetz, J.A.; Singh, S.; Suber, L.M.; Kull, F.J.; Robbins, D.J. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J. Biol. Chem. 2006, 281, 4087–4093. [Google Scholar] [CrossRef]
- Kohtz, J.D.; Lee, H.Y.; Gaiano, N.; Segal, J.; Ng, E.; Larson, T.; Baker, D.P.; Garber, E.A.; Williams, K.P.; Fishell, G. N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development 2001, 128, 2351–2363. [Google Scholar] [CrossRef]
- Qi, C.; Di Minin, G.; Vercellino, I.; Wutz, A.; Korkhov, V.M. Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci. Adv. 2019, 5, eaaw6490. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Schmiege, P.; Coutavas, E.; Li, X. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 2018, 362, eaas8843. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Schmiege, P.; Coutavas, E.; Wang, J.; Li, X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 2018, 560, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Cao, P.; Hu, M.; Gao, S.; Yan, N.; Gong, X. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat. Commun. 2019, 10, 2320. [Google Scholar] [CrossRef] [PubMed]
- Ehring, K.; Ehlers, S.F.; Froese, J.; Gude, F.; Puschmann, J.; Grobe, K. Two-way Dispatched function in Sonic hedgehog shedding and transfer to high-density lipoproteins. eLife 2023, 12, RP86920. [Google Scholar] [CrossRef]
- Bateman, J.R.; Lee, A.M.; Wu, C.T. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 2006, 173, 769–777. [Google Scholar] [CrossRef]
- Manikowski, D.; Kastl, P.; Schürmann, S.; Ehring, K.; Steffes, G.; Jakobs, P.; Grobe, K. C-Terminal Peptide Modifications Reveal Direct and Indirect Roles of Hedgehog Morphogen Cholesteroylation. Front. Cell Dev. Biol. 2021, 8, 615698. [Google Scholar] [CrossRef]
- Nakamura, T.; Aikawa, T.; Iwamoto-Enomoto, M.; Iwamoto, M.; Higuchi, Y.; Pacifici, M.; Kinto, N.; Yamaguchi, A.; Noji, S.; Kurisu, K.; et al. Induction of osteogenic differentiation by hedgehog proteins. Biochem. Biophys. Res. Commun. 1997, 237, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Ericson, J.; Morton, S.; Kawakami, A.; Roelink, H.; Jessell, T.M. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 1996, 87, 661–673. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Kuzmickas, R.P.; Jao, C.Y.; Liu, J.; Salic, A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012, 2, 308–320. [Google Scholar] [CrossRef]
- Wierbowski, B.M.; Petrov, K.; Aravena, L.; Gu, G.; Xu, Y.; Salic, A. Hedgehog Pathway Activation Requires Coreceptor-Catalyzed, Lipid-Dependent Relay of the Sonic Hedgehog Ligand. Dev. Cell 2020, 55, 450–467.e458. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, P.; Exner, S.; Schurmann, S.; Pickhinke, U.; Bandari, S.; Ortmann, C.; Kupich, S.; Schulz, P.; Hansen, U.; Seidler, D.G.; et al. Scube2 enhances proteolytic Shh processing from the surface of Shh-producing cells. J. Cell Sci. 2014, 127, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, P.; Schulz, P.; Schurmann, S.; Niland, S.; Exner, S.; Rebollido-Rios, R.; Manikowski, D.; Hoffmann, D.; Seidler, D.G.; Grobe, K. Calcium coordination controls sonic hedgehog structure and Scube2-cubulin domain regulated release. J. Cell Sci. 2017, 130, 3261–3271. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Rhim, J.; Kwon, Y.; Choi, S.Y.; Shin, S.; Ha, C.W.; Lee, C. Comparative analysis of differentially secreted proteins in serum-free and serum-containing media by using BONCAT and pulsed SILAC. Sci. Rep. 2019, 9, 3096. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.; Rudling, M.; Angelin, B. Interstitial fluid lipoproteins. Curr. Opin. Lipidol. 2013, 24, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Ehring, K.; Manikowski, D.; Goretzko, J.; Froese, J.; Gude, F.; Jakobs, P.; Rescher, U.; Kirchhefer, U.; Grobe, K. Conserved cholesterol-related activities of Dispatched 1 drive Sonic hedgehog shedding from the cell membrane. J. Cell Sci. 2022, 135, jcs258672. [Google Scholar] [CrossRef] [PubMed]
- Gallet, A.; Ruel, L.; Staccini-Lavenant, L.; Therond, P.P. Cholesterol modification is necessary for controlled planar long-range activity of Hedgehog in Drosophila epithelia. Development 2006, 133, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.A.; Ekker, S.C.; Park, W.J.; von Kessler, D.P.; Young, K.E.; Chen, C.H.; Ma, Y.; Woods, A.S.; Cotter, R.J.; Koonin, E.V.; et al. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 1996, 86, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.M.; Dunn, M.P.; McMahon, J.A.; Logan, M.; Martin, J.F.; St-Jacques, B.; McMahon, A.P. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 2001, 105, 599–612. [Google Scholar] [CrossRef]
- Huang, X.; Litingtung, Y.; Chiang, C. Region-specific requirement for cholesterol modification of sonic hedgehog in patterning the telencephalon and spinal cord. Development 2007, 134, 2095–2105. [Google Scholar] [CrossRef]
- Wa, Q.; Liu, Y.; Huang, S.; He, P.; Zuo, J.; Li, X.; Li, Z.; Dong, L.; Peng, J.; Wu, S.; et al. miRNA-140 inhibits C3H10T1/2 mesenchymal stem cell proliferation by targeting CXCL12 during transforming growth factor-beta3-induced chondrogenic differentiation. Mol. Med. Rep. 2017, 16, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Otto, T.C.; Lane, M.D. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 2004, 101, 9607–9611. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, A.; Nojima, Y.; Toyoda, A.; Takahoko, M.; Satoh, M.; Tanaka, H.; Wada, H.; Masai, I.; Terasaki, H.; Sakaki, Y.; et al. The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr. Biol. 2005, 15, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Marigo, V.; Tabin, C.J. Regulation of patched by sonic hedgehog in the developing neural tube. Proc. Natl. Acad. Sci. USA 1996, 93, 9346–9351. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Platt, K.A.; Censullo, P.; Ruiz i Altaba, A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 1997, 124, 2537–2552. [Google Scholar] [CrossRef] [PubMed]
- Maity, T.; Fuse, N.; Beachy, P.A. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc. Natl. Acad. Sci. USA 2005, 102, 17026–17031. [Google Scholar] [CrossRef] [PubMed]
- Cardin, A.D.; Weintraub, H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 1989, 9, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.K.; Porter, J.A.; Young, K.E.; Beachy, P.A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 1998, 280, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Maun, H.R.; Wen, X.; Lingel, A.; de Sauvage, F.J.; Lazarus, R.A.; Scales, S.J.; Hymowitz, S.G. The hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site. J. Biol. Chem. 2010, 285, 26570–26580. [Google Scholar] [CrossRef]
- Ma, C.; Zhou, Y.; Beachy, P.A.; Moses, K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 1993, 75, 927–938. [Google Scholar] [CrossRef]
- Rogers, E.M.; Brennan, C.A.; Mortimer, N.T.; Cook, S.; Morris, A.R.; Moses, K. Pointed regulates an eye-specific transcriptional enhancer in the Drosophila hedgehog gene, which is required for the movement of the morphogenetic furrow. Development 2005, 132, 4833–4843. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- Torroja, C.; Gorfinkiel, N.; Guerrero, I. Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development 2004, 131, 2395–2408. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.; Goswami, D.; Manonmani, A.; Sharma, P.; Ranganath, H.A.; VijayRaghavan, K.; Shashidhara, L.S.; Sowdhamini, R.; Mayor, S. Nanoscale organization of hedgehog is essential for long-range signaling. Cell 2008, 133, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Petrov, K.; Watanabe, M.; Salic, A. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. USA 2016, 113, E5866–E5875. [Google Scholar] [CrossRef]
- Hardy, R.Y.; Resh, M.D. Identification of N-terminal Residues of Sonic Hedgehog Important for Palmitoylation by Hedgehog Acyltransferase. J. Biol. Chem. 2012, 287, 42881–42889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bulkley, D.P.; Xin, Y.; Roberts, K.J.; Asarnow, D.E.; Sharma, A.; Myers, B.R.; Cho, W.; Cheng, Y.; Beachy, P.A. Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell 2018, 175, 1352–1364.e1314. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Swierczynska, M.M.; Kumari, V.; Ehrhart-Bornstein, M.; Bornstein, S.R.; Eaton, S. Secretion and signaling activities of lipoprotein-associated hedgehog and non-sterol-modified hedgehog in flies and mammals. PLoS Biol. 2013, 11, e1001505. [Google Scholar] [CrossRef]
- Li, W.; Wang, L.; Wierbowski, B.M.; Lu, M.; Dong, F.; Liu, W.; Li, S.; Wang, P.; Salic, A.; Gong, X. Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release. Nat. Commun. 2021, 12, 6966. [Google Scholar] [CrossRef]
- Stewart, D.P.; Marada, S.; Bodeen, W.J.; Truong, A.; Sakurada, S.M.; Pandit, T.; Pruett-Miller, S.M.; Ogden, S.K. Cleavage activates dispatched for Sonic Hedgehog ligand release. Elife 2018, 7, e31678. [Google Scholar] [CrossRef]
- Panakova, D.; Sprong, H.; Marois, E.; Thiele, C.; Eaton, S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 2005, 435, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.T.; Cleverdon, E.R.; Ogden, S.K. Dispatching Sonic Hedgehog: Molecular Mechanisms Controlling Deployment. Trends Cell Biol. 2019, 29, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, P.E.; Labouesse, M. The sterol-sensing domain: Multiple families, a unique role? Trends Genet. 2002, 18, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Cao, P.; Song, W.; Qian, H.; Du, X.; Coates, H.W.; Zhao, X.; Li, Y.; Gao, S.; Gong, X.; et al. A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols. Science 2021, 371, eabb2224. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Asarnow, D.E.; Ding, K.; Mann, R.K.; Hatakeyama, J.; Zhang, Y.; Ma, Y.; Cheng, Y.; Beachy, P.A. Dispatched uses Na(+) flux to power release of lipid-modified Hedgehog. Nature 2021, 599, 320–324. [Google Scholar] [CrossRef]
- Eugster, C.; Panakova, D.; Mahmoud, A.; Eaton, S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev. Cell 2007, 13, 57–71. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehlers, S.F.; Manikowski, D.; Steffes, G.; Ehring, K.; Gude, F.; Grobe, K. A Residual N-Terminal Peptide Enhances Signaling of Depalmitoylated Hedgehog to the Patched Receptor. J. Dev. Biol. 2024, 12, 11. https://doi.org/10.3390/jdb12020011
Ehlers SF, Manikowski D, Steffes G, Ehring K, Gude F, Grobe K. A Residual N-Terminal Peptide Enhances Signaling of Depalmitoylated Hedgehog to the Patched Receptor. Journal of Developmental Biology. 2024; 12(2):11. https://doi.org/10.3390/jdb12020011
Chicago/Turabian StyleEhlers, Sophia F., Dominique Manikowski, Georg Steffes, Kristina Ehring, Fabian Gude, and Kay Grobe. 2024. "A Residual N-Terminal Peptide Enhances Signaling of Depalmitoylated Hedgehog to the Patched Receptor" Journal of Developmental Biology 12, no. 2: 11. https://doi.org/10.3390/jdb12020011
APA StyleEhlers, S. F., Manikowski, D., Steffes, G., Ehring, K., Gude, F., & Grobe, K. (2024). A Residual N-Terminal Peptide Enhances Signaling of Depalmitoylated Hedgehog to the Patched Receptor. Journal of Developmental Biology, 12(2), 11. https://doi.org/10.3390/jdb12020011





