1. Introduction
Retinal ganglion cells (RGCs) are the projection neurons of the retina, extending their axons from the eye to the dorsal midbrain. During RGC differentiation, axons exit the eye at the optic nerve head and form the optic nerve, which transverses the ventral diencephalon toward the midline. In the zebrafish, the two nerves cross over each other to form the optic chiasm. After crossing, the optic axons form the optic tracts and continue through the diencephalon until they reach their targets in the optic tectum [
1]. In certain species with binocular vision, a subset of axons does not cross at the chiasm but instead turns ipsilaterally, as is the case, for example, for mammals. When comparing different mammalian species, a correlation is found between the fraction of ipsilateral RGC projections and the degree of binocular vision: the mouse has few, while primates have many [
2]. In the zebrafish, on the other hand, all RGCs project contralaterally [
1].
Axon crossing at the optic chiasm is a finely regulated process, which relies on the interplay of a wide range of signaling molecules, including Slits, Ephrins and Semaphorins. Slit molecules constitute a family of secreted glycoproteins of ≈200 kDa, with various forms present across animals with bilateral symmetry [
3]. In the zebrafish, four genes have been identified (Slit1a, 1b, 2 and 3) [
4,
5]. Although Slit ligands are secreted, their diffusion is limited due to their strong association with extracellular matrix components [
6,
7,
8]. These ligands act through the Robo family of proteins, which are encoded by four genes in vertebrates, including the zebrafish (Robo1–4; [
9,
10,
11]). In addition, increasing evidence suggests that Slits can bind to other molecules, receptors or co-receptors, such as heparan sulfate proteoglycan (HSPG) [
12] or PlexinA1, a Semaphorin receptor [
13].
The Slit-Robo signaling pathway has been proven instrumental for commissural axon crossing at the midline [
14]. Slit2 is the most studied in this regard, and was shown to be essential in guiding axons in a wide variety of neuronal types, including RGCs [
15,
16]. In the zebrafish, we previously showed that loss of
slit2 causes defects in axon organization at the optic nerve, optic chiasm and the proximal portion of the optic tract, as well as minor guidance defects and a reduction in growth cone velocity around the midline [
17]. However, this can be considered a relatively “mild” phenotype when compared to that observed in
astray embryos, mutant for the putative receptor expressed in RGCs, Robo2 [
18]. This suggests the presence of other Slits regulating the midline crossing of retinal axons. A strong candidate for this is Slit3, whose mRNA is expressed around the optic chiasm at the same time as that of Slit2, albeit with a different spatial distribution [
19]. Even though the role of Slit3, acting either by itself or together with Slit2, has been previously analyzed in zebrafish commissural axons, such as those of the supraoptic tract (SOT; [
20]) or the postoptic commissure (POC; [
21]), the functional interaction of these two ligands at the optic chiasm remains unexplored.
In this work, we analyzed the hypothesis that both Slit2 and Slit3 might cooperate to organize retinal axons at the optic chiasm by generating zebrafish embryos deficient for these factors using CRISPR/Cas9 technology. Interestingly, we were able to partially reproduce the phenotype of Robo2-deficient embryos when disrupting
slit2 and
slit3 simultaneously. This was evidenced as the appearance of severe axon guidance errors around the chiasm, including invasion of extra-optic regions or misdirections in the optic pathway, and innervation of the ipsilateral optic tectum with the formation of a secondary commissure. Time-lapse imaging of growing axons also revealed some apparent functional differences of
slit2 and
slit3 in regulating RGC axon crossing at the midline. The milder defects seen in the single-deficient embryos could be partly due to these functional differences, probably based on their different expression patterns, as mentioned above [
17,
19], and partly to a modest but significant compensation of
slit3 deficiency by an increase in
slit2 expression. Altogether, our observations support the “repulsive channel” model for the action of Slits at the brain commissures [
22], and suggest that both Slit2 and Slit3 could be acting through the Robo2 receptor to guide retinal axons at the optic chiasm.
2. Materials and Methods
2.1. Fish Breeding and Care
Zebrafish were maintained and bred in a stand-alone system (Tecniplast, Buguggiate, Italy), with controlled temperature (28 °C), conductivity (500 μS/cm
2) and pH (7.5), under live and pellet dietary regime. Embryos were raised at temperatures ranging from 28.5 to 32 °C and staged in hours post-fertilization (hpf) according to Kimmel and collaborators [
23]. Fish lines used: wild-type (Tab5), Tg(
atoh7:gap43-EGFP)
cu1 [
24],
robo2 mutant
astrayti272z [
25] and the CRISPR-generated mutant line NM_131753.1:g.30_39del, or
slit2−/−ipm1 [
17]. Since the homozygotes are viable in this mutant line, for the present report we have used only maternal-zygotic mutants, hereafter referred to as
slit2−/−mz.
2.2. Embryo Microinjection
We designed four single-guide RNAs (sgRNA) against the
slit3 gene using the CRISPRscan tool [
26] and injected them together with mRNA for the zfCas9 flanked by two nuclear localization signal sequences (“nCas9n”), previously reported as highly efficient [
27]. We first tried each sgRNA individually to assess their toxicity and efficiency. We recognized one sgRNA complementary to a sequence in the second exon of the
slit3 gene (
slit3 206, indicating the position of its binding site in the coding sequence;
Table S1) as having no toxic effects and being highly efficient based both on microscopic inspection of the phenotype and genotyping of the expected mutation site in 72 hpf embryos (as described below). We injected one-cell stage Tab5 or
slit2−/−mz embryos with this sgRNA, together with nCas9n mRNA. Alternatively, we co-injected the
slit3 sgRNA together with a
slit2 sgRNA previously reported by us (
slit2 71;
Table S1 [
17]). For mosaic transgenic labeling, 2.5 pg of DNA coding for
atoh7:EGFP-CAAX [
28] were injected, together with 6 pg of Tol2 transposase mRNA.
2.3. Embryo Genotyping
For genotyping, genomic DNA from single embryos was extracted at 72 hpf. Individual embryos were placed in tubes and 20 μL of 50 mM NaOH was added. After a 15 min incubation at 95 °C, the tubes were put on ice and briefly homogenized using a P-20 micropipette. Finally, 2 μL of 1 M Tris-HCl (pH 8) were added, followed by a 1 min centrifugation at 20,000×
g. The resulting supernatant was used for the PCR reaction (2.5 μL per 10 μL reaction), with the specific primers listed in
Table S2, flanking the target sequence of the
slit3 206 sgRNA. PCR conditions were as follows: initial denaturation step at 95 °C for 2 min, followed by 30 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 2 min, and a final extension at 72 °C for 7 min. The PCR products obtained were analyzed through electrophoresis on either 3% agarose or 8% polyacrylamide gels.
2.4. RNA Isolation and Real-Time Quantitative RT-PCR
Total RNA was extracted from the cephalic region of 30 and 48 hpf embryos. For each stage, four conditions were analyzed: non-injected wild-type,
slit2−/−mz,
slit3 crispants, and
slit2−/−mz;
slit3 crispant embryos. 40 embryos (coming from at least three different crosses) corresponding to each condition were dissected, lysed and the heads were stored in Trizol at −80 °C. Total RNA was isolated using the Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA), following the manufacturer’s protocol. cDNA was synthesized from normalized RNA amounts using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA), with a mixture of oligo dT and random primers. Quantitative PCR was performed using the SensiFAST Sybr Low Rox kit (Meridian Bioscience, Cincinnati, OH, USA) with specific primers listed in
Table S2. The qPCR was done in a 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA), and the conditions were as follows: initial denaturation step at 95 °C for 2 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 15 s and 72 °C for 30 s.
Two biological samples and two technical replicas were used. Expression fold-changes between groups were calculated relative to internal control expression (
eif1a), according to Pfaffl et al. [
29]. Statistical significance was determined using a permutation-based procedure, the non-parametric pairwise fixed reallocation randomization test, implemented in the Relative Expression Software Tool (REST, v. 2; Pfaffl and Horgan, Munich, Germany).
p value < 0.05 was considered statistically significant.
2.5. Whole-Mount Immunofluorescence
Embryos were grown in 0.003% phenylthiourea (Sigma, St. Louis, MO, USA) from 10 hpf onwards to delay pigmentation, and fixed overnight at 4 °C, by immersion in 4% paraformaldehyde in phosphate-buffered saline, pH 7.4 (PFA-PBS). For whole-mount immunostaining all subsequent washes were performed in PBS containing 1% Triton X-100. Further permeability was achieved by incubating the embryos in 0.25% trypsin-EDTA for 10–15 min at 0 °C. Blocking was for 30 min in 0.1% bovine serum albumin (BSA), 1% Triton X-100 in PBS. The primary antibody zn8 (ZIRC, Eugene, OR, USA), recognizing the adhesion molecule neurolin/DM-grasp, was used 1/100 in blocking solution. The secondary antibody used was anti-mouse IgG-Alexa 488 (Thermo Fisher Scientific, Waltham, MA, USA), 1/1000 in blocking solution. Nuclei were fluorescently stained with methyl green [
30]. All antibody incubations were performed overnight at 4 °C. Embryos were mounted in 1.5% agarose-50% glycerol in 20 mM Tris buffer (pH 8.0) and stored at 4 °C or −20 °C. Observation of whole embryos was performed using an LSM 880 laser confocal microscope (Zeiss, Oberkochen, Germany), with a 25 × 0.8 NA glycerol immersion objective.
2.6. Lipophilic Dye Labeling
Phenylthiourea-treated embryos were fixed at 48 hpf or 5 dpf as described above. They were then immobilized on glass slides using 2% agarose and injected with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI; Thermo Fisher Scientific, Waltham, MA, USA) or 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO; Thermo Fisher Scientific, Waltham, MA, USA) dissolved in chloroform. For optic chiasm observation in 48 hpf embryos, DiI was injected into the vitreous chamber of one eye, whereas DiO was injected into the contralateral eye. For optic tectum observation in 5 dpf larvae, DiI was injected into the vitreous chamber of one eye. In all of the cases, after the injection the embryos or larvae were incubated for 48 h at room temperature and the dissected brains were mounted in 1.5% agarose-50% glycerol in 20 mM Tris buffer (pH 8.0) and stored at 4 °C or −20 °C. Observation was performed using a Zeiss LSM 880 laser confocal microscope, with a 25 × 0.8 NA glycerol immersion objective.
2.7. Time-Lapse Imaging
Embryos were selected around 30 hpf, anesthetized using 0.04 mg/mL MS222 (Sigma, St. Louis, MO, USA) and mounted in 1% low-melting point agarose (Sigma, St. Louis, MO, USA) over glass bottom dishes. After agarose gelification and during image acquisition, embryos were kept in Ringer’s solution (116 mM NaCl, 2.9 mM KCl, 1.8 mM CaCl2, 5 mM HEPES, pH 7.2) with 0.04 mg/mL MS222. Live acquisitions were made using a Zeiss LSM 880 laser confocal microscope, with a 40×, 1.2 NA silicone oil immersion objective. Stacks around 40 µm-thick were acquired in bidirectional scanning mode, at 1 µm spacing and 512 × 512 pixel resolution every 15 min, for 2.5–16.5 h. The acquisition time per embryo was approximately 1 min, and up to 8 embryos were imaged in each experiment. The embryos were fixed in 4% PFA immediately after the end of the time-lapse, and processed for further confocal microscopy, labeling nuclei with methyl green.
2.8. Microscopy Data Analysis
Images were analyzed using Fiji [
31]. Tracking of axons to obtain distance and velocity measurements was performed with the Manual Tracking plugin, using the site of pioneer axon crossing at the midline as a reference point. For all these analyses, the embryo midline was set as 0 in “x” (the medio-lateral dimension) and the ventral surface of the diencephalon as 0 in “y” (the ventro-dorsal dimension). The instantaneous velocity of the growth cone was determined for each point and expressed in μm/min, and the turn angle (difference between consecutive axon growth direction angles with respect to the “x” axis) was calculated and expressed in degrees (°). In order to better compare between these angles, we applied a correction for the curvature based on the averaged angles in the control embryos (subtracting 0.5° to each value). To visualize and quantify the eventual “axon turn errors”, we used the absolute values of the turn angles, and only analyzed those values that were larger than the mean value in control embryos (10.4°).
Statistical analysis was performed using GraphPad Prism software, v. 8.0.2 (Graphpad Software, Inc., San Diego, CA, USA). As a routine, the data sets were checked for normality using Kolmogorov–Smirnoff normality test. For multiple sets of data, in the case of normal distributions we used either uncorrected one-way ANOVA or Brown-Forsythe and Welch test, for equal or unequal SD values, respectively, or the Kruskal–Wallis test for non-normal distributions. For the comparison of proportions, we used the Fisher’s exact test.
4. Discussion
We report a cooperative mechanism involving a simultaneous action of Slit2 and Slit3 around the optic chiasm in the zebrafish. Redundancy and complementarity of genetic pathways are expected to be widespread mechanisms in developmental processes, since they would ensure robustness [
33]. An example of this is axon growth and guidance along the optic pathway, which depends on the interplay of a number of signals, including the Slit-Robo pathway. Differentiating RGCs only express Robo2 [
18], but their axons traverse a long path, in which they might encounter different, if not all, Slit secreted molecules, which in turn may also act through other receptor types. Arborization of retinal axons at the zebrafish optic tectum, for example, requires Slit1a acting both in a Robo2-dependent and -independent manner [
34]. At the level of the optic chiasm, where axon crossing is finely regulated, a cooperation between Slit1 and Slit2 was described in mice [
35].
Our previous observations indicated an important function of Slit2 in this region, but surprisingly, the
slit2 null mutant phenotype appeared much less severe regarding RGC axon guidance defects than the
robo2 mutant phenotype [
18,
25]. In situ hybridization studies have demonstrated that two
slit genes are expressed in the area immediately surrounding the optic chiasm in the zebrafish:
slit2 and
slit3. Two previous reports have shown
slit2 mRNA expression very tightly surrounding the optic nerve and chiasm area [
17,
19], while
slit3 was shown by Chalasani et al. [
19] to be highly expressed in a restricted area just caudal to the optic chiasm, with some lower expression anterior to the optic pathway (summarized in
Figure 7A). Hence, it could be possible that Slit3 is either complementing the function of Slit2 (i.e.: also having a function in this process in physiological conditions) or compensating for the lack of Slit2 (i.e.: increasing its expression in response to
slit2 gene deficiency). The quantitative mRNA expression results we presented here indicate that there are no evident changes in the expression of
slit3 in response to the null mutation of
slit2, ruling out the compensation hypothesis. Remarkably, however, there is a small but significant increase in the expression of
slit2 in
slit3 crispant embryos. Therefore, Slit3 would not compensate for a lack of Slit2, but Slit2 could at least partially compensate for the loss of Slit3. Interestingly, this change was detected at 30 hpf, the initial stage of optic chiasm formation, and not at 48 hpf, when the process is largely completed.
A role for Slit3 in axon guidance at the zebrafish forebrain midline, both acting by itself and together with Slit2, has been previously demonstrated for other forebrain commissures. In the zebrafish supraoptic tract (SOT), for example, Slit3, and to a lesser extent Slit2, promote ipsilateral axon growth downstream of Wnt activation [
20]. Moreover, knockdown of
slit2,
slit3, and
slit2/
slit3 together resulted in defasciculation of the postoptic commissure (POC; [
21]), suggesting a channeling mechanism similar to the one we later proposed for
slit2 at the optic chiasm [
17]. In the research presented here, we observed that
slit3 expression disruption through the generation of mutations by the co-injection of a specific sgRNA and nCas9n, caused by itself a phenotype comparable to that of
slit2 null mutation, with the appearance of a similar proportion of cases of nerve bifurcation at the optic chiasm. In addition,
slit3 crispants displayed very minor guidance (“turn angle”) defects along the pathway, only detectable by time-lapse analysis, as was the case for
slit2 crispants. Interestingly, this increase in axon turn errors was more conspicuous in these embryos, along with a reduction in instantaneous velocity particularly in the proximal optic tract. This observation was consistent with our previous report of a severe disruption in axon segregation in this region for the
slit2 null mutant line [
17].
The gross phenotype we observed in
slit2/
slit3 double-deficient embryos included retinal axons projecting towards the ipsilateral optic tract and anteriorly into the telencephalon. Moreover, some axons appeared to follow the path of the contralateral optic nerve. Similar defects were reported for the
astray/
robo2 mutant [
18,
22], which we reproduced in this work. Interestingly, turn angle errors made by RGC axons were equally frequent in all situations, including the wild-type embryos, while ipsilateral turns (in the direction opposite to normal growth, and evidenced here as negative instant velocity values), were present in
slit2,
slit3 and
slit2 +
slit3 crispants, but virtually absent in wild-types. A similar phenomenon was described by Hutson and Chien [
22] when comparing the trajectory of RGC axons at the chiasm in wild-type and
astray/
robo2 mutants by time-lapse microscopy. In addition, by following axon trajectories through lipophilic dye tracing we found ipsilateral optic tectum innervation only in
slit2 +
slit3 crispants, indicating that most, if not all, errors were corrected in the
slit2 or
slit3 deficient embryos. Projections to the ipsilateral optic tectum are also a hallmark of
astray/
robo2 mutants [
18,
25]. Finally, many retinal axons in
astray/
robo2 mutants were observed to reach the ipsilateral tectum via the posterior commissure instead of the optic tract [
18], which could correspond to an ectopic crossing site we observed for a number of axons in
slit2 +
slit3 crispants, slightly posterior to the optic chiasm. Although these correlations point to the possibility of a combined action of Slit2 and Slit3 on Robo2 receptors, we cannot exclude an action on other receptors or the occurrence of more complex interactions. These could include the eventual modulation by other pathways, such as was demonstrated for SDF1-CRXR4b [
19].
In addition to a collaborative role of Slit2 and Slit3 in assembling axon organization at the optic chiasm during the early stages of its development, our results indicate some differences in their individual functions. There are two main pieces of evidence to support this: first, there is a differential distribution of minor axon turn errors along the analyzed area (evidenced both in the spatial organization and in the dispersion of instantaneous velocity values), with errors appearing more frequently in the optic tract portion for
slit2 crispants and in the optic nerve for
slit2/3 double crispants; second, some specific phenotypic defects in the single crispants appeared to be corrected in the double deficient embryos, like the distal turn errors in
slit2 crispants. The different expression patterns for these two genes could at least partly explain these differences (see
Figure 7A). Since retinal axons are known to respond differently to guidance cues depending on their position, it would be interesting to determine whether the observed differential defects are related to actions on specific axon populations. A role like this was proposed for HSPGs in regulating dorso-ventral sorting in the zebrafish optic tract through selective axon degeneration [
36]. Similarly, Slit molecules could be regulating naso-temporal axon segregation at the optic chiasm and proximal tract, like we described for
slit2 morphants [
17], albeit most probably through error correction.
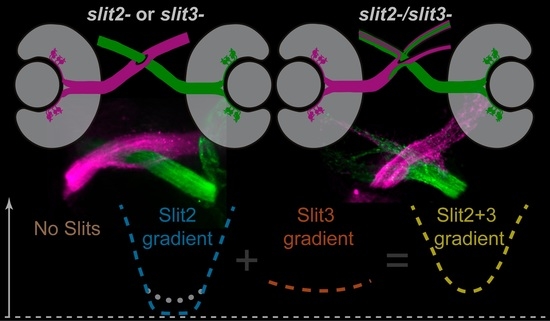
Based on these previous data and our present observations, we suggest a hypothetical model by which Slit2 would have a “channeling” role, as proposed by Chi-Bin Chien and colleagues several years ago [
1,
22], with steep gradients surrounding the path of optic axons before, across and just after the optic chiasm. Slit3, on the other hand, would generate a smoother and wider gradient, eventually preventing the axons from entering into the brain as they cross at the optic chiasm, and nevertheless collaborating with Slit2 in preventing axon turn errors at the midline (see a diagram in
Figure 7B). Together, they would generate a “U” shaped Slit gradient to keep retinal axons on track. Loss of Slit2 would only leave the shallow Slit3 gradient, leading to relatively important defects in axon organization particularly in the proximal optic tract; loss of Slit3 would, in turn, leave the marked Slit2 gradient intact, with the possibility of an increased local expression due to genetic compensation, and leading to milder defects overall; the simultaneous loss of Slit2 and Slit3 would lead to a Robo2-like phenotype, due to a complete absence of Slits in the area surrounding the optic chiasm (schematic representation in
Figure 7C).
It is interesting to note that Slit2 and Slit3 in the zebrafish could be acting in ways comparable to those described for Slit1 and Slit2, respectively, in mice [
35], as in this species Slit1 is expressed in a pattern tightly surrounding the optic pathway proximal to the chiasm, and Slit2 is expressed in a larger area located in the anterior midline, at some distance [
37]. In mice, like in most mammalian species, there is some degree of binocular vision, and some retinal axons do not cross at the optic chiasm, but project ipsilaterally. It is tempting to speculate that the use of different Slit factors, with differential expression patterns, all acting on Robo2, could be part of an evolutionary mechanism for adaptation to different degrees of binocular or non-binocular vision, which appears more related to the particular vertebrate species habits than to phylogenetic constraints.