Feedback Regulation of Signaling Pathways for Precise Pre-Placodal Ectoderm Formation in Vertebrate Embryos
Abstract
1. Introduction
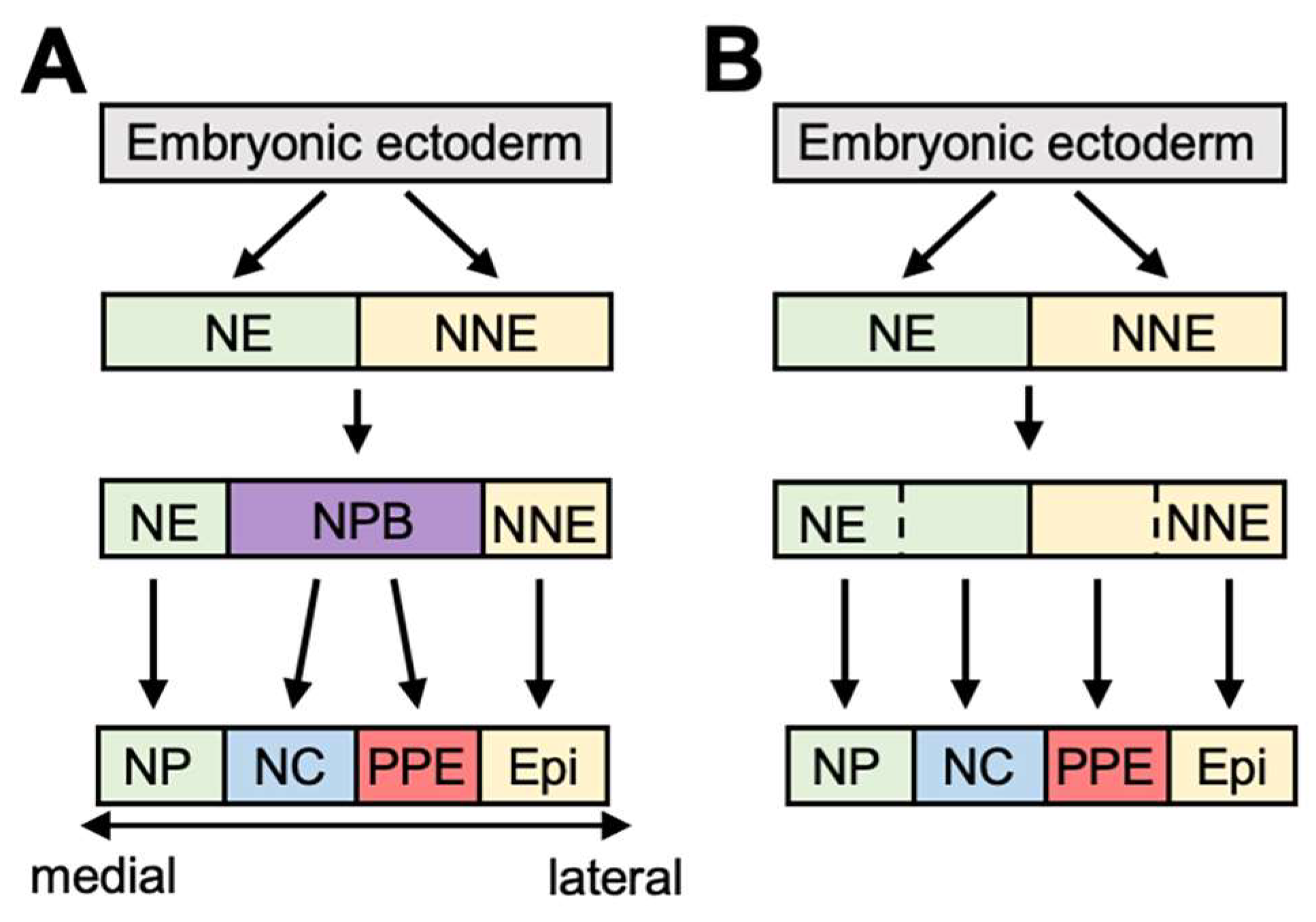
2. An Outline of PPE Formation
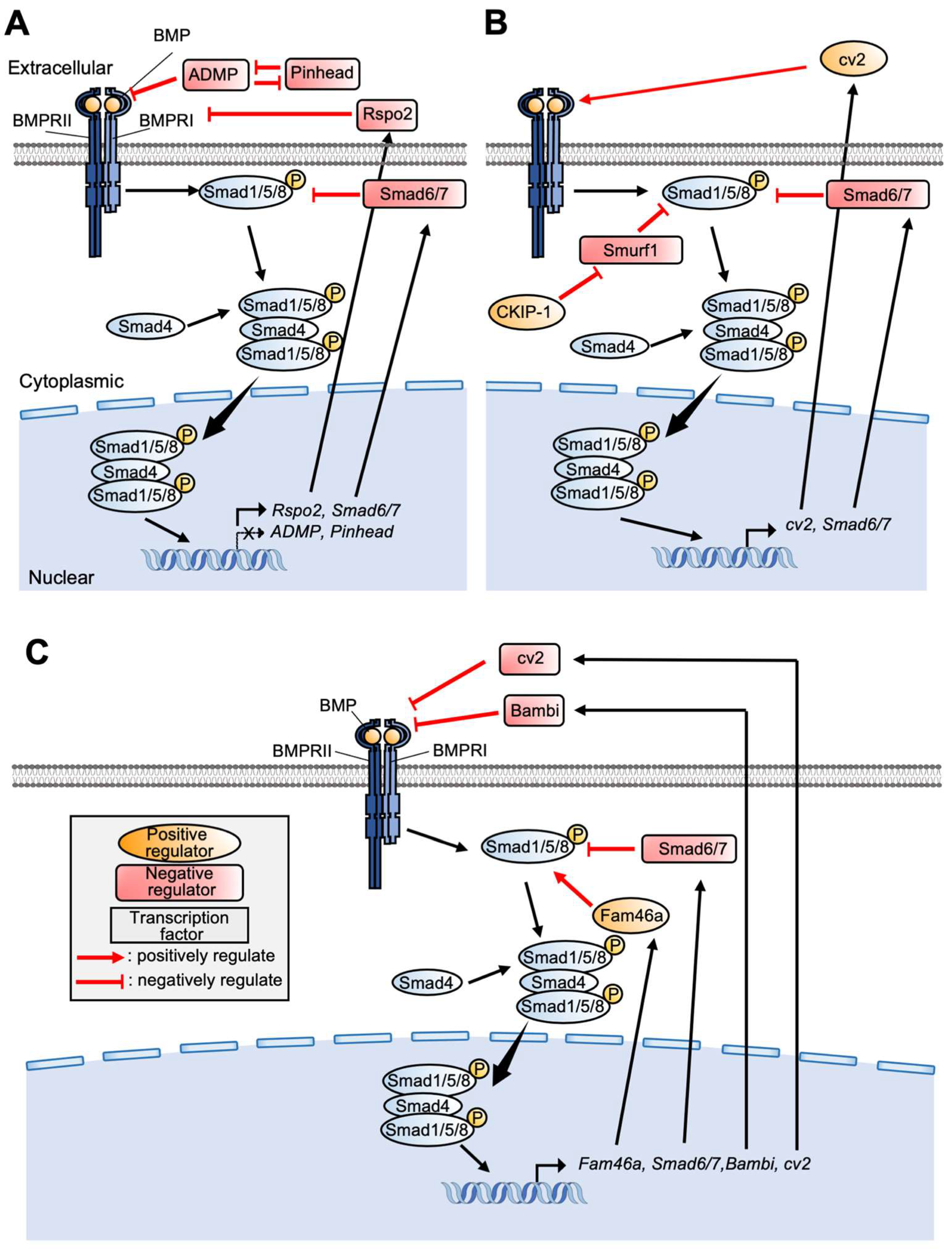
3. Control of BMP Signaling in PPE Formation
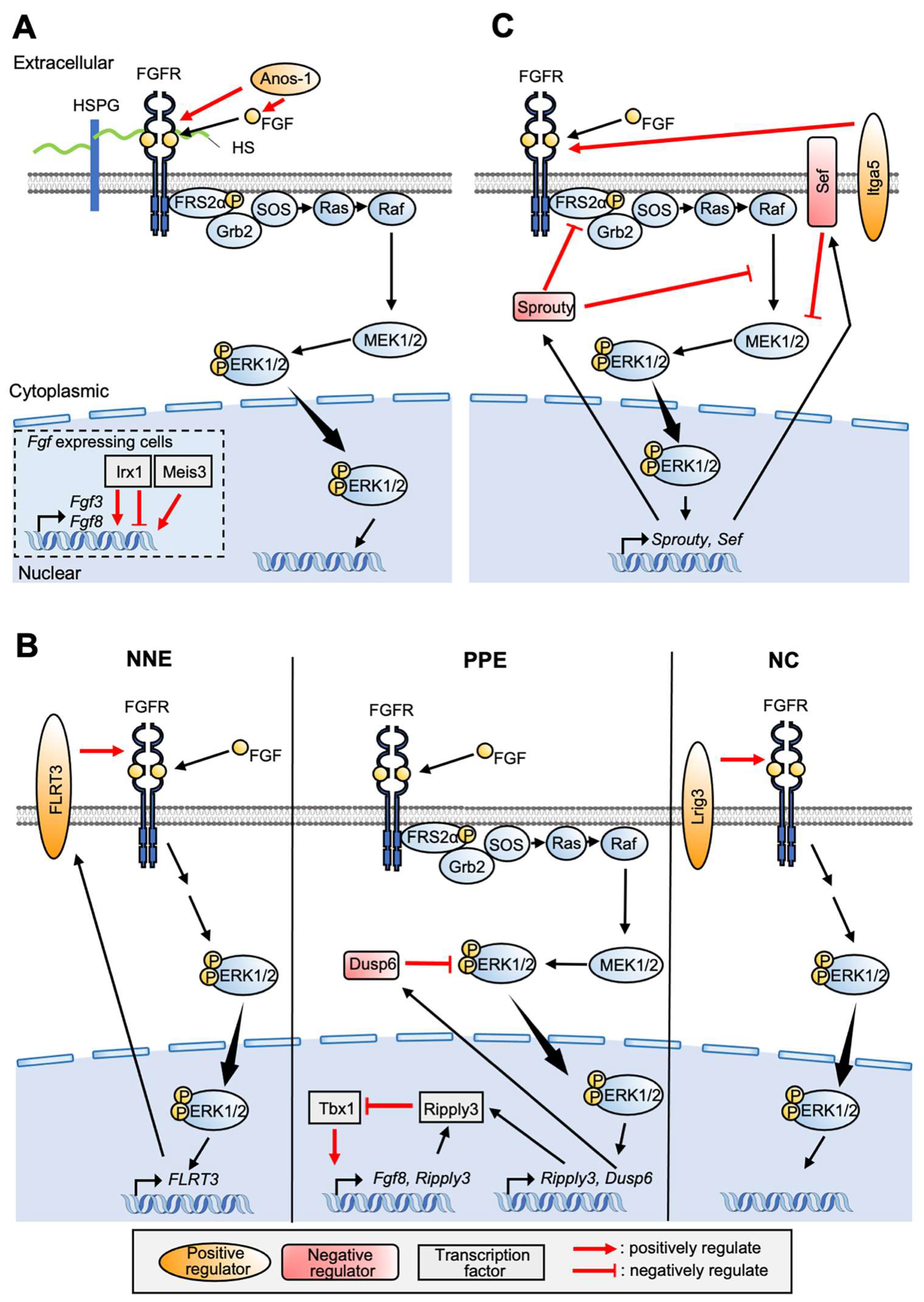
4. Involvement of FGF Signaling for PPE Formation
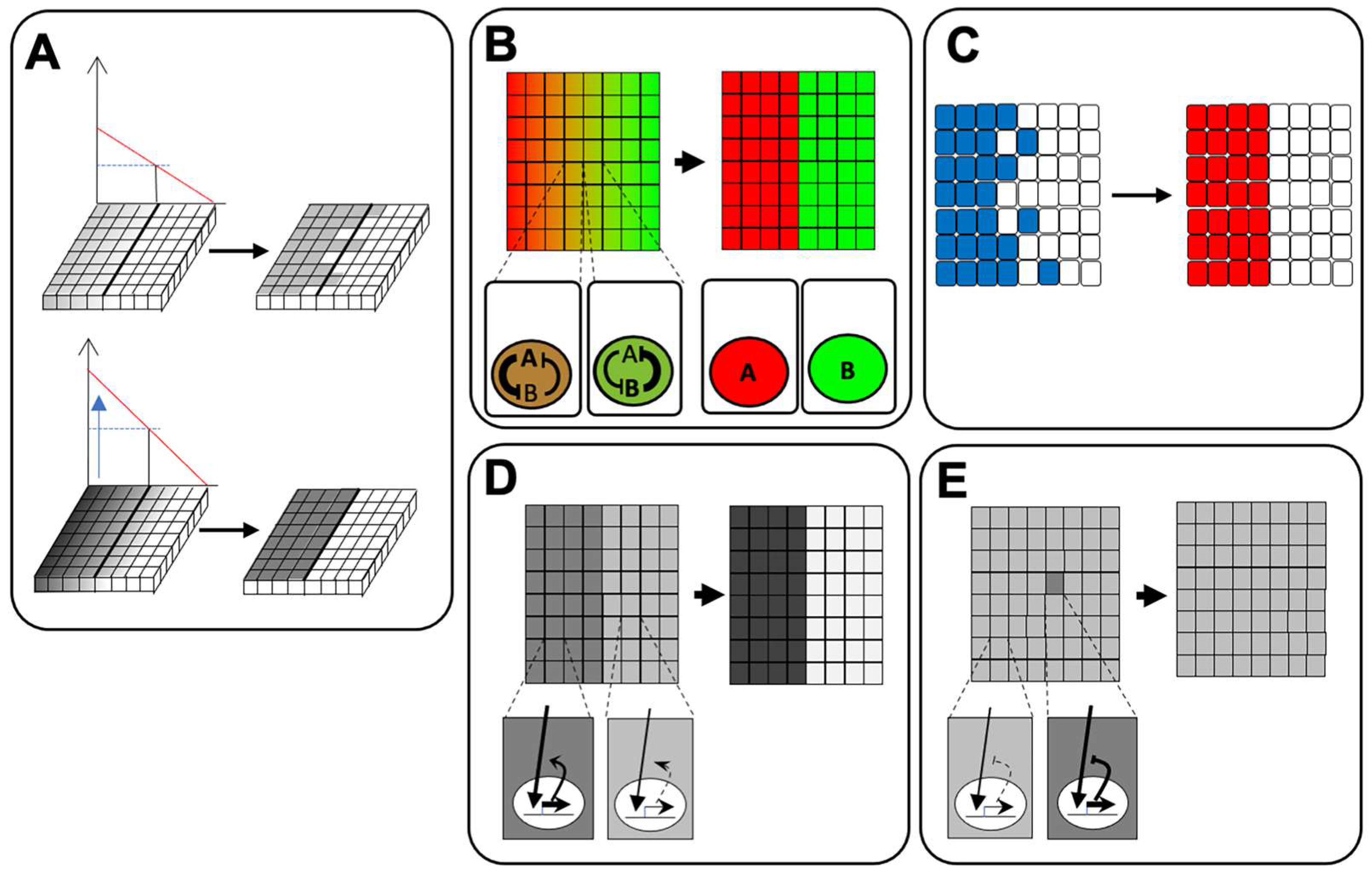
5. Feedback Regulation of Signaling Pathways for Ectodermal Patterning
5.1. BMP Signaling
5.2. FGF Signaling
6. Conclusions
7. Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolpert, L. One Hundred Years of Positional Information. Trends Genet. 1996, 12, 359–364. [Google Scholar] [CrossRef]
- Lander, A.D. Pattern, Growth, and Control. Cell 2011, 144, 955–969. [Google Scholar] [CrossRef] [PubMed]
- Turing, A.M. The Chemical Basis of Morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1952, 237, 37–72. [Google Scholar] [CrossRef]
- Kondo, S.; Asal, R. A Reaction-Diffusion Wave on the Skin of the Marine Angelfish Pomacanthus. Nature 1995, 376, 765–768. [Google Scholar] [CrossRef]
- Bier, E.; De Robertis, E.M. BMP Gradients: A Paradigm for Morphogen-Mediated Developmental Patterning. Science 2015, 348, aaa5838. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, K.; Schlosser, G. Tissues and Signals Involved in the Induction of Placodal Six1 Expression in Xenopus Laevis. Dev. Biol. 2005, 288, 40–59. [Google Scholar] [CrossRef] [PubMed]
- Litsiou, A.; Hanson, S.; Streit, A. A Balance of FGF, BMP and WNT Signalling Positions the Future Placode Territory in the Head. Development 2005, 132, 4051–4062. [Google Scholar] [CrossRef]
- Lander, A.D.; Lo, W.-C.; Nie, Q.; Wan, F.Y.M. The Measure of Success: Constraints, Objectives, and Tradeoffs in Morphogen-Mediated Patterning. Cold Spring Harb. Perspect. Biol. 2009, 1, a002022. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Bronner-Fraser, M. Hierarchy of Regulatory Events in Sensory Placode Development. Curr. Opin. Genet. Dev. 2004, 14, 520–526. [Google Scholar] [CrossRef]
- Streit, A. The Preplacodal Region: An Ectodermal Domain with Multipotential Progenitors That Contribute to Sense Organs and Cranial Sensory Ganglia. Int. J. Dev. Biol. 2007, 51, 447–461. [Google Scholar] [CrossRef]
- Pla, P.; Monsoro-Burq, A.H. The Neural Border: Induction, Specification and Maturation of the Territory That Generates Neural Crest Cells. Dev. Biol. 2018, 444, S36–S46. [Google Scholar] [CrossRef] [PubMed]
- Steventon, B.; Carmona-Fontaine, C.; Mayor, R. Genetic Network during Neural Crest Induction: From Cell Specification to Cell Survival. Semin. Cell Dev. Biol. 2005, 16, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Mayor, R.; Aybar, M.J. Induction and Development of Neural Crest in Xenopus Laevis. Cell Tissue Res. 2001, 305, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Milet, C.; Monsoro-Burq, A.H. Neural Crest Induction at the Neural Plate Border in Vertebrates. Dev. Biol. 2012, 366, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.V.H.; Bronner-Fraser, M. Vertebrate Cranial Placodes I. Embryonic Induction. Dev. Biol. 2001, 232, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, G. Induction and Specification of Cranial Placodes. Dev. Biol. 2006, 294, 303–351. [Google Scholar] [CrossRef]
- Schlosser, G. Making Senses: Development of Vertebrate Cranial Placodes. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 283, pp. 129–234. [Google Scholar]
- Grocott, T.; Tambalo, M.; Streit, A. The Peripheral Sensory Nervous System in the Vertebrate Head: A Gene Regulatory Perspective. Dev. Biol. 2012, 370, 3–23. [Google Scholar] [CrossRef]
- Saint-Jeannet, J.-P.; Moody, S.A. Establishing the Pre-Placodal Region and Breaking It into Placodes with Distinct Identities. Dev. Biol. 2014, 389, 13–27. [Google Scholar] [CrossRef]
- Singh, S.; Groves, A.K. The Molecular Basis of Craniofacial Placode Development. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 363–376. [Google Scholar] [CrossRef]
- Breau, M.A.; Schneider-Maunoury, S. Cranial Placodes: Models for Exploring the Multi-Facets of Cell Adhesion in Epithelial Rearrangement, Collective Migration and Neuronal Movements. Dev. Biol. 2015, 401, 25–36. [Google Scholar] [CrossRef]
- Streit, A. Specification of Sensory Placode Progenitors: Signals and Transcription Factor Networks. Int. J. Dev. Biol. 2018, 62, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Pandur, P.D.; Moody, S.A. Xenopus Six1 Gene Is Expressed in Neurogenic Cranial Placodes and Maintained in the Differentiating Lateral Lines. Mech. Dev. 2000, 96, 253–257. [Google Scholar] [CrossRef]
- Schlosser, G.; Ahrens, K. Molecular Anatomy of Placode Development in Xenopus Laevis. Dev. Biol. 2004, 271, 439–466. [Google Scholar] [CrossRef] [PubMed]
- David, R.; Ahrens, K.; Wedlich, D.; Schlosser, G. Xenopus Eya1 Demarcates All Neurogenic Placodes as Well as Migrating Hypaxial Muscle Precursors. Mech. Dev. 2001, 103, 189–192. [Google Scholar] [CrossRef]
- Maharana, S.K.; Schlosser, G. A Gene Regulatory Network Underlying the Formation of Pre-Placodal Ectoderm in Xenopus Laevis. BMC Biol. 2018, 16, 79. [Google Scholar] [CrossRef]
- Brugmann, S.A.; Pandur, P.D.; Kenyon, K.L.; Pignoni, F.; Moody, S.A. Six1 Promotes a Placodal Fate within the Lateral Neurogenic Ectoderm by Functioning as Both a Transcriptional Activator and Repressor. Development 2004, 131, 5871–5881. [Google Scholar] [CrossRef]
- Schlosser, G. Early Embryonic Specification of Vertebrate Cranial Placodes. Wiley Interdiscip. Rev. Dev. Biol. 2014, 3, 349–363. [Google Scholar] [CrossRef]
- Pieper, M.; Ahrens, K.; Rink, E.; Peter, A.; Schlosser, G. Differential Distribution of Competence for Panplacodal and Neural Crest Induction to Non-Neural and Neural Ectoderm. Development 2012, 139, 1175–1187. [Google Scholar] [CrossRef]
- Kwon, H.-J.; Bhat, N.; Sweet, E.M.; Cornell, R.A.; Riley, B.B. Identification of Early Requirements for Preplacodal Ectoderm and Sensory Organ Development. PLoS Genet. 2010, 6, e1001133. [Google Scholar] [CrossRef]
- Hong, C.-S.; Saint-Jeannet, J.-P. The Activity of Pax3 and Zic1 Regulates Three Distinct Cell Fates at the Neural Plate Border. Mol. Biol. Cell 2007, 18, 2192–2202. [Google Scholar] [CrossRef]
- Garnett, A.T.; Square, T.A.; Medeiros, D.M. BMP, Wnt and FGF Signals Are Integrated through Evolutionarily Conserved Enhancers to Achieve Robust Expression of Pax3 and Zic Genes at the Zebrafish Neural Plate Border. Development 2012, 139, 4220–4231. [Google Scholar] [CrossRef] [PubMed]
- Plouhinec, J.L.; Roche, D.D.; Pegoraro, C.; Figueiredo, A.L.; Maczkowiak, F.; Brunet, L.J.; Milet, C.; Vert, J.P.; Pollet, N.; Harland, R.M.; et al. Pax3 and Zic1 Trigger the Early Neural Crest Gene Regulatory Network by the Direct Activation of Multiple Key Neural Crest Specifiers. Dev. Biol. 2014, 386, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Roellig, D.; Tan-Cabugao, J.; Esaian, S.; Bronner, M.E. Dynamic Transcriptional Signature and Cell Fate Analysis Reveals Plasticity of Individual Neural Plate Border Cells. eLife 2017, 6, e21620. [Google Scholar] [CrossRef] [PubMed]
- Thiery, A.; Buzzi, A.L.; Hamrud, E.; Cheshire, C.; Luscombe, N.; Briscoe, J.; Streit, A. A Gradient Border Model for Cell Fate Decisions at the Neural Plate Border. bioRxiv 2022. [Google Scholar] [CrossRef]
- Miyazono, K.; Kamiya, Y.; Morikawa, M. Bone Morphogenetic Protein Receptors and Signal Transduction. J. Biochem. 2010, 147, 35–51. [Google Scholar] [CrossRef]
- Hill, C.S. Establishment and Interpretation of NODAL and BMP Signaling Gradients in Early Vertebrate Development, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2022; Volume 149, ISBN 9780128170977. [Google Scholar]
- Fainsod, A.; Steinbeisser, H.; De Robertis, E.M. On the Function of BMP-4 in Patterning the Marginal Zone of the Xenopus Embryo. EMBO J. 1994, 13, 5015–5025. [Google Scholar] [CrossRef]
- Hemmati-Brivanlou, A.; Thomsen, G.H. Ventral Mesodermal Patterning InXenopus Embryos: Expression Patterns and Activities of BMP-2 and BMP-4. Dev. Genet. 1995, 17, 78–89. [Google Scholar] [CrossRef]
- Liem, K.F.; Tremml, G.; Roelink, H.; Jessell, T.M. Dorsal Differentiation of Neural Plate Cells Induced by BMP-Mediated Signals from Epidermal Ectoderm. Cell 1995, 82, 969–979. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Suzuki, A.; Ueno, N.; Kimelman, D. Localized BMP-4 Mediates Dorsal/Ventral Patterning in the Early Xenopus Embryo. Dev. Biol. 1995, 169, 37–50. [Google Scholar] [CrossRef]
- Streit, A.; Stern, C.D. Establishment and Maintenance of the Border of the Neural Plate in the Chick: Involvement of FGF and BMP Activity. Mech. Dev. 1999, 82, 51–66. [Google Scholar] [CrossRef]
- Ogita, J.; Isogai, E.; Sudo, H.; Sakiyama, S.; Nakagawara, A.; Koseki, H. Expression of the Dan Gene during Chicken Embryonic Development. Mech. Dev. 2001, 109, 363–365. [Google Scholar] [CrossRef]
- Esterberg, R.; Fritz, A. Dlx3b/4b Are Required for the Formation of the Preplacodal Region and Otic Placode through Local Modulation of BMP Activity. Dev. Biol. 2009, 325, 189–199. [Google Scholar] [CrossRef]
- Wilson, P.A.; Lagna, G.; Suzuki, A.; Hemmati-Brivanlou, A. Concentration-Dependent Patterning of the Xenopus Ectoderm by BMP4 and Its Signal Transducer Smad1. Development 1997, 124, 3177–3184. [Google Scholar] [CrossRef] [PubMed]
- Marchant, L.; Linker, C.; Ruiz, P.; Guerrero, N.; Mayor, R. The Inductive Properties of Mesoderm Suggest That the Neural Crest Cells Are Specified by a BMP Gradient. Dev. Biol. 1998, 198, 319–329. [Google Scholar] [CrossRef]
- Steventon, B.; Araya, C.; Linker, C.; Kuriyama, S.; Mayor, R. Differential Requirements of BMP and Wnt Signalling during Gastrulation and Neurulation Define Two Steps in Neural Crest Induction. Development 2009, 136, 771–779. [Google Scholar] [CrossRef]
- Selleck, M.A.J.; García-Castro, M.I.; Artinger, K.B.; Bronner-Fraser, M. Effects of Shh and Noggin on Neural Crest Formation Demonstrate That BMP Is Required in the Neural Tube but Not Ectoderm. Development 1998, 125, 4919–4930. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Osumi, N.; Wakamatsu, Y. Bimodal Functions of Notch-Mediated Signaling Are Involved in Neural Crest Formation during Avian Ectoderm Development. Development 2002, 129, 863–873. [Google Scholar] [CrossRef]
- Schumacher, J.A.; Hashiguchi, M.; Nguyen, V.H.; Mullins, M.C. An Intermediate Level of Bmp Signaling Directly Specifies Cranial Neural Crest Progenitor Cells in Zebrafish. PLoS ONE 2011, 6, e27403. [Google Scholar] [CrossRef]
- Faure, S.; De Santa Barbara, P.; Roberts, D.J.; Whitman, M. Endogenous Patterns of BMP Signaling during Early Chick Development. Dev. Biol. 2002, 244, 44–65. [Google Scholar] [CrossRef]
- Watanabe, T.; Kanai, Y.; Matsukawa, S.; Michiue, T. Specific Induction of Cranial Placode Cells from Xenopus Ectoderm by Modulating the Levels of BMP, Wnt, and FGF Signaling. Genesis 2015, 53, 652–659. [Google Scholar] [CrossRef]
- Luo, T.; Matsuo-Takasaki, M.; Lim, J.H.; Sargent, T.D. Differential Regulation of Dlx Gene Expression by a BMP Morphogenetic Gradient. Int. J. Dev. Biol. 2001, 45, 681–684. [Google Scholar] [PubMed]
- Nguyen, V.H.; Schmid, B.; Trout, J.; Connors, S.A.; Ekker, M.; Mullins, M.C. Ventral and Lateral Regions of the Zebrafish Gastrula, Including the Neural Crest Progenitors, Are Established by Abmp2b/SwirlPathway of Genes. Dev. Biol. 1998, 199, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Neave, B.; Holder, N.; Patient, R. A Graded Response to BMP-4 Spatially Coordinates Patterning of the Mesoderm and Ectoderm in the Zebrafish. Mech. Dev. 1997, 62, 183–195. [Google Scholar] [CrossRef]
- Bhat, N.; Kwon, H.-J.; Riley, B.B. A Gene Network That Coordinates Preplacodal Competence and Neural Crest Specification in Zebrafish. Dev. Biol. 2013, 373, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Sjödal, M.; Edlund, T.; Gunhaga, L. Time of Exposure to BMP Signals Plays a Key Role in the Specification of the Olfactory and Lens Placodes Ex Vivo. Dev. Cell 2007, 13, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Guimond, S.E.; Travers, P.; Cadman, S.; Hohenester, E.; Tumbull, J.E.; Kim, S.H.; Bouloux, P.M. Novel Mechanisms of Fibroblast Growth Factor Receptor 1 Regulation by Extracellular Matrix Protein Anosmin-1. J. Biol. Chem. 2009, 284, 29905–29920. [Google Scholar] [CrossRef]
- Endo, Y.; Ishiwata-Endo, H.; Yamada, K.M. Extracellular Matrix Protein Anosmin Promotes Neural Crest Formation and Regulates FGF, BMP, and WNT Activities. Dev. Cell 2012, 23, 305–316. [Google Scholar] [CrossRef]
- Hu, Y.; González-Martínez, D.; Kim, S.H.; Bouloux, P.M.G. Cross-Talk of Anosmin-1, the Protein Implicated in X-Linked Kallmann’s Syndrome, with Heparan Sulphate and Urokinase-Type Plasminogen Activator. Biochem. J. 2004, 384, 495–505. [Google Scholar] [CrossRef]
- Korsensky, L.; Ron, D. Regulation of FGF Signaling: Recent Insights from Studying Positive and Negative Modulators. Semin. Cell Dev. Biol. 2016, 53, 101–114. [Google Scholar] [CrossRef]
- Bae, C.-J.; Hong, C.-S.; Saint-Jeannet, J.-P. Anosmin-1 Is Essential for Neural Crest and Cranial Placodes Formation in Xenopus. Biochem. Biophys. Res. Commun. 2018, 495, 2257–2263. [Google Scholar] [CrossRef]
- Gutkovich, Y.E.; Ofir, R.; Elkouby, Y.M.; Dibner, C.; Gefen, A.; Elias, S.; Frank, D. Xenopus Meis3 Protein Lies at a Nexus Downstream to Zic1 and Pax3 Proteins, Regulating Multiple Cell-Fates during Early Nervous System Development. Dev. Biol. 2010, 338, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Tanegashima, K.; Ro, H.; Dawid, I.B. Lrig3 Regulates Neural Crest Formation in Xenopus by Modulating Fgf and Wnt Signaling Pathways. Development 2008, 135, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Dinsmore, C.J.; Soriano, P. MAPK and PI3K Signaling: At the Crossroads of Neural Crest Development. Dev. Biol. 2018, 444, S79–S97. [Google Scholar] [CrossRef] [PubMed]
- Geary, L.; LaBonne, C. FGF Mediated MAPK and PI3K/Akt Signals Make Distinct Contributions to Pluripotency and the Establishment of Neural Crest. eLife 2018, 7, e33845. [Google Scholar] [CrossRef]
- Monsoro-Burq, A.H.; Fletcher, R.B.; Harland, R.M. Neural Crest Induction by Paraxial Mesoderm in Xenopus Embryos Requires FGF Signals. Development 2003, 130, 3111–3124. [Google Scholar] [CrossRef]
- Lawson, A.; Colas, J.F.; Schoenwolf, G.C. Classification Scheme for Genes Expressed during Formation and Progression of the Avian Primitive Streak. Anat. Rec. 2001, 262, 221–226. [Google Scholar] [CrossRef]
- Fletcher, R.B.; Baker, J.C.; Harland, R.M. FGF8 Spliceforms Mediate Early Mesoderm and Posterior Neural Tissue Formation in Xenopus. Development 2006, 133, 1703–1714. [Google Scholar] [CrossRef]
- Tereshina, M.B.; Ermakova, G.V.; Ivanova, A.S.; Zaraisky, A.G. Ras-Dva1 Small GTPase Regulates Telencephalon Development in Xenopus Laevis Embryos by Controlling Fgf8 and Agr Signaling at the Anterior Border of the Neural Plate. Biol. Open 2014, 3, 192–203. [Google Scholar] [CrossRef]
- Sullivan, C.H.; Majumdar, H.D.; Neilson, K.M.; Moody, S.A. Six1 and Irx1 Have Reciprocal Interactions during Cranial Placode and Otic Vesicle Formation. Dev. Biol. 2019, 446, 68–79. [Google Scholar] [CrossRef]
- Schimmang, T. Expression and Functions of FGF Ligands during Early Otic Development. Int. J. Dev. Biol. 2007, 51, 473–481. [Google Scholar] [CrossRef]
- Wright, T.J.; Mansour, S.L. Fgf3 and Fgf10 Are Required for Mouse Otic Placode Induction. Development 2003, 130, 3379–3390. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Frutos, E.; Vendrell, V.; Alvarez, Y.; Zelarayan, L.C.; López-Hernández, I.; Ros, M.; Schimmang, T. Tissue-Specific Requirements for FGF8 during Early Inner Ear Development. Mech. Dev. 2009, 126, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Bhat, N.; Riley, B.B. Integrin-A5 Coordinates Assembly of Posterior Cranial Placodes in Zebrafish and Enhances Fgf-Dependent Regulation of Otic/Epibranchial Cells. PLoS ONE 2011, 6, e27778. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Ning, G.; Li, L.; Liu, J.; Yang, S.; Cao, Y.; Wang, Q. The BMP Ligand Pinhead Together with Admp Supports the Robustness of Embryonic Patterning. Sci. Adv. 2019, 5, eaau6455. [Google Scholar] [CrossRef]
- Moos, M.; Wang, S.; Krinks, M. Anti-Dorsalizing Morphogenetic Protein Is a Novel TGF-Beta Homolog Expressed in the Spemann Organizer. Development 1995, 121, 4293–4301. [Google Scholar] [CrossRef]
- Lele, Z.; Nowak, M.; Hammerschmidt, M. Zebrafish Admp Is Required to Restrict the Size of the Organizer and to Promote Posterior and Ventral Development. Dev. Dyn. 2001, 222, 681–687. [Google Scholar] [CrossRef]
- Lee, H.; Seidl, C.; Sun, R.; Glinka, A.; Niehrs, C. R-Spondins Are BMP Receptor Antagonists in Xenopus Early Embryonic Development. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Onichtchouk, D.; Chen, Y.G.; Dosch, R.; Gawantka, V.; Delius, H.; Massagué, J.; Niehrs, C. Silencing of TGF-β Signalling by the Pseudoreceptor BAMBI. Nature 1999, 401, 480–485. [Google Scholar] [CrossRef]
- Paulsen, M.; Legewie, S.; Eils, R.; Karaulanov, E.; Niehrs, C. Negative Feedback in the Bone Morphogenetic Protein 4 (BMP4) Synexpression Group Governs Its Dynamic Signaling Range and Canalizes Development. Proc. Natl. Acad. Sci. USA 2011, 108, 10202–10207. [Google Scholar] [CrossRef]
- Coffinier, C.; Ketpura, N.; Tran, U.; Geissert, D.; De Robertis, E.M. Mouse Crossveinless-2 Is the Vertebrate Homolog of a Drosophila Extracellular Regulator of BMP Signaling. Mech. Dev. 2002, 119, S179–S184. [Google Scholar] [CrossRef]
- Ambrosio, A.L.; Taelman, V.F.; Lee, H.X.; Metzinger, C.A.; Coffinier, C.; De Robertis, E.M. Crossveinless-2 Is a BMP Feedback Inhibitor That Binds Chordin/BMP to Regulate Xenopus Embryonic Patterning. Dev. Cell 2008, 15, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, F.; Zhang, J.; Kramer, C.; Sebald, W.; Hammerschmidt, M. Crossveinless 2 Is an Essential Positive Feedback Regulator of Bmp Signaling during Zebrafish Gastrulation. Development 2006, 133, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Reichert, S.; Randall, R.A.; Hill, C.S. A BMP Regulatory Network Controls Ectodermal Cell Fate Decisions at the Neural Plate Border. Development 2013, 140, 4435–4444. [Google Scholar] [CrossRef] [PubMed]
- Piacentino, M.L.; Bronner, M.E. Intracellular Attenuation of BMP Signaling via CKIP-1/Smurf1 Is Essential during Neural Crest Induction. PLoS Biol. 2018, 16, e2004425. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Yamamoto, T.; Tsukano, K.; Hirano, S.; Horikawa, A.; Michiue, T. Fam46a Regulates BMP-Dependent Pre-Placodal Ectoderm Differentiation in Xenopus. Development 2018, 145, dev166710. [Google Scholar] [CrossRef]
- Janesick, A.; Shiotsugu, J.; Taketani, M.; Blumberg, B. RIPPLY3 Is a Retinoic Acid-Inducible Repressor Required for Setting the Borders of the Pre-Placodal Ectoderm. Development 2012, 139, 1213–1224. [Google Scholar] [CrossRef]
- Böttcher, R.T.; Pollet, N.; Delius, H.; Niehrs, C. The Transmembrane Protein XFLRT3 Forms a Complex with FGF Receptors and Promotes FGF Signalling. Nat. Cell Biol. 2004, 6, 38–44. [Google Scholar] [CrossRef]
- Cho, G.S.; Choi, S.C.; Han, J.K. BMP Signal Attenuates FGF Pathway in Anteroposterior Neural Patterning. Biochem. Biophys. Res. Commun. 2013, 434, 509–515. [Google Scholar] [CrossRef]
- Cho, G.-S.; Park, D.-S.; Choi, S.-C.; Han, J.-K. Tbx2 Regulates Anterior Neural Specification by Repressing FGF Signaling Pathway. Dev. Biol. 2017, 421, 183–193. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Tan, T.-H. DUSPs, to MAP Kinases and Beyond. Cell Biosci. 2012, 2, 24. [Google Scholar] [CrossRef]
- Muhammad, K.A.; Nur, A.A.; Nurul, H.S.; Narazah, M.Y.; Siti, R.A.R. Dual-Specificity Phosphatase 6 (DUSP6): A Review of Its Molecular Characteristics and Clinical Relevance in Cancer. Cancer Biol. Med. 2018, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, O.; Pagès, G.; Gimond, C. The Dual-Specificity MAP Kinase Phosphatases: Critical Roles in Development and Cancer. Am. J. Physiol. Physiol. 2010, 299, C189–C202. [Google Scholar] [CrossRef] [PubMed]
- Gómez, A.R.; López-Varea, A.; Molnar, C.; de la Calle-Mustienes, E.; Ruiz-Gómez, M.; Gómez-Skarmeta, J.L.; de Celis, J.F. Conserved Cross-Interactions in Drosophila and Xenopus between Ras/MAPK Signaling and the Dual-Specificity Phosphatase MKP3. Dev. Dyn. 2005, 232, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Tsukano, K.; Yamamoto, T.; Watanabe, T.; Michiue, T. Xenopus Dusp6 Modulates FGF Signaling to Precisely Pattern Pre-Placodal Ectoderm. Dev. Biol. 2022, 488, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Morrison, D.J.; Basson, M.A.; Licht, J.D. Sprouty Proteins: Multifaceted Negative-Feedback Regulators of Receptor Tyrosine Kinase Signaling. Trends Cell Biol. 2006, 16, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, M.A.; Christofori, G. Sprouty Proteins, Masterminds of Receptor Tyrosine Kinase Signaling. Angiogenesis 2008, 11, 53–62. [Google Scholar] [CrossRef]
- Kawazoe, T.; Taniguchi, K. The Sprouty/Spred Family as Tumor Suppressors: Coming of Age. Cancer Sci. 2019, 110, 1525–1535. [Google Scholar] [CrossRef]
- Sasaki, A.; Taketomi, T.; Wakioka, T.; Kato, R.; Yoshimura, A. Identification of a Dominant Negative Mutant of Sprouty That Potentiates Fibroblast Growth Factor-but Not Epidermal Growth Factor-Induced ERK Activation. J. Biol. Chem. 2001, 276, 36804–36808. [Google Scholar] [CrossRef]
- Ozaki, K.; Kadomoto, R.; Asato, K.; Tanimura, S.; Itoh, N.; Kohno, M. Erk Pathway Positively Regulates the Expression of Sprouty Genes. Biochem. Biophys. Res. Commun. 2001, 285, 1084–1088. [Google Scholar] [CrossRef]
- Yang, X.; Kilgallen, S.; Andreeva, V.; Spicer, D.B.; Pinz, I.; Friesel, R. Conditional Expression of Spry1 in Neural Crest Causes Craniofacial and Cardiac Defects. BMC Dev. Biol. 2010, 10, 48. [Google Scholar] [CrossRef]
- Wright, K.D.; Mahoney Rogers, A.A.; Zhang, J.; Shim, K. Cooperative and Independent Functions of FGF and Wnt Signaling during Early Inner Ear Development Organogenesis. BMC Dev. Biol. 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mahoney Rogers, A.A.; Zhang, J.; Shim, K. Sprouty1 and Sprouty2 Limit Both the Size of the Otic Placode and Hindbrain Wnt8a by Antagonizing FGF Signaling. Dev. Biol. 2011, 353, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Simrick, S.; Lickert, H.; Basson, M.A. Sprouty Genes Are Essential for the Normal Development of Epibranchial Ganglia in the Mouse Embryo. Dev. Biol. 2011, 358, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.B.; Ng, C.K.D.; Wasserman, S.M.; Kömüves, L.G.; Gerritsen, M.E.; Topper, J.N. A Novel Interleukin-17 Receptor-like Protein Identified in Human Umbilical Vein Endothelial Cells Antagonizes Basic Fibroblast Growth Factor-Induced Signaling. J. Biol. Chem. 2003, 278, 33232–33238. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Friesel, R.; Kudoh, T.; Dawid, I.B. Identification of Sef, a Novel Modulator of FGF Signalling. Nat. Cell Biol. 2002, 4, 165–169. [Google Scholar] [CrossRef]
- Harduf, H.; Halperin, E.; Reshef, R.; Ron, D. Sef Is Synexpressed with FGFs during Chick Embryogenesis and Its Expression Is Differentially Regulated by FGFs in the Developing Limb. Dev. Dyn. 2005, 233, 301–312. [Google Scholar] [CrossRef]
- Dubey, A.; Yu, J.; Liu, T.; Kane, M.A.; Saint-Jeannet, J.-P. Retinoic Acid Production, Regulation and Containment through Zic1, Pitx2c and Cyp26c1 Control Cranial Placode Specification. Development 2021, 148, dev193227. [Google Scholar] [CrossRef]
- Maier, E.C.; Whitfield, T.T. RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities. PLoS Genet. 2014, 10, e1004858. [Google Scholar] [CrossRef]
- Jaurena, M.B.; Juraver-Geslin, H.; Devotta, A.; Saint-Jeannet, J.P. Zic1 Controls Placode Progenitor Formation Non-Cell Autonomously by Regulating Retinoic Acid Production and Transport. Nat. Commun. 2015, 6, 7476. [Google Scholar] [CrossRef]
- Swinburne, I.A.; Miguez, D.G.; Landgraf, D.; Silver, P.A. Intron Length Increases Oscillatory Periods of Gene Expression in Animal Cells. Genes Dev. 2008, 22, 2342–2346. [Google Scholar] [CrossRef]
- Xue, X.; Sun, Y.; Resto-Irizarry, A.M.; Yuan, Y.; Aw Yong, K.M.; Zheng, Y.; Weng, S.; Shao, Y.; Chai, Y.; Studer, L.; et al. Mechanics-Guided Embryonic Patterning of Neuroectoderm Tissue from Human Pluripotent Stem Cells. Nat. Mater. 2018, 17, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Shellard, A.; Mayor, R. Collective Durotaxis along a Self-Generated Stiffness Gradient in Vivo. Nature 2021, 600, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Tsuboi, T.; Ishinabe, N.; Kitaguchi, T.; Michiue, T. Wide and High Resolution Tension Measurement Using FRET in Embryo. Sci. Rep. 2016, 6, 28535. [Google Scholar] [CrossRef] [PubMed]
- Hirano, S.; Yamamoto, T.; Michiue, T. FRET-Based Tension Measurement across Actin-Associated Mechanotransductive Structures Using Lima1. Int. J. Dev. Biol. 2018, 62, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Soussi-Yanicostas, N.; Hardelin, J.P.; Arroyo-Jimenez, M.D.M.; Ardouin, O.; Legouis, R.; Levilliers, J.; Traincard, F.; Betton, J.M.; Cabanié, L.; Petit, C. Initial Characterization of Anosmin-1, a Putative Extracellular Matrix Protein Synthesized by Definite Neuronal Cell Populations in the Central Nervous System. J. Cell Sci. 1996, 109, 1749–1757. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michiue, T.; Tsukano, K. Feedback Regulation of Signaling Pathways for Precise Pre-Placodal Ectoderm Formation in Vertebrate Embryos. J. Dev. Biol. 2022, 10, 35. https://doi.org/10.3390/jdb10030035
Michiue T, Tsukano K. Feedback Regulation of Signaling Pathways for Precise Pre-Placodal Ectoderm Formation in Vertebrate Embryos. Journal of Developmental Biology. 2022; 10(3):35. https://doi.org/10.3390/jdb10030035
Chicago/Turabian StyleMichiue, Tatsuo, and Kohei Tsukano. 2022. "Feedback Regulation of Signaling Pathways for Precise Pre-Placodal Ectoderm Formation in Vertebrate Embryos" Journal of Developmental Biology 10, no. 3: 35. https://doi.org/10.3390/jdb10030035
APA StyleMichiue, T., & Tsukano, K. (2022). Feedback Regulation of Signaling Pathways for Precise Pre-Placodal Ectoderm Formation in Vertebrate Embryos. Journal of Developmental Biology, 10(3), 35. https://doi.org/10.3390/jdb10030035






