Abstract
Protein kinase CK2 (CK2) is a ubiquitous holoenzyme involved in a wide array of developmental processes. The involvement of CK2 in events such as neurogenesis, cardiogenesis, skeletogenesis, and spermatogenesis is essential for the viability of almost all organisms, and its role has been conserved throughout evolution. Further into adulthood, CK2 continues to function as a key regulator of pathways affecting crucial processes such as osteogenesis, adipogenesis, chondrogenesis, neuron differentiation, and the immune response. Due to its vast role in a multitude of pathways, aberrant functioning of this kinase leads to embryonic lethality and numerous diseases and disorders, including cancer and neurological disorders. As a result, CK2 is a popular target for interventions aiming to treat the aforementioned diseases. Specifically, two CK2 inhibitors, namely CX-4945 and CIBG-300, are in the early stages of clinical testing and exhibit promise for treating cancer and other disorders. Further, other researchers around the world are focusing on CK2 to treat bone disorders. This review summarizes the current understanding of CK2 in development, the structure of CK2, the targets and signaling pathways of CK2, the implication of CK2 in disease progression, and the recent therapeutics developed to inhibit the dysregulation of CK2 function in various diseases.
1. Introduction
During development, the phosphorylation and dephosphorylation of proteins is essential for the proper formation of organisms [1,2,3]. This process is catalyzed primarily by enzymes that function as kinases, phosphatases, or both [4,5]. Currently, over 530 protein kinases have been identified in humans, many of them involved in embryonic and developmental processes [6,7,8,9]. One of the proteins, protein kinase CK2 (CK2), formerly known as casein kinase II, regulates many of these processes and is expressed during embryogenesis and adulthood [10,11]. CK2 is a holoenzyme consisting of two catalytic subunits (two CK2α, CK2α’, or one CK2α and one CK2α’) and two regulatory beta subunits (CK2β) [12,13,14]. This kinase has over 200 substrates and targets more than 280 phosphorylation sites, indicating its ubiquitous expression and involvement in many cell signaling pathways and molecular processes [10]. It is present in every eukaryotic cell and has been highly conserved throughout evolution. CK2 regulates developmental pathways such as neurogenesis and cardiogenesis, as well as processes in adulthood such as chondrogenesis, osteogenesis, and adipogenesis [15,16,17,18,19,20,21,22]. Further, CK2 is implicated in the progression of diseases including cancer, Alzheimer’s disease, senescence, diabetes, obesity, infectious disease, and cardiovascular diseases [23,24,25]. Due to the vast involvement of CK2 in embryogenesis, development, adulthood, and disease progression, this review aims to summarize the current understanding of this enzyme.
2. CK2 and Development
CK2 is highly expressed and prominent throughout many stages of development. During early embryogenesis, CK2 activity was identified first at embryonic day 11 (E11) in mice, rats, and chickens [26,27,28]. Continuously after this day, CK2 regulates the formation of various organ systems. Specifically, during neurogenesis and regarding the negative regulation of neuron differentiation, CK2 phosphorylates Groucho/Transducin-like enhancer (TLE1) to limit the formation of neurons [29]. CK2α null mice display improper neural tube formation, emphasizing the essential role of CK2 in this process [30]. Furthermore, CK2 contributes to memory formation, and the overexpression of this kinase is correlated with improved long-term memory [31]. In cardiogenesis, CK2, specifically CK2α’, is essential for the proper formation of the heart, and knockouts of this subunit lead to irregular cardiogenesis, as well as death in mid-gestation [30]. Similarly, CK2 is critical for proper spermatogenesis and the fertility of organisms [32]. While CK2α’−/− mice remain viable, male mice are infertile and incapable of producing offspring, as germ cells do not form properly [30,33,34]. Moreover, CK2α’ is highly expressed in testis, and a lack of CK2α’ also leads to oligozoospermia [33]. CK2 is also required for the formation of the skeleton during development, specifically at E17.5 [35]. It has also been demonstrated that the conditional deletion of the gene encoding beta subunit of CK2 (Csnk2b) leads to the shortening of the limbs, improper endochondral ossification, and lethality in mice [35]. Therefore, CK2 is critical for many stages of development and the production of viable organisms (Figure 1). The evolutionary conservation of this enzyme is also evident from the effects of its knockout mutations in yeast (Table 1) [36]. In addition, during the third trimester of pregnancy, feto–maternal circulation is established through a mechanism involving trophoblast invasion. This process requires mitochondrial modifications, and CK2 is critical throughout the process. Therefore, this kinase is one of the central enzymes during placental development [37].
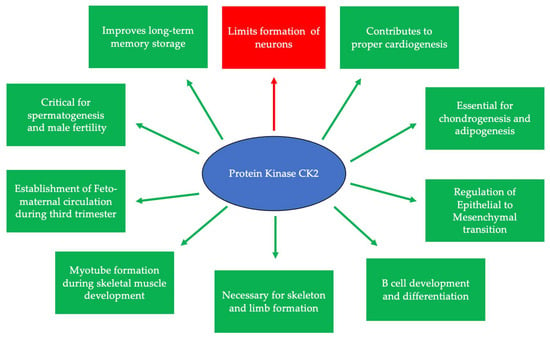
Figure 1.
CK2 is expressed at embryonic day 11 and is critical for many developmental processes. Specifically, CK2 is necessary for limiting neurogenesis and preventing the excessive differentiation of neurons. In addition, CK2 expression promotes long-term memory storage. Further, this protein is essential for skeletogenesis, chondrogenesis, adipogenesis, and proper limb formation. CK2 contributes to spermatogenesis, and the inhibition of its expression leads to infertility. Finally, CK2 is important for B cell differentiation and development, myotube formation, the regulation of the epithelial-to-mesenchymal transition (EMT), and the establishment of circulation between the fetus and the mother during the third trimester.

Table 1.
Phenotypes associated with CK2-specific subunit knockouts.
There are three classifications of substrates for CK2. Class I substrates are equally phosphorylated by the holoenzyme and free catalytic subunits, whereas Class II substrates are phosphorylated by only free catalytic subunits but not by the holoenzyme. Further, substrates of Class III are phosphorylated preferentially by the holoenzyme but not by free catalytic subunits. Thus, the phosphorylation of these different classes varies during development (Figure 2) [45]. To study the role of the individual subunits during development, specific subunit knockouts were induced. As described in Table 1, the involvement of specific subunits in developmental processes is evident from the conditional knockouts and the resulting associated phenotype or molecular consequence (Table 1). Specifically, CK2β knockouts in mice display developmental arrest after implantation [46]. Furthermore, the balance between CK2α and CK2β subunits is also important. The ratio of CK2α:CK2β is critical for maintaining epithelial morphology [47].
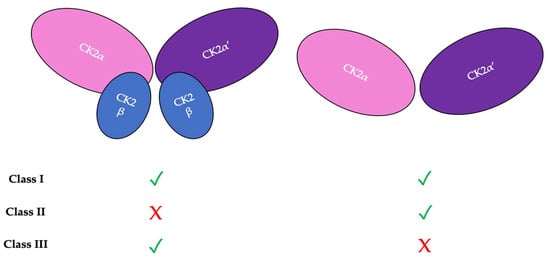
Figure 2.
There are three classifications of CK2 substrates. Class I substrates are identified as proteins that are equally phosphorylated by the holoenzyme and individually by the catalytic subunits. Class II substrates are specifically phosphorylated by the catalytic submits of CK2 but not by the holoenzyme. Class III substrates are preferentially targeted and phosphorylated by the holoenzyme but not by the catalytic subunits of CK2.
3. The Substrates and Signaling Pathways Regulated by CK2
The implication of CK2 in many developmental processes is due to its multifunctionality. Specifically, over 400 substates are identified as targets of CK2 [19,21]. While these substrates are involved in a myriad of pathways, some are critical for the survival and maintenance of cells. A notable pathway of the CK2 is Wnt-signaling, as it binds directly to both β-catenin and T cell-specific transcription factor/lymphoid enhancer-binding factor (TCF/LEF) [17,48,49,50,51]. Furthermore, CK2 phosphorylates other substrates such as dishevelled (Dvl) and adenomatous polyposis coli (APC) [17,52,53]. Regarding β-catenin, CK2 phosphorylates this protein at threonine 393, which interacts with Axin to stabilize β-catenin and prevent it being marked for destruction by protein kinase CK1 [54,55].
A second binding site of CK2 is the bone morphogenetic protein receptor type Ia (BMPRIa), which regulates Smad and non-Smad signaling pathways. Within cells, such as osteoblasts, chondrocytes, and adipocytes, CK2 typically binds to BMPRIa and prevents the downstream activation of Smad signaling [56,57,58,59,60,61]. However, once members of the transforming growth factor beta (TGFβ) superfamily, i.e., BMP-2 and BMP-4, bind to BMPRIa, CK2 is released from BMPRIa, leading to the increased differentiation and activity in osteoblasts, osteoclasts, adipocytes, and chondrocytes [18,19,22,58,62,63,64,65,66,67,68,69,70,71,72,73,74].
Aside from Wnts and Bone morphogenetic proteins (BMPs), CK2 is involved in signaling pathways responsible for proliferation and survival, such as Akt/Phosphatidylinositol-3-Kinase (PI3K) signaling. Specifically, Akt is phosphorylated by CK2 at serine 129, which promotes cell viability and proliferation [75,76,77]. The activation of Akt/PI3K by CK2 has been well documented in cancer progression, which will be discussed later in this review. Similarly, CK2 directly interacts with extracellular signal-regulated kinase (Erk) 1/2, thereby activating the mitogen-activated protein (MAP) kinase signaling pathway. After the phosphorylation of Erk1/2 by CK2 at serine residues within the nuclear translocation signal (NTS) region, Erk1/2 is imported into the nucleus by importin7 to function as a transcription factor in promoting cellular processes [78,79,80,81,82].
Next, CK2 activates the nuclear factor kappa B (NF-κB) signaling pathways. In this context, CK2 phosphorylates the RelA/p65 subunit of NF-κB at serine 529, leading to the activation of this protein [83,84,85]. Further, as CK2 phosphorylates NF-κB inhibitor alpha (IκBα) at its C-terminal sites; this leads to increased cellular proliferation and protection against ultraviolet (UV) radiation and has been implicated in cancer progression [86,87,88,89].
In addition, CK2 activates the Janus Kinase/signal transducer and activator of transcription (JAK/STAT) pathway, leading to enhanced cellular survival, differentiation, and proliferation [90]. Here, CK2 binds to the JAKs, which subsequently activate STATs, such as STAT3, to initiate downstream signaling [91,92]. Additionally, CK2 is essential for the activation of the JAK/STAT pathway by interleukins (ILs), such as IL-6, to initiate transient signaling in the immune system [93,94,95].
CK2 also has a critical role in mammalian target of rapamycin (mTOR) signaling. Specifically, the inhibition of CK2 expression is associated with an increased activity of Ikaros, which binds to the mTOR gene to suppress its activity and prevent tumor formation [96,97,98,99,100]. However, the overexpression of CK2 subsequently increases mTOR signaling, leading to poor outcomes such as tumorigenesis and cancer progression [101,102,103,104]. Therefore, by targeting CK2 activity with inhibitors such as CX-4945 (described later in this review), the mTOR pathway can be diminished, leading to improved prognoses in patients.
Furthermore, CK2 plays a role in signaling pathways governing the DNA damage and the repair response. Specifically, CK2 responds to DNA damage and increases the repair response to accelerate the process [105,106]. Here, CK2 phosphorylates X-ray repair, cross complementing protein 4 (XRCC4), XRCC1, DNA-dependent protein kinase, and apurinic/apyrimidinic endonuclease (APE), among others [107,108,109]. Its ability to bind to a multitude of substrates during DNA damage repair aids in the survival and viability of cells.
Finally, an additional pathway in which CK2 is interestingly involved includes zinc signaling. As zinc can serve as a secondary messenger, it is involved in various signaling pathways. Specifically, CK2 can regulate the concentration of zinc through the phosphorylation of one of its channels, zinc channel ZIP7, located on the Golgi apparatus and endoplasmic reticulum (ER) [110,111]. Once this channel is phosphorylated, there is an influx of zinc into the cytoplasm that leads to continued epidermal growth factor receptor (EGFR) signaling and activation of other proteins, such as Erk1/2 [110,111,112]. While the precise mechanism of CK2 and zinc signaling is not fully delineated, targeting this pathway for future therapeutics may be promising. These pathways are summarized precisely and effectively in the publication written by Borgo et al. [106]. Taken together, CK2 is largely involved in the regulation of multiple cellular signaling pathways, cell survival, and regulation at transcriptional and translational levels.
4. CK2 Structure and Kinase Activity
To understand the multifunctionality of CK2, its structure and kinase activity must be observed. CK2 is comprised of α and β subunits, both contributing to the activity of this kinase. This enzyme contains 391 amino acids with a molecular mass of 45.1 kDa [113,114,115]. The formation of a heterotetrametric complex contributes to the stability and activity of CK2 [116]. Specifically, the formation of the CK2 holoenzyme may take the form of α2β2, αα’β2, or α’2β2 subunits [117,118,119]. The interaction of the regulatory and catalytic subunits contributes to the function and enzymatic activity of CK2. In 2001, Niefind et al. crystallized CK2 to determine the precise molecular spatial localizations of its subunits [113]. Here, the β2 dimer interacts slightly with the α subunits to connect them and stabilize the molecule [113]. Similarly, the β2 subunits interact strongly, whereas the α subunits do not interact at all. This strong interaction is attributed to the formation of salt bridges and hydrogen bonding, the zinc (Zn2+) binding motif, and a hydrophobic core [113]. Further, the active site is between the C-terminus, which is helical, and the N-terminus, which has the architecture of many β-sheets [113]. The structure of this active site aids in CK2′s ability to bind to a myriad of substrates and maintain its catalytic activity. When targeting substrates, CK2 typically phosphorylates serine/threonine residues that share the S/T-X-X-D/E/pS/pY consensus sequence [120]. Further, in other circumstances, CK2 can phosphorylate substrates that do not share the consensus sequence, such as serine 392 of p53 or the tyrosine residues of Fpr3 in yeast [120,121,122]. Due to the versatility of this enzyme, CK2 activates numerous interacting proteins, such as nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 1 and transient receptor potential cation channel subfamily M (TRPM) 3 ion channels [123,124]. As CK2 targets many substrates, it has increasingly become a candidate for novel therapeutics, as it is implicated in the progression of many diseases, such as breast and colon cancer [23,125,126,127].
5. CK2 Expression and the Progression of Diseases
The ubiquitous expression of CK2 occurs within almost all cellular compartments [128]. The continued identification of its newer substrates broadens the understanding of the processes in which this protein is involved [129]. Alternative forms of this holoenzyme, such as single catalytic or regulatory subunits, have differential expression and functions within tissues. The CK2α’ subunit has enhanced mRNA expression in the testis, skin, and across brain tissue. Further, the CKα and CK2β subunits have a more ubiquitous expression within tissues [130]. Oligomeric super-structures of the CK2 holoenzyme also exist, in which this protein controls the exposure of the catalytic active sites and subsequent enzymatic activity [131]. The balance between the number of catalytic CK2α and regulatory CK2β subunits is maintained by a mechanism whereby CK2α activates the transcription factors for CK2β when its levels are depleted. Once sufficient levels of CK2β are obtained, transcription is inhibited by holoenzyme formation, which then disables the interaction between CK2α and its own transcription factors [132].
Several micro ribonucleic acids (miRNAs) are trans-regulating elements of CK2 protein expression. Small noncoding RNAs targeting CK2 include miR-760, miR-186, miR-337-3p, and miR-216b. These miRNAs reduce CK2α protein expression by binding at the 3′ untranslated region (UTR) of the transcript and inhibiting translation [117,133,134]. During senescence, miR-186, miR-216b, miR-337-3p, and miR-760 have been shown to target the CK2α subunit at the 3′UTR region and cause its degradation [135,136]. Long noncoding RNA KCNQ10T1 and its interaction with miR-760 is a positive regulator of CK2 to enhance its activity [137].
Protein interactors of CK2 also regulate its expression and activity. In chondrocytes, the downregulation of αB-crystallin by CK2 is shown to protect cells against apoptosis. The pharmacological inhibition of CK2 also cause changes in the localization of this protein [138]. Further, HSP90 interacts with CK2 to prevent its self-aggregation and loss of activity [139].
5.1. Dynamic Localization of CK2
The subcellular localization of CK2 determines its regulation, which predominantly includes the cytoplasm, plasma membrane, and nucleus. Interactor proteins and substrates of CK2 are located predominantly in these three cellular compartments [140]. The localization of CK2 varies during cell cycle progression and is sensitive to external stimuli, such as radiation, hypoxia, and stress. Exposure to ionizing radiation (IR) was identified to alter CK2α subunit localization from the cytoplasm to the nucleus in human non-small cell lung cancer cell lines, including H460, A549, and PC9 (Figure 3). The nuclear localization is reported to be transient. In addition, there is an increase in the kinase activity of CK2 during the S phase when cells are exposed to IR. Furthermore, with the pharmacological inhibition of CK2, there is an increase in IR sensitivity [141,142]. This emphasizes the role of CK2 in transmitting radiation stimuli to the nucleus and its involvement in the DNA damage response. Similarly, in M059K human glioma cells, it colocalizes with the IR-induced DNA damage sites [108]. Resistance to IR treatment could be attributed to the dynamic localization of this kinase. During hypoxia, CK2 activity is seen to increase without an increase in its expression, but rather by the transport of catalytic α and α’ subunits to the nucleus (Figure 4). CK2 also plays a role in activating a histone-modifying enzyme, histone deacetylase 2 (HDAC2), in tumors under hypoxic conditions. Here, both catalytic subunits of this kinase are important for the activation of HDAC2 in tumors, and, further, their downregulation impairs the hypoxia-induced activation of histone deacetylase [143]. The nuclear status of CK2 is important for the regulation of several nuclear proteins involved in cell proliferation, survival, and cell death. B23 (also known as nucleophosmin or numatrin) is a nuclear matrix-associated protein that regulates ribosomal RNA (rRNA) synthesis. Its recruitment to specific nucleolar compartments during cellular growth is dependent on its phosphorylation by CK2 [144]. The downregulation of CK2 in prostate cancer cells results in a loss of the nuclear localization of this protein [145].
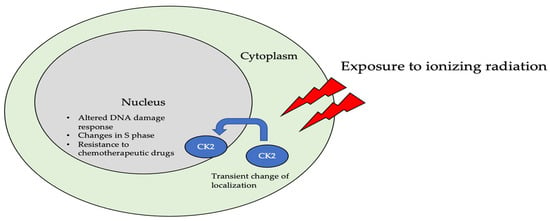
Figure 3.
The localization of CK2 changes when cells are exposed to ionizing radiation (IR). Specifically, CK2α is translocated from the cytoplasm to the nucleus in multiple cell lines, including A549, H460, PC9, and M059K. Further, upon radiation exposure, the overall kinase activity of CK2 increases, emphasizing its role in the DNA-damage-repair response, as it colocalizes with DNA in the nucleus of cells.
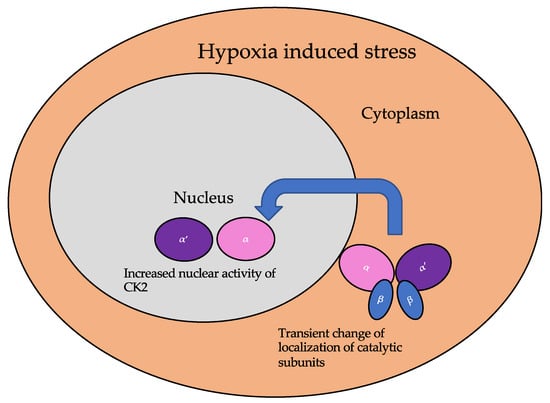
Figure 4.
The localization of the catalytic subunits of CK2 is altered during hypoxia. Specifically, these subunits are translocated to the nucleus transiently to aid the cellular response to hypoxia.
5.2. CK2 in Diseases
CK2 is associated with the etiology of many diseases due to its pleiotropic nature and involvement in almost all cellular process. The diseases associated with the aberrant expression or function of this kinase include cancer, multiple sclerosis, cystic fibrosis, diabetes mellitus, neurological disorders, cardiac hypertrophy, and inflammation [102]. The examples delineated below describe the involvement of CK2 in disease progression based on unique mechanisms specific for the disease. The implication of aberrant CK2 expression and/or function is apparent in several studies.
5.2.1. CK2 in Cancer Progression
CK2 regulates the progression of at least 15 cancer-related proteins including the tumor suppressor p53, histone-modification enzymes such as HDAC1 and HDAC2, and NF-κB subunit RelA [146]. The interaction of additional direct interactors and indirectly regulated proteins remains unclear. There is an upregulation of CK2 during the tumor progression of many cancers and leukemia [147,148]. CK2 itself can be used as a prognostic marker for some cancer types (Table 2) [149]. Proteomic analyses of cell lines indicates that the catalytic CK2α subunit is predominantly expressed in several cancerous cell lines, including U2OS from human osteosarcoma, epidermoid squamous cell carcinoma, brain glioblastoma A549 from lung carcinoma, GAMG from glioblastoma, HEK293 from embryonic kidney cells, HeLa from cervical carcinoma, HepG2 from hepatoma, Jurkat from Acute T cell lymphoma, LnCap from prostate carcinoma, MCF from mammary carcinoma, and RKO from colon carcinoma [131,150,151,152]. Contrasting observations exist for the expression status of CK2. For example, in breast cancer, the nuclear localization of CK2α is emerging as an independent prognostic factor [153]. However, in esophageal cancer, CK2 expression is downregulated [154]. This downregulation resulted from the overexpression of Smad4-mediated transcriptional repression under the influence of the sex-determining region Y-box 2 (SOX-2) [154].

Table 2.
CK2-subunit overexpression is a prognostic marker for several types of tumors.
Even further, because a gain-of-function mutation associated with CK2 has yet to be revealed, this enzyme is classified as a tumor-promoting non-oncogene [162]. However, the deletion of CSNK2B, which codes for the CK2β subunit, is present in diffuse large B-cell lymphoma (DLBCL) [163]. Additionally, a recent case of the CSNK2A1 fusion gene is present in a myeloid neoplasm, indicating the diverse roles of CK2 implications in cancer [164].
As CK2 regulates the activation of several proteins required for the cell cycle, the consequences of its overexpression are exacerbated in solid tumors and leukemias [165]. CK2 overexpression and/or overactivity results in the abnormal phosphorylation of its targets. One example is tumor suppressor Ikaros, which is a transcriptional repressor of the genes encoding for Bcl-2-like protein 1 (BCL2L1, PIK3CD, and PIKFYVE) [98,100].
The regulation by second messengers, such as cyclic nucleotides, is important for many oncogenic kinases [166]. However, unlike other kinases except for CK2, there is no regulation by the concentration of second messenger molecules (such as cyclic nucleotides, inositol phosphate, and calcium) or its own phosphorylation status for kinase activity. Instead, the high basal activity of this kinase is demonstrated by its crystallographic structure. The interaction between helix c of the N-terminal region is in tight interaction within the activation loop [167]. Thus, the constitutive activity of CK2 due to its structure adds to the mystery of how this kinase is regulated. The following table classifies general mechanisms by which CK2 is involved in cancer pathogenesis (Table 3) [131].

Table 3.
The involvement of CK2 in tumorigenic mechanisms.
The central role of CK2 in pathway networks for cell cycle regulation and proliferation makes this protein an interesting pharmacological target for treatment of cancer.
5.2.2. CK2 and Diabetes and Obesity
In pancreatic β cells, a G-protein coupled-receptor muscarinic 3 receptor (M3R) regulates insulin release and homeostasis. CK2 is a negative modulator of M3R. Its phosphorylation of the M3Rs on β-cells inhibits insulin release [171]. Therefore, by phosphorylating the M3Rs, CK2 prevents the release of insulin. However, the abnormal activity of CK2 is linked to diabetes due to its role with these receptors.
Sirtuin1 (SIRT1) deacetylase is a substrate of CK2 and is a regulator for hepatic lipid metabolism. This protein modulates transcription as an energy sensor molecule with its deacetylase function. In diet-induced obesity, CK2 is highly overexpressed and therefore increases the activity of SIRT1. Aberrant phosphorylation of SIRT1 at serine 164 by CK2 inhibits the entry of the former into the nucleus and mildly impairs deacetylase function, thus hampering its regulatory function in hepatocytes, and has been linked to the onset of obesity [172].
5.2.3. CK2 and Heterotrophic Bone Formation
Heterotrophic bone formation in soft tissue due to injury, medication, or as a symptom of another disease can be a painful condition. CK2 phosphorylation stabilizes runt-related transcription factor 2 (RUNX2) by deubiquitylation through the CK2/herpesvirus-associated ubiquitin-specific protease (HAUSP)/RUNX2 pathway. This regulation is important during skeletal development for the fine-tuning of RUNX2, which is a master transcription factor during osteoblastogenesis. However, the overexpression or overactivity of CK2 during heterotrophic bone formation causes ectopic osteoblastogenesis from skeletal stem cells by the same mechanism for RUNX2 stabilization [35].
5.2.4. CK2 and Cardiovascular Diseases
Within the cardiovascular system, CK2α interacts with p27 to prevent its ubiquitination or degradation. Here, the stabilization of p27, which is an inhibitor of cell progression, leads to its accumulation in cytoplasm and the subsequent apoptosis of the cell. Further, CK2α is responsive to external growth stimuli to regulate its interaction with p27. The interaction between CK2α and p27 is interrupted during cardiac hypertrophy [173]. Here, transcriptional reprogramming leading to fetal gene expression causes cardiac hypertrophy. In addition, this reprogramming is related to HDAC2 activity. Specifically, CK2α phosphorylates HDAC2 at serine 394, causing its activation, and leads to cardiac hypertrophy [174].
Patients with cardiac desynchrony are associated with higher risk for morbidity and mortality from heart failure compared to patients with synchronic heart function. Cardiac resynchronization therapy (CRT, pacemaker) is the only known non-pharmaceutical intervention that has long-term effects on this disorder. However, some patients do not respond to this intervention. To study molecular mechanisms for dyssynchronous heart failure (HFdys) and the impact of CRT, proteomic analysis was performed. The comparison of phosphoproteome of HFdys before and after CRT led to the discovery that CK2 signaling is activated during HFdys and that CRT reverses it [175]. The contribution of CK2 signaling in cardiac desynchrony is still an ongoing area of research.
5.2.5. CK2 and Neurodegenerative Disorders
The expression and activity of CK2 in brain cells is involved in neurodegenerative disorders [176]. Here, the overexpression of CK2 is associated with neurodegeneration [177]. Further, there are numerous CK2 substrates associated with brain development and function. In Alzheimer’s disease, CK2 colocalizes with the neurofibrillary tangles. The hyperphosphorylation of the SET protein at serine 9 leads to its improper localization in the cytoplasm caused by the activation of CK2 by tau or β-amyloid (Aβ). The overexpression of CK2 also causes cognitive deficits by impairing synaptic plasticity and synaptogenesis [178]. In Parkinson’s disease (PD), CK2 is localized to Lewy bodies and phosphorylates α-synuclein and synphilin-1 [179]. The treatment of PD using levodopa (L-DOPA) is known to induce L-DOPA-induced dyskinesis (LID). CK2 is important for the regulation of pathways leading to LID. Specifically, CK2α downregulation reduces the severity of LID [176].
Regarding dopaminergic signaling, CK2 is a negative regulator of the D1 receptor (D1R). It binds directly to the Gαs or Gαolf (Golf) subunits, and its knockout leads to the elevated expression of D1R on the plasma membrane. This leads to an increased response to D1 agonists [40]. With the CK2 conditional knockouts in medium spiny neurons, which express dopaminergic receptors, CK2α regulates the D1 signaling pathway in vivo [180].
5.2.6. CK2 and Neurological Disorders
Further, in the neuropsychiatric disorder Tourette syndrome (TS), a mutation in the SLITRK1 gene is present. The gene product is a membrane-bound protein that regulates synapse formation. The symptoms of TS arise early in the childhood, and affected individuals often suffer from attention-deficit/hyperactivity disorder (ADHD) and obsessive–compulsive disorder (OCD). CK2 phosphorylates the SLITRK1 protein at its 14-3-3 binding site, and a mutation at this phosphorylation site inhibits neurite formation [181].
5.2.7. CK2 in Infectious Diseases
Viral proteins are substrates of CK2. The presence of host kinase phosphorylation sites within these proteins can be useful for the virus to identify host cell status [182]. In the proteome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a total of 25 phosphorylation sites have been identified. CK2 is one of the main kinases identified to regulate these sites. There is also activation of CK2 signaling observed during SARS-CoV-2 infection [182].
The hepatitis C virus (HCV) nonstructural protein 5A (NS5A) plays an important role in viral particle assembly. Structural studies of the NS5A and CK2 complex using time-resolved nuclear magnetic resonance (NMR) indicate that there are four possible CK2 phosphorylation sites in the NS5A D3 domain [183]. The expression of hepatitis B virus (HBV) is regulated by the Human La protein (hLa). The phosphorylation of hLa at serine 366 by CK2 activates the protein and increases HBV expression. The pharmacological inhibition of CK2 by tetrabromobenzimidazole (TBBz) or a knockdown its gene results in a reduction of HBV expression [184].
In prion diseases, the pathogenic form of the prion protein (Prp) interferes with cellular processes, such as fast axonal transport (FAT), which causes an onset of neurodegenerative disorders. The intracellular accumulation of cellular prion protein (PrPC) and full-length PrP (PrP-FL) induces neuronal toxicity, causing severe ataxia in mice, and further, FAT is often inhibited. CK2 inhibits FAT in a similar pattern as in PrP-FL-induced symptoms [48,185,186,187].
5.2.8. Regulation of Immune Response by CK2
The regulation of the immune cell response is targeted for the use of CK2 inhibitors. However, delineating its role during development in the immune response under disease progression has been a challenge. The involvement of CK2 is being investigated during this disease response in ongoing studies.
During Listeria monocytogenes (Lm) infection, the adaptive and innate immune responses are involved. In myeloid cell-specific conditional knockout of CK2α in mice, host resistance to Lm infection is significantly increased without affecting myeloid cell development. Myeloid cell recruitment is also seen to be negatively regulated by CK2α [188].
Further, the expression of CK2 is increased in the inflamed mucosa of ulcerative colitis (UC) patients. It is important for maintaining reciprocal balance between Th17 and Treg cells [189]. Interestingly, CK2 activity is reduced, along with an increase in reactive oxygen species (ROS) generation, during acute colitis. NADPH oxidase 1 (NOX1), which is an important regulator of mucosal immunity, is dysregulated during inflammatory bowel disease. Further, CK2 is a suppressor of NOX1 activity [123].
5.2.9. CK2 and Senescence
CK2 activity and expression is downregulated in older animals. Further, the pharmacological inhibition of CK2 induces cellular senescence [190,191]. With artificial downregulation of CK2 by the activity of histone trimethylases, such as histone-lysine N-methyltransferase SUV39H1(SUV39h1), its activity is upregulated in senescence and leads to an increase in tri-methylation at the 9th lysine residue of the histone H3 (H3K9me3) and senescence-associated heterochromatin formation (SAHF). An increase in SAHF leads to the suppression of genes associated with cell cycle progression, such as cyclin D1 [192].
Next, in CK2α knockdown cell lines (MCF-7 and HCT116 cells), the expression of histone demethylases [JmjC domain-containing histone demethylase (JMJD2/KDM4) and lysine-specific demethylase 1 (LSD1/KDM1a)] is downregulated at the translational level. The ectopic expression of these histone demethylases suppressed SAHF and senescence-associated β-galactosidase activity. The effects of the translational downregulation of LSD1 due to the reduction in CK2 activity are complex, since it has non-histone substrates such as p53. The p53/p21Cip1/WAF1 pathway, which is an upstream regulator of SUV39h1 and H3K9me3, is also upregulated in CK2α knockdown cells. [193,194]. Therefore, this pathway is important for CK2-downregulation-mediated senescence.
6. Clinical Applications of CK2
The implication of CK2 in the progression of many diseases has led to its popularity as a therapeutic target. Specifically, researchers have attempted to prevent the kinase activity of CK2, thereby to prevent pathological activity. In cells, CK2 contains pocket domains for adenosine triphosphate (ATP)-binding [195]. The pocket domain is essential for the activation and subsequent enzymatic activity of CK2. Therefore, blocking this domain may deactivate CK2. One such inhibitor under investigation is CX-4945, known as silmitasertib in the clinic. CX-4945 is currently in clinical trials to treat cancer and is being tested as an oral administration to patients to limit the activity of CK2. Specifically, CX-4945 functions as an ATP-competitive inhibitor, leading to cell cycle arrest and apoptosis [196]. Here, CX-4945 inhibits the activity of both CK2α and CK2α’ by binding to the ATP-binding site of these subunits with a higher affinity than ATP [195,197,198,199]. This competitive binding leads to the decreased expression of CK2 and its subsequent signaling, such as Smad and PI3K/Akt pathways [196,199] (Figure 5A). This decreased activation of these pathways by CX-4945 has inhibited the progressions of several cancers, including gastric cancer, renal cancer, hematological cancer, cholangiocarcinoma, basal cell carcinoma, and medulla blastoma [24,197,200,201]. CX-4945 is a promising therapeutic agent and has displayed minimal side-effects, such as diarrhea, nausea, and anemia, suggesting its role as a safe treatment [201,202].
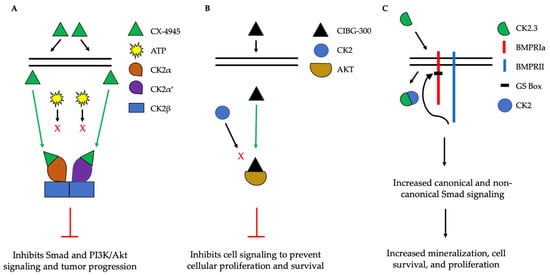
Figure 5.
Simplified schematic demonstrating three therapeutics targeting the activity of CK2. (A). CX-4945 is a competitive ATP inhibitor that binds preferentially to ATP-pocket domains. Here, it binds to both CK2α and CK2α’, thereby preventing ATP from binding and activating CK2. CX-4945 has prevented tumor progression by inhibiting signaling pathways such as Smad and PI3K/AKT. (B). CIBG-300 functions by binding to conserved phosphorylation sequences on the substrates of CK2, such as Akt. This association prevents the phosphorylation of these substrates by CK2, leading to decreased cell signaling, proliferation, and survival. (C). CK2.3 is uptaken by cells and binds to CK2, therefore preventing its association with BMPRIa. As a result, BMPRII can phosphorylate BMPRIa, leading to the downstream activation of signaling pathways that induce mineralization and cell survival.
Another CK2 inhibitor, known as CIBG-300, is also currently in clinical trials as a therapeutic to treat cancer [106,203]. This inhibitor binds specifically to CK2 to prevent cellular proliferation; leads to decreased cell adhesion and migration; and promotes the apoptosis of the cells, specifically in cervical cancer, breast cancer, and colorectal cancer, both in vivo and in vitro [203,204,205,206,207,208,209,210]. Here, CIBG-300 functions by binding to the conserved phosphorylation sequences of CK2 substrates [207,211]. Subsequently, CK2 is unable to phosphorylate its downstream target; CIBG-300 thereby prevents the activity of various proteins involved in cancer progression and inhibits angiogenesis [207,208,209,212,213]. More interestingly, CIBG-300 inhibits the activation of Akt, PI3K, PTEN, and NF-κB signaling pathways, therefore decreasing cell proliferation and survival [95,214,215] (Figure 5B). Further, CIBG-300 treatment leads to limited side-effects including edema, hot flashes, tachycardia, lower abdominal pain, and bleeding [213]. Thus, this potential therapeutic has been efficient in clinical trials and holds promise to treat a wide array of cancers.
In addition, inhibiting the enzymatic activity of CK2 can be used to treat other disorders, such as osteoporosis (OP) and osteoarthritis (OA). CK2 is expressed in bone cells, such as osteoblasts and osteoclasts. Within these cells, CK2 is bound to the intracellular domain of BMPRIa at its phosphorylation sites. As a result, the BMP-signaling pathway is inactive until an extracellular ligand, such as BMP-2, binds to BMPRIa to cause a conformational change of BMPRIa and release CK2. This release allows BMPRII to phosphorylate BMPRIa, leading to the downstream activation of Smad and non-Smad signaling to increase bone and cartilage formation. However, as humans age, bone mineral density and cartilage decrease; these changes lead to osteoporosis and osteoarthritis, even in the absence of extracellular ligands, respectfully. The interaction between CK2 and BMPRIa can be disrupted intracellularly to increase Smad and non-Smad signaling. Here, the Nohe lab constructed mimetic peptides named CK2.1, 2.2, and 2.3, each of which contain a phosphorylation site sequence of BMPRIa. This sequence is flanked by other amino acids, and the peptides contain the antennapedia homeodomain for cellular uptake. Once endocytosed by cells, CK2.X binds to CK2 and prevents it from associating with BMPRIa. CK2.1 (phosphorylation site SYED) leads to chondrogenesis; CK2.2 (phosphorylation site SLYD) leads to adipogenesis, and CK2.3 (SLKD) leads to osteogenesis. Further, CK2.3 upregulates signaling and leads to increased mineralization in primary osteoblasts isolated from OP patients. Further, this peptide enhances bone mineralization in both in vivo and in vitro models in the absence of BMP-2 (Figure 5C). These peptides, along with the BMP-signaling pathway, may be useful in treating bone disorders such as OP.
7. Discussion
This review aimed to describe the current roles of CK2 in development and adulthood, as well as describing its role in signaling pathways and disease progression. CK2 is essential for a myriad of developmental processes and the regulation of signaling pathways throughout adulthood. CK2 is involved in neurogenesis, cardiogenesis, spermatogenesis, and limb formation. Further, this enzyme has over 200 known phosphorylation substrates and is involved in pathways including cellular proliferation and survival, osteogenesis, angiogenesis, and chondrogenesis. However, due to the ubiquitous expression and constitutive activity of CK2, it has been well documented to be involved in the progression of many cancers, bone disorders, neurological and neurodegenerative disorders, immune disorders, and infectious diseases. Further, as the role of CK2 in various diseases is established, it has also become a popular target to treat the aforementioned disorders. With the novel findings about the role of the individual subunits of CK2 during disease progression, the targeting of CK2 activity may become more effective. Further, studies with its cell-lineage-specific function in contrast to its ubiquitous expression and constitutive activity can identify specific pathways for therapeutics to target. Specifically, two peptide-derived therapeutics are currently in clinical trials, known as CX-4945 and CIBG-300. These peptides have been successful in inhibiting the proliferation and progression of cancers and tumors. Further, other peptides currently being investigated in research are CK2.1 and CK2.3, which may be utilized to treat osteoarthritis and osteoporosis, respectively. Therefore, targeting CK2 and further elucidating its role in disease progression may be useful in treating many different disorders.
Author Contributions
Conceptualization, A.N., D.H. and V.P. methodology, A.N. and D.H.; software, A.N., D.H. and V.P.; validation, A.N., D.H. and V.P.; formal analysis, D.H.; investigation, A.N., D.H. and V.P.; resources, A.N.; data curation, D.H.; writing—original draft preparation, D.H.; writing—review and editing, A.N., D.H. and V.P.; visualization, A.N., D.H. and V.P.; supervision, A.N.; project administration, A.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to acknowledge all members of the Nohe lab, the University of Delaware, and the Delaware Biotechnology Institute for their continued support during this project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Presler, M.; Van Itallie, E.; Klein, A.M.; Kunz, R.; Coughlin, M.L.; Peshkin, L.; Gygi, S.P.; Wühr, M.; Kirschner, M.W. Proteomics of phosphorylation and protein dynamics during fertilization and meiotic exit in the. Proc. Natl. Acad. Sci. USA 2017, 114, E10838–E10847. [Google Scholar] [CrossRef] [PubMed]
- Kurochkina, N.; Bhaskar, M.; Yadav, S.P.; Pant, H.C. Phosphorylation, Dephosphorylation, and Multiprotein Assemblies Regulate Dynamic Behavior of Neuronal Cytoskeleton: A Mini-Review. Front. Mol. Neurosci. 2018, 11, 373. [Google Scholar] [CrossRef]
- Yamazaki, H.; Kosako, H.; Yoshimura, S.H. Quantitative proteomics indicate a strong correlation of mitotic phospho-/dephosphorylation with non-structured regions of substrates. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140295. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Shacter, E.; Chock, P.B.; Stadtman, E.R. Energy consumption in a cyclic phosphorylation/dephosphorylation cascade. J. Biol. Chem. 1984, 259, 12260–12264. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, X.; Tang, M.; Zhong, G.; Si, Y.; Li, H.; Zhu, F.; Liao, Q.; Li, L.; Zhao, J.; et al. A subcellular map of the human kinome. eLife 2021, 10, e64943. [Google Scholar] [CrossRef]
- Lee, J.; Park, Y.J.; Jung, H. Protein Kinases and Their Inhibitors in Pluripotent Stem Cell Fate Regulation. Stem Cells Int. 2019, 2019, 1569740. [Google Scholar] [CrossRef]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Rusin, S.F.; Adamo, M.E.; Kettenbach, A.N. Identification of Candidate Casein Kinase 2 Substrates in Mitosis by Quantitative Phosphoproteomics. Front. Cell Dev. Biol. 2017, 5, 97. [Google Scholar] [CrossRef]
- Pyerin, W. Human casein kinase II: Structures, genes, expression and requirement in cell growth stimulation. Adv. Enzym. Regul. 1994, 34, 225–246. [Google Scholar] [CrossRef]
- Tripodi, F.; Nicastro, R.; Busnelli, S.; Cirulli, C.; Maffioli, E.; Tedeschi, G.; Alberghina, L.; Coccetti, P. Protein kinase CK2 holoenzyme promotes start-specific transcription in Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Lolli, G.; Ranchio, A.; Battistutta, R. Active form of the protein kinase CK2 α2β2 holoenzyme is a strong complex with symmetric architecture. ACS Chem. Biol. 2014, 9, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, F.; Allende, C.C.; Allende, J.E. Protein kinase casein kinase 2 holoenzyme produced ectopically in human cells can be exported to the external side of the cellular membrane. Proc. Natl. Acad. Sci. USA 2005, 102, 4718–4723. [Google Scholar] [CrossRef] [PubMed]
- Bender, M.; Schwind, L.; Grundmann, D.; Martin, M.; Klotz, M.; Götz, C.; Montenarh, M.; Schäfer, K.H. Impact of protein kinase CK2 inhibitors on proliferation and differentiation of neural stem cells. Heliyon 2017, 3, e00318. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.S.; Jung, J.E.; Niizuma, K.; Chan, P.H. CK2 is a novel negative regulator of NADPH oxidase and a neuroprotectant in mice after cerebral ischemia. J. Neurosci. 2009, 29, 14779–14789. [Google Scholar] [CrossRef]
- Dominguez, I.; Sonenshein, G.E.; Seldin, D.C. Protein kinase CK2 in health and disease: CK2 and its role in Wnt and NF-kappaB signaling: Linking development and cancer. Cell Mol. Life Sci. 2009, 66, 1850–1857. [Google Scholar] [CrossRef]
- Halloran, D.; Durbano, H.W.; Nohe, A. Bone Morphogenetic Protein-2 in Development and Bone Homeostasis. J. Dev. Biol. 2020, 8, 19. [Google Scholar] [CrossRef]
- Bragdon, B.; Thinakaran, S.; Bonor, J.; Underhill, T.M.; Petersen, N.O.; Nohe, A. FRET reveals novel protein-receptor interaction of bone morphogenetic proteins receptors and adaptor protein 2 at the cell surface. Biophys. J. 2009, 97, 1428–1435. [Google Scholar] [CrossRef] [PubMed]
- Bragdon, B.; Thinakaran, S.; Moseychuk, O.; King, D.; Young, K.; Litchfield, D.W.; Petersen, N.O.; Nohe, A. Casein kinase 2 beta-subunit is a regulator of bone morphogenetic protein 2 signaling. Biophys. J. 2010, 99, 897–904. [Google Scholar] [CrossRef]
- Bragdon, B.; Thinakaran, S.; Moseychuk, O.; Gurski, L.; Bonor, J.; Price, C.; Wang, L.; Beamer, W.G.; Nohe, A. Casein kinase 2 regulates in vivo bone formation through its interaction with bone morphogenetic protein receptor type Ia. Bone 2011, 49, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Moseychuk, O.; Akkiraju, H.; Dutta, J.; D’Angelo, A.; Bragdon, B.; Duncan, R.L.; Nohe, A. Inhibition of CK2 binding to BMPRIa induces C2C12 differentiation into osteoblasts and adipocytes. J. Cell Commun. Signal. 2013, 7, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Chua, M.M.; Ortega, C.E.; Sheikh, A.; Lee, M.; Abdul-Rassoul, H.; Hartshorn, K.L.; Dominguez, I. CK2 in Cancer: Cellular and Biochemical Mechanisms and Potential Therapeutic Target. Pharmaceuticals 2017, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Drygin, D.; Streiner, N.; Chua, P.; Pierre, F.; O’Brien, S.E.; Bliesath, J.; Omori, M.; Huser, N.; Ho, C.; et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [Google Scholar] [CrossRef]
- Rosenberger, A.F.; Morrema, T.H.; Gerritsen, W.H.; van Haastert, E.S.; Snkhchyan, H.; Hilhorst, R.; Rozemuller, A.J.; Scheltens, P.; van der Vies, S.M.; Hoozemans, J.J. Increased occurrence of protein kinase CK2 in astrocytes in Alzheimer’s disease pathology. J. Neuroinflamm. 2016, 13, 4. [Google Scholar] [CrossRef]
- Schneider, H.R.; Reichert, G.H.; Issinger, O.G. Enhanced casein kinase II activity during mouse embryogenesis. Identification of a 110-kDa phosphoprotein as the major phosphorylation product in mouse embryos and Krebs II mouse ascites tumor cells. Eur. J. Biochem. 1986, 161, 733–738. [Google Scholar] [CrossRef]
- Perez, M.; Grande, J.; Itarte, E. Developmental changes in rat hepatic casein kinases 1 and 2. Eur. J. Biochem. 1987, 170, 493–498. [Google Scholar] [CrossRef]
- Maridor, G.; Park, W.; Krek, W.; Nigg, E.A. Casein kinase II. cDNA sequences, developmental expression, and tissue distribution of mRNAs for alpha, alpha’, and beta subunits of the chicken enzyme. J. Biol. Chem. 1991, 266, 2362–2368. [Google Scholar] [CrossRef]
- Nuthall, H.N.; Joachim, K.; Stifani, S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol. Cell Biol. 2004, 24, 8395–8407. [Google Scholar] [CrossRef]
- Lou, D.Y.; Dominguez, I.; Toselli, P.; Landesman-Bollag, E.; O’Brien, C.; Seldin, D.C. The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol. Cell Biol. 2008, 28, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Ma, Y.L.; Lee, E.H. Protein kinase CK2 impairs spatial memory formation through differential cross talk with PI-3 kinase signaling: Activation of Akt and inactivation of SGK1. J. Neurosci. 2007, 27, 6243–6248. [Google Scholar] [CrossRef] [PubMed]
- Mannowetz, N.; Kartarius, S.; Wennemuth, G.; Montenarh, M. Protein kinase CK2 and new binding partners during spermatogenesis. Cell Mol. Life Sci. 2010, 67, 3905–3913. [Google Scholar] [CrossRef] [PubMed]
- Escalier, D.; Silvius, D.; Xu, X. Spermatogenesis of mice lacking CK2alpha’: Failure of germ cell survival and characteristic modifications of the spermatid nucleus. Mol. Reprod. Dev. 2003, 66, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Schwind, L.; Wilhelm, N.; Kartarius, S.; Montenarh, M.; Gorjup, E.; Götz, C. Protein kinase CK2 is necessary for the adipogenic differentiation of human mesenchymal stem cells. Biochim. Biophys. Acta 2015, 1853, 2207–2216. [Google Scholar] [CrossRef]
- Kim, J.M.; Yang, Y.S.; Park, K.H.; Ge, X.; Xu, R.; Li, N.; Song, M.; Chun, H.; Bok, S.; Charles, J.F.; et al. A RUNX2 stabilization pathway mediates physiologic and pathologic bone formation. Nat. Commun. 2020, 11, 2289. [Google Scholar] [CrossRef] [PubMed]
- Snell, V.; Nurse, P. Genetic analysis of cell morphogenesis in fission yeast—A role for casein kinase II in the establishment of polarized growth. EMBO J. 1994, 13, 2066–2074. [Google Scholar] [CrossRef]
- Abi Nahed, R.; Reynaud, D.; Lemaitre, N.; Lartigue, S.; Roelants, C.; Vaiman, D.; Benharouga, M.; Cochet, C.; Filhol, O.; Alfaidy, N. Protein kinase CK2 contributes to placental development: Physiological and pathological implications. J. Mol. Med. 2020, 98, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yang, W.; Hong, H.; Yan, Z.; Qin, H.; Benveniste, E.N. Protein Kinase CK2 Regulates B Cell Development and Differentiation. J. Immunol. 2021, 207, 799–808. [Google Scholar] [CrossRef]
- Salizzato, V.; Zanin, S.; Borgo, C.; Lidron, E.; Salvi, M.; Rizzuto, R.; Pallafacchina, G.; Donella-Deana, A. Protein kinase CK2 subunits exert specific and coordinated functions in skeletal muscle differentiation and fusogenic activity. FASEB J. 2019, 33, 10648–10667. [Google Scholar] [CrossRef]
- Rebholz, H.; Nishi, A.; Liebscher, S.; Nairn, A.C.; Flajolet, M.; Greengard, P. CK2 negatively regulates Galphas signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 14096–14101. [Google Scholar] [CrossRef]
- Lettieri, A.; Borgo, C.; Zanieri, L.; D’Amore, C.; Oleari, R.; Paganoni, A.; Pinna, L.A.; Cariboni, A.; Salvi, M. Protein Kinase CK2 Subunits Differentially Perturb the Adhesion and Migration of GN11 Cells: A Model of Immature Migrating Neurons. Int. J. Mol. Sci. 2019, 20, 5951. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, E.; Vilardell, J.; Borgo, C.; Sarró, E.; Plana, M.; Marin, O.; Pinna, L.A.; Bayascas, J.R.; Meseguer, A.; Salvi, M.; et al. Effects of CK2β subunit down-regulation on Akt signalling in HK-2 renal cells. PLoS ONE 2020, 15, e0227340. [Google Scholar] [CrossRef] [PubMed]
- Cheusova, T.; Khan, M.A.; Schubert, S.W.; Gavin, A.C.; Buchou, T.; Jacob, G.; Sticht, H.; Allende, J.; Boldyreff, B.; Brenner, H.R.; et al. Casein kinase 2-dependent serine phosphorylation of MuSK regulates acetylcholine receptor aggregation at the neuromuscular junction. Genes Dev. 2006, 20, 1800–1816. [Google Scholar] [CrossRef] [PubMed]
- Kravic, B.; Harbauer, A.B.; Romanello, V.; Simeone, L.; Vögtle, F.N.; Kaiser, T.; Straubinger, M.; Huraskin, D.; Böttcher, M.; Cerqua, C.; et al. In mammalian skeletal muscle, phosphorylation of TOMM22 by protein kinase CSNK2/CK2 controls mitophagy. Autophagy 2018, 14, 311–335. [Google Scholar] [CrossRef]
- Pinna, L.A. Protein kinase CK2: A challenge to canons. J. Cell Sci. 2002, 115, 3873–3878. [Google Scholar] [CrossRef] [PubMed]
- Buchou, T.; Vernet, M.; Blond, O.; Jensen, H.H.; Pointu, H.; Olsen, B.B.; Cochet, C.; Issinger, O.G.; Boldyreff, B. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol. 2003, 23, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Deshiere, A.; Duchemin-Pelletier, E.; Spreux, E.; Ciais, D.; Combes, F.; Vandenbrouck, Y.; Couté, Y.; Mikaelian, I.; Giusiano, S.; Charpin, C.; et al. Unbalanced expression of CK2 kinase subunits is sufficient to drive epithelial-to-mesenchymal transition by Snail1 induction. Oncogene 2013, 32, 1373–1383. [Google Scholar] [CrossRef]
- Wang, S.; Jones, K.A. CK2 controls the recruitment of Wnt regulators to target genes in vivo. Curr. Biol. 2006, 16, 2239–2244. [Google Scholar] [CrossRef]
- Song, D.H.; Sussman, D.J.; Seldin, D.C. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J. Biol. Chem. 2000, 275, 23790–23797. [Google Scholar] [CrossRef]
- Miravet, S.; Piedra, J.; Miró, F.; Itarte, E.; García de Herreros, A.; Duñach, M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. J. Biol. Chem. 2002, 277, 1884–1891. [Google Scholar] [CrossRef] [PubMed]
- Pukrop, T.; Gradl, D.; Henningfeld, K.A.; Knochel, W.; Wedlich, D.; Kuhl, M. Identification of two regulatory elements within the high mobility group box transcription factor XTCF-4. J. Biol. Chem. 2001, 276, 8968–8978. [Google Scholar] [CrossRef]
- Willert, K.; Brink, M.; Wodarz, A.; Varmus, H.; Nusse, R. Casein kinase 2 associates with and phosphorylates dishevelled. EMBO J. 1997, 16, 3089–3096. [Google Scholar] [CrossRef]
- Homma, M.K.; Li, D.; Krebs, E.G.; Yuasa, Y.; Homma, Y. Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. USA 2002, 99, 5959–5964. [Google Scholar] [CrossRef]
- Song, D.H.; Dominguez, I.; Mizuno, J.; Kaut, M.; Mohr, S.C.; Seldin, D.C. CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J. Biol. Chem. 2003, 278, 24018–24025. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Symes, K.; Seldin, D.C.; Dominguez, I. Threonine 393 of beta-catenin regulates interaction with Axin. J. Cell Biochem. 2009, 108, 52–63. [Google Scholar] [CrossRef]
- Bragdon, B.; D’Angelo, A.; Gurski, L.; Bonor, J.; Schultz, K.L.; Beamer, W.G.; Rosen, C.J.; Nohe, A. Altered plasma membrane dynamics of bone morphogenetic protein receptor type Ia in a low bone mass mouse model. Bone 2012, 50, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Deng, C.; Li, Y.P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Durbano, H.W.; Halloran, D.; Nguyen, J.; Stone, V.; McTague, S.; Eskander, M.; Nohe, A. Aberrant BMP2 Signaling in Patients Diagnosed with Osteoporosis. Int. J. Mol. Sci. 2020, 21, 6909. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Puerto, M.C.; Iyengar, P.V.; García de Vinuesa, A.; Ten Dijke, P.; Sanchez-Duffhues, G. Bone morphogenetic protein receptor signal transduction in human disease. J. Pathol. 2019, 247, 9–20. [Google Scholar] [CrossRef]
- Hartung, A.; Bitton-Worms, K.; Rechtman, M.M.; Wenzel, V.; Boergermann, J.H.; Hassel, S.; Henis, Y.I.; Knaus, P. Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 2006, 26, 7791–7805. [Google Scholar] [CrossRef]
- Nohe, A.; Keating, E.; Knaus, P.; Petersen, N.O. Signal transduction of bone morphogenetic protein receptors. Cell. Signal. 2004, 16, 291–299. [Google Scholar] [CrossRef]
- Akkiraju, H.; Bonor, J.; Olli, K.; Bowen, C.; Bragdon, B.; Coombs, H.; Donahue, L.R.; Duncan, R.; Nohe, A. Systemic injection of CK2.3, a novel peptide acting downstream of bone morphogenetic protein receptor BMPRIa, leads to increased trabecular bone mass. J. Orthop. Res. 2015, 33, 208–215. [Google Scholar] [CrossRef]
- Akkiraju, H.; Bonor, J.; Nohe, A. CK2.1, a novel peptide, induces articular cartilage formation in vivo. J. Orthop. Res. 2017, 35, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Bragdon, B.; Moseychuk, O.; Saldanha, S.; King, D.; Julian, J.; Nohe, A. Bone morphogenetic proteins: A critical review. Cell. Signal. 2011, 23, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Halloran, D.; Vrathasha, V.; Durbano, H.W.; Nohe, A. Bone Morphogenetic Protein-2 Conjugated to Quantum Dot. Nanomaterials 2020, 10, 1208. [Google Scholar] [CrossRef] [PubMed]
- Heubel, B.; Nohe, A. The Role of BMP Signaling in Osteoclast Regulation. J. Dev. Biol. 2021, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.; Weidner, H.; Schell, L.M.; Sequeira, L.; Kabrick, R.; Dharmadhikari, S.; Coombs, H.; Duncan, R.L.; Wang, L.; Nohe, A. Synthetic Peptide CK2.3 Enhances Bone Mineral Density in Senile Mice. J. Bone Res. 2018, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.; Kelly, S.; Wood, R.; Heubel, B.; Nohe, A. A Synthetic Peptide, CK2.3, Inhibits RANKL-Induced Osteoclastogenesis through BMPRIa and ERK Signaling Pathway. J. Dev. Biol. 2020, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Vrathasha, V.; Weidner, H.; Nohe, A. Mechanism of CK2.3, a Novel Mimetic Peptide of Bone Morphogenetic Protein Receptor Type IA, Mediated Osteogenesis. Int. J. Mol. Sci. 2019, 20, 2500. [Google Scholar] [CrossRef]
- Weidner, H.; Yuan Gao, V.; Dibert, D.; McTague, S.; Eskander, M.; Duncan, R.; Wang, L.; Nohe, A. CK2.3, a Mimetic Peptide of the BMP Type I Receptor, Increases Activity in Osteoblasts over BMP2. Int. J. Mol. Sci. 2019, 20, 5877. [Google Scholar] [CrossRef]
- Huang, H.; Song, T.J.; Li, X.; Hu, L.; He, Q.; Liu, M.; Lane, M.D.; Tang, Q.Q. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 2009, 106, 12670–12675. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Komori, T.; Suda, T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr. Rev. 2000, 21, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Li, Q.; Lin, X.; Hu, N.; Liao, J.Y.; Lin, L.B.; Zhao, C.; Hu, Z.M.; Liang, X.; Xu, W.; et al. BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell Tissue Res. 2016, 366, 101–111. [Google Scholar] [CrossRef]
- Halloran, D.R.; Heubel, B.; MacMurray, C.; Root, D.; Eskander, M.; McTague, S.P.; Pelkey, H.; Nohe, A. Differentiation of Cells Isolated from Human Femoral Heads into Functional Osteoclasts. J. Dev. Biol. 2022, 10, 6. [Google Scholar] [CrossRef]
- Di Maira, G.; Salvi, M.; Arrigoni, G.; Marin, O.; Sarno, S.; Brustolon, F.; Pinna, L.A.; Ruzzene, M. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005, 12, 668–677. [Google Scholar] [CrossRef]
- Girardi, C.; James, P.; Zanin, S.; Pinna, L.A.; Ruzzene, M. Differential phosphorylation of Akt1 and Akt2 by protein kinase CK2 may account for isoform specific functions. Biochim. Biophys. Acta 2014, 1843, 1865–1874. [Google Scholar] [CrossRef]
- Gibson, S.A.; Yang, W.; Yan, Z.; Liu, Y.; Rowse, A.L.; Weinmann, A.S.; Qin, H.; Benveniste, E.N. Protein Kinase CK2 Controls the Fate between Th17 Cell and Regulatory T Cell Differentiation. J. Immunol. 2017, 198, 4244–4254. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.; Chuderland, D.; Karamansha, Y.; Livnah, O.; Seger, R. Nuclear extracellular signal-regulated kinase 1 and 2 translocation is mediated by casein kinase 2 and accelerated by autophosphorylation. Mol. Cell. Biol. 2011, 31, 3515–3530. [Google Scholar] [CrossRef]
- Plotnikov, A.; Chuderland, D.; Karamansha, Y.; Livnah, O.; Seger, R. Nuclear ERK Translocation is Mediated by Protein Kinase CK2 and Accelerated by Autophosphorylation. Cell. Physiol. Biochem. 2019, 53, 366–387. [Google Scholar] [CrossRef]
- Whitmarsh, A.J. Casein kinase 2 sends extracellular signal-regulated kinase nuclear. Mol. Cell. Biol. 2011, 31, 3512–3514. [Google Scholar] [CrossRef] [PubMed]
- James, B.P.; Bunch, T.A.; Krishnamoorthy, S.; Perkins, L.A.; Brower, D.L. Nuclear localization of the ERK MAP kinase mediated by Drosophila alphaPS2betaPS integrin and importin-7. Mol. Biol. Cell 2007, 18, 4190–4199. [Google Scholar] [CrossRef] [PubMed]
- Maik-Rachline, G.; Hacohen-Lev-Ran, A.; Seger, R. Nuclear ERK: Mechanism of Translocation, Substrates, and Role in Cancer. Int. J. Mol. Sci. 2019, 20, 1194. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yeh, J.; Van Waes, C. Protein kinase casein kinase 2 mediates inhibitor-kappaB kinase and aberrant nuclear factor-kappaB activation by serum factor(s) in head and neck squamous carcinoma cells. Cancer Res. 2006, 66, 6722–6731. [Google Scholar] [CrossRef]
- Wang, D.; Baldwin, A.S. Activation of nuclear factor-kappaB-dependent transcription by tumor necrosis factor-alpha is mediated through phosphorylation of RelA/p65 on serine 529. J. Biol. Chem. 1998, 273, 29411–29416. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Westerheide, S.D.; Hanson, J.L.; Baldwin, A.S. Tumor necrosis factor alpha-induced phosphorylation of RelA/p65 on Ser529 is controlled by casein kinase II. J. Biol. Chem. 2000, 275, 32592–32597. [Google Scholar] [CrossRef]
- Kato, T.; Delhase, M.; Hoffmann, A.; Karin, M. CK2 Is a C-Terminal IkappaB Kinase Responsible for NF-kappaB Activation during the UV Response. Mol. Cell 2003, 12, 829–839. [Google Scholar] [CrossRef]
- Romieu-Mourez, R.; Landesman-Bollag, E.; Seldin, D.C.; Sonenshein, G.E. Protein kinase CK2 promotes aberrant activation of nuclear factor-kappaB, transformed phenotype, and survival of breast cancer cells. Cancer Res. 2002, 62, 6770–6778. [Google Scholar]
- Schwarz, E.M.; Van Antwerp, D.; Verma, I.M. Constitutive phosphorylation of IkappaBalpha by casein kinase II occurs preferentially at serine 293: Requirement for degradation of free IkappaBalpha. Mol. Cell. Biol. 1996, 16, 3554–3559. [Google Scholar] [CrossRef][Green Version]
- McElhinny, J.A.; Trushin, S.A.; Bren, G.D.; Chester, N.; Paya, C.V. Casein kinase II phosphorylates I kappa B alpha at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol. Cell. Biol. 1996, 16, 899–906. [Google Scholar] [CrossRef]
- Zheng, Y.; Qin, H.; Frank, S.J.; Deng, L.; Litchfield, D.W.; Tefferi, A.; Pardanani, A.; Lin, F.T.; Li, J.; Sha, B.; et al. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathway. Blood 2011, 118, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Zeinalzadeh, E.; Valerievich Yumashev, A.; Rahman, H.S.; Marofi, F.; Shomali, N.; Kafil, H.S.; Solali, S.; Sajjadi-Dokht, M.; Vakili-Samiani, S.; Jarahian, M.; et al. The Role of Janus Kinase/STAT3 Pathway in Hematologic Malignancies With an Emphasis on Epigenetics. Front. Genet. 2021, 12, 703883. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, U.; Kasembeli, M.M.; Robinson, P.; Tweardy, D.J. Targeting Janus Kinases and Signal Transducer and Activator of Transcription 3 to Treat Inflammation, Fibrosis, and Cancer: Rationale, Progress, and Caution. Pharmacol. Rev. 2020, 72, 486–526. [Google Scholar] [CrossRef] [PubMed]
- Aparicio-Siegmund, S.; Sommer, J.; Monhasery, N.; Schwanbeck, R.; Keil, E.; Finkenstädt, D.; Pfeffer, K.; Rose-John, S.; Scheller, J.; Garbers, C. Inhibition of protein kinase II (CK2) prevents induced signal transducer and activator of transcription (STAT) 1/3 and constitutive STAT3 activation. Oncotarget 2014, 5, 2131–2148. [Google Scholar] [CrossRef]
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun. Signal. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Husain, K.; Williamson, T.T.; Nelson, N.; Ghansah, T. Protein kinase 2 (CK2): A potential regulator of immune cell development and function in cancer. Immunol. Med. 2021, 44, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Song, C.; Ding, Y.; Tan, B.H.; Desai, D.; Sharma, A.; Gowda, R.; Yue, F.; Huang, S.; Spiegelman, V.; et al. Dual targeting of MTOR as a novel therapeutic approach for high-risk B-cell acute lymphoblastic leukemia. Leukemia 2021, 35, 1267–1278. [Google Scholar] [CrossRef]
- Dhanyamraju, P.K.; Iyer, S.; Smink, G.; Bamme, Y.; Bhadauria, P.; Payne, J.L.; Dovat, E.; Klink, M.; Ding, Y. Transcriptional Regulation of Genes by Ikaros Tumor Suppressor in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1377. [Google Scholar] [CrossRef] [PubMed]
- Dovat, E.; Song, C.; Hu, T.; Rahman, M.A.; Dhanyamraju, P.K.; Klink, M.; Bogush, D.; Soliman, M.; Kane, S.; McGrath, M.; et al. Transcriptional Regulation of PIK3CD and PIKFYVE in T-Cell Acute Lymphoblastic Leukemia by IKAROS and Protein Kinase CK2. Int. J. Mol. Sci. 2021, 22, 819. [Google Scholar] [CrossRef]
- Gowda, C.; Song, C.; Kapadia, M.; Payne, J.L.; Hu, T.; Ding, Y.; Dovat, S. Regulation of cellular proliferation in acute lymphoblastic leukemia by Casein Kinase II (CK2) and Ikaros. Adv. Biol. Regul. 2017, 63, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Ge, Z.; Ding, Y.; Tan, B.H.; Desai, D.; Gowda, K.; Amin, S.; Gowda, R.; Robertson, G.P.; Yue, F.; et al. IKAROS and CK2 regulate expression of BCL-XL and chemosensitivity in high-risk B-cell acute lymphoblastic leukemia. Blood 2020, 136, 1520–1534. [Google Scholar] [CrossRef]
- Olsen, B.B.; Svenstrup, T.H.; Guerra, B. Downregulation of protein kinase CK2 induces autophagic cell death through modulation of the mTOR and MAPK signaling pathways in human glioblastoma cells. Int. J. Oncol. 2012, 41, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Silva-Pavez, E.; Tapia, J.C. Protein Kinase CK2 in Cancer Energetics. Front. Oncol. 2020, 10, 893. [Google Scholar] [CrossRef] [PubMed]
- Rowse, A.L.; Gibson, S.A.; Meares, G.P.; Rajbhandari, R.; Nozell, S.E.; Dees, K.J.; Hjelmeland, A.B.; McFarland, B.C.; Benveniste, E.N. Protein kinase CK2 is important for the function of glioblastoma brain tumor initiating cells. J. Neurooncol. 2017, 132, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; Ruzzene, M. Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Cancer Res. 2019, 38, 287. [Google Scholar] [CrossRef] [PubMed]
- Rabalski, A.J.; Gyenis, L.; Litchfield, D.W. Molecular Pathways: Emergence of Protein Kinase CK2 (CSNK2) as a Potential Target to Inhibit Survival and DNA Damage Response and Repair Pathways in Cancer Cells. Clin. Cancer Res. 2016, 22, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; D’Amore, C.; Sarno, S.; Salvi, M.; Ruzzene, M. Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021, 6, 183. [Google Scholar] [CrossRef]
- Loizou, J.I.; El-Khamisy, S.F.; Zlatanou, A.; Moore, D.J.; Chan, D.W.; Qin, J.; Sarno, S.; Meggio, F.; Pinna, L.A.; Caldecott, K.W. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 2004, 117, 17–28. [Google Scholar] [CrossRef]
- Olsen, B.B.; Wang, S.Y.; Svenstrup, T.H.; Chen, B.P.; Guerra, B. Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage. BMC Mol. Biol. 2012, 13, 7. [Google Scholar] [CrossRef]
- Yacoub, A.; Kelley, M.R.; Deutsch, W.A. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res. 1997, 57, 5457–5459. [Google Scholar] [PubMed]
- Taylor, K.M.; Hiscox, S.; Nicholson, R.I.; Hogstrand, C.; Kille, P. Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7. Sci. Signal. 2012, 5, ra11. [Google Scholar] [CrossRef]
- Bafaro, E.; Liu, Y.; Xu, Y.; Dempski, R.E. The emerging role of zinc transporters in cellular homeostasis and cancer. Signal Transduct. Target. Ther. 2017, 2, 17029. [Google Scholar] [CrossRef]
- Taylor, K.M.; Vichova, P.; Jordan, N.; Hiscox, S.; Hendley, R.; Nicholson, R.I. ZIP7-mediated intracellular zinc transport contributes to aberrant growth factor signaling in antihormone-resistant breast cancer Cells. Endocrinology 2008, 149, 4912–4920. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Guerra, B.; Ermakowa, I.; Issinger, O.G. Crystal structure of human protein kinase CK2: Insights into basic properties of the CK2 holoenzyme. EMBO J. 2001, 20, 5320–5331. [Google Scholar] [CrossRef]
- Thornburg, W.; Lindell, T.J. Purification of rat liver nuclear protein kinase NII. J. Biol. Chem. 1977, 252, 6660–6665. [Google Scholar] [CrossRef]
- Filhol, O.; Martiel, J.L.; Cochet, C. Protein kinase CK2: A new view of an old molecular complex. EMBO Rep. 2004, 5, 351–355. [Google Scholar] [CrossRef]
- Gratz, A.; Bollacke, A.; Stephan, S.; Nienberg, C.; Le Borgne, M.; Götz, C.; Jose, J. Functional display of heterotetrameric human protein kinase CK2 on Escherichia coli: A novel tool for drug discovery. Microb. Cell Fact. 2015, 14, 74. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.K.; Son, S.H.; Jin, B.S.; Na, J.H.; Kim, S.Y.; Kim, K.H.; Kim, E.E.; Yu, Y.G.; Lee, H.H. Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc. Natl. Acad. Sci. USA 2013, 110, 19360–19365. [Google Scholar] [CrossRef]
- Gietz, R.D.; Graham, K.C.; Litchfield, D.W. Interactions between the subunits of casein kinase II. J. Biol. Chem. 1995, 270, 13017–13021. [Google Scholar] [CrossRef]
- Chester, N.; Yu, I.J.; Marshak, D.R. Identification and characterization of protein kinase CKII isoforms in HeLa cells. Isoform-specific differences in rates of assembly from catalytic and regulatory subunits. J. Biol. Chem. 1995, 270, 7501–7514. [Google Scholar] [CrossRef] [PubMed]
- Vilk, G.; Weber, J.E.; Turowec, J.P.; Duncan, J.S.; Wu, C.; Derksen, D.R.; Zien, P.; Sarno, S.; Donella-Deana, A.; Lajoie, G.; et al. Protein kinase CK2 catalyzes tyrosine phosphorylation in mammalian cells. Cell. Signal. 2008, 20, 1942–1951. [Google Scholar] [CrossRef]
- Meek, D.W.; Simon, S.; Kikkawa, U.; Eckhart, W. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990, 9, 3253–3260. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.K.; Dhillon, N.; Thorner, J.; Martin, G.S. Casein kinase II catalyzes tyrosine phosphorylation of the yeast nucleolar immunophilin Fpr3. J. Biol. Chem. 1997, 272, 12961–12967. [Google Scholar] [CrossRef]
- Liu, D.; Marie, J.C.; Pelletier, A.L.; Song, Z.; Ben-Khemis, M.; Boudiaf, K.; Pintard, C.; Leger, T.; Terrier, S.; Chevreux, G.; et al. Protein Kinase CK2 Acts as a Molecular Brake to Control NADPH Oxidase 1 Activation and Colon Inflammation. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1073–1093. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Götz, C.; Montenarh, M.; Philipp, S.E. Control of TRPM3 Ion Channels by Protein Kinase CK2-Mediated Phosphorylation in Pancreatic β-Cells of the Line INS-1. Int. J. Mol. Sci. 2021, 22, 13133. [Google Scholar] [CrossRef] [PubMed]
- Trembley, J.H.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. Protein kinase CK2 in health and disease: CK2: A key player in cancer biology. Cell. Mol. Life Sci. 2009, 66, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Turowec, J.P.; Vilk, G.; Gabriel, M.; Litchfield, D.W. Characterizing the convergence of protein kinase CK2 and caspase-3 reveals isoform-specific phosphorylation of caspase-3 by CK2α’: Implications for pathological roles of CK2 in promoting cancer cell survival. Oncotarget 2013, 4, 560–571. [Google Scholar] [CrossRef]
- Chua, M.M.J.; Lee, M.; Dominguez, I. Cancer-type dependent expression of CK2 transcripts. PLoS ONE 2017, 12, e0188854. [Google Scholar] [CrossRef]
- Faust, M.; Montenarh, M. Subcellular localization of protein kinase CK2. A key to its function? Cell Tissue Res. 2000, 301, 329–340. [Google Scholar] [CrossRef]
- Götz, C.; Montenarh, M. Protein kinase CK2 in development and differentiation. Biomed. Rep. 2017, 6, 127–133. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Roffey, S.E.; Litchfield, D.W. CK2 Regulation: Perspectives in 2021. Biomedicines 2021, 9, 1361. [Google Scholar] [CrossRef]
- Robitzki, A.; Bodenbach, L.; Voss, H.; Pyerin, W. Human casein kinase II. The subunit alpha protein activates transcription of the subunit beta gene. J. Biol. Chem. 1993, 268, 5694–5702. [Google Scholar] [CrossRef]
- Lee, Y.H.; Yuk, H.J.; Park, K.H.; Bae, Y.S. Coumestrol induces senescence through protein kinase CKII inhibition-mediated reactive oxygen species production in human breast cancer and colon cancer cells. Food Chem. 2013, 141, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, A.; Castellvi, J.; Artero-Castro, A.; Leal, J.A.; Romagosa, C.; Hernández-Losa, J.; Peg, V.; Fabra, A.; Vidal, F.; Kondoh, H.; et al. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS ONE 2013, 8, e76247. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lee, Y.H.; Bae, Y.S. MiR-186, miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting α subunit of protein kinase CKII in human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2012, 429, 173–179. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, H.S.; Kim, N.; Shin, C.M.; Lee, S.H.; Park, Y.S.; Hwang, J.H.; Kim, J.W.; Jeong, S.H.; Lee, D.H.; et al. Prevalence and clinicopathologic characteristics of gastric cardia cancer in South Korea. Helicobacter 2012, 17, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Bae, Y.S. Long Non-Coding RNA. Int. J. Mol. Sci. 2022, 23, 1888. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Rho, J.H.; Lee, S.Y.; Yoo, S.H.; Kim, H.Y.; Chung, W.T.; Yoo, Y.H. Alpha B-Crystallin Protects Rat Articular Chondrocytes against Casein Kinase II Inhibition-Induced Apoptosis. PLoS ONE 2016, 11, e0166450. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Yamagishi, N.; Hatayama, T. Protein kinase CK2 phosphorylates Hsp105 alpha at Ser509 and modulates its function. Biochem. J. 2003, 371, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Filhol, O.; Cochet, C. Protein kinase CK2 in health and disease: Cellular functions of protein kinase CK2: A dynamic affair. Cell. Mol. Life Sci. 2009, 66, 1830–1839. [Google Scholar] [CrossRef]
- Li, S.; Chen, P.; Yang, Q. Denosumab versus zoledronic acid in cases of surgically unsalvageable giant cell tumor of bone: A randomized clinical trial. J. Bone Oncol. 2019, 15, 100217. [Google Scholar] [CrossRef]
- Li, Q.; Li, K.; Zhang, S.; Zhou, Y.; Hong, J.; Zhou, X.; Li, Z.; Wu, B.; Wu, G.; Meng, R. The effect of ionizing radiation on the subcellular localization and kinase activity of protein kinase CK2 in human non-small cell lung cancer cells. Int. J. Radiat. Biol. 2019, 95, 1462–1471. [Google Scholar] [CrossRef]
- Pluemsampant, S.; Safronova, O.S.; Nakahama, K.; Morita, I. Protein kinase CK2 is a key activator of histone deacetylase in hypoxia-associated tumors. Int. J. Cancer 2008, 122, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Louvet, E.; Junéra, H.R.; Berthuy, I.; Hernandez-Verdun, D. Compartmentation of the nucleolar processing proteins in the granular component is a CK2-driven process. Mol. Biol. Cell 2006, 17, 2537–2546. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, G.; Pan, Y.; Ahmad, K.A.; Ahmed, K. Protein B23/nucleophosmin/numatrin nuclear dynamics in relation to protein kinase CK2 and apoptotic activity in prostate cells. Biochemistry 2010, 49, 3842–3852. [Google Scholar] [CrossRef] [PubMed]
- Nuñez de Villavicencio-Diaz, T.; Rabalski, A.J.; Litchfield, D.W. Protein Kinase CK2: Intricate Relationships within Regulatory Cellular Networks. Pharmaceuticals 2017, 10, 27. [Google Scholar] [CrossRef]
- Daya-Makin, M.; Sanghera, J.S.; Mogentale, T.L.; Lipp, M.; Parchomchuk, J.; Hogg, J.C.; Pelech, S.L. Activation of a tumor-associated protein kinase (p40TAK) and casein kinase 2 in human squamous cell carcinomas and adenocarcinomas of the lung. Cancer Res. 1994, 54, 2262–2268. [Google Scholar]
- Faust, R.A.; Gapany, M.; Tristani, P.; Davis, A.; Adams, G.L.; Ahmed, K. Elevated protein kinase CK2 activity in chromatin of head and neck tumors: Association with malignant transformation. Cancer Lett. 1996, 101, 31–35. [Google Scholar] [CrossRef]
- Ahmad, K.A.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. Protein kinase CK2—A key suppressor of apoptosis. Adv. Enzym. Regul. 2008, 48, 179–187. [Google Scholar] [CrossRef]
- Beck, M.; Schmidt, A.; Malmstroem, J.; Claassen, M.; Ori, A.; Szymborska, A.; Herzog, F.; Rinner, O.; Ellenberg, J.; Aebersold, R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011, 7, 549. [Google Scholar] [CrossRef]
- Lundberg, E.; Fagerberg, L.; Klevebring, D.; Matic, I.; Geiger, T.; Cox, J.; Algenäs, C.; Lundeberg, J.; Mann, M.; Uhlen, M. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 2010, 6, 450. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Wehner, A.; Schaab, C.; Cox, J.; Mann, M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteom. 2012, 11, M111.014050. [Google Scholar] [CrossRef] [PubMed]
- Homma, M.K.; Kiko, Y.; Hashimoto, Y.; Nagatsuka, M.; Katagata, N.; Masui, S.; Homma, Y.; Nomizu, T. Intracellular localization of CK2α as a prognostic factor in invasive breast carcinomas. Cancer Sci. 2021, 112, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Ren, Y.; Wu, K.; Tian, X.; Wen, F.; Liu, D.; Fan, Y.; Zhao, S. SOX2-Upregulated microRNA-30e Promotes the Progression of Esophageal Cancer via Regulation of the USP4/SMAD4/CK2 Axis. Mol. Ther. Nucleic Acids 2021, 23, 200–214. [Google Scholar] [CrossRef]
- Kim, J.S.; Eom, J.I.; Cheong, J.W.; Choi, A.J.; Lee, J.K.; Yang, W.I.; Min, Y.H. Protein kinase CK2alpha as an unfavorable prognostic marker and novel therapeutic target in acute myeloid leukemia. Clin. Cancer Res. 2007, 13, 1019–1028. [Google Scholar] [CrossRef]
- Zhang, H.X.; Jiang, S.S.; Zhang, X.F.; Zhou, Z.Q.; Pan, Q.Z.; Chen, C.L.; Zhao, J.J.; Tang, Y.; Xia, J.C.; Weng, D.S. Protein kinase CK2α catalytic subunit is overexpressed and serves as an unfavorable prognostic marker in primary hepatocellular carcinoma. Oncotarget 2015, 6, 34800–34817. [Google Scholar] [CrossRef]
- Lin, K.Y.; Fang, C.L.; Chen, Y.; Li, C.F.; Chen, S.H.; Kuo, C.Y.; Tai, C.; Uen, Y.H. Overexpression of nuclear protein kinase CK2 Beta subunit and prognosis in human gastric carcinoma. Ann. Surg. Oncol. 2010, 17, 1695–1702. [Google Scholar] [CrossRef]
- Gapany, M.; Faust, R.A.; Tawfic, S.; Davis, A.; Adams, G.L.; Ahmed, K. Association of elevated protein kinase CK2 activity with aggressive behavior of squamous cell carcinoma of the head and neck. Mol. Med. 1995, 1, 659–666. [Google Scholar] [CrossRef]
- Rabjerg, M.; Guerra, B.; Oliván-Viguera, A.; Mikkelsen, M.L.; Köhler, R.; Issinger, O.G.; Marcussen, N. Nuclear localization of the CK2α-subunit correlates with poor prognosis in clear cell renal cell carcinoma. Oncotarget 2017, 8, 1613–1627. [Google Scholar] [CrossRef]
- Lin, K.Y.; Tai, C.; Hsu, J.C.; Li, C.F.; Fang, C.L.; Lai, H.C.; Hseu, Y.C.; Lin, Y.F.; Uen, Y.H. Overexpression of nuclear protein kinase CK2 α catalytic subunit (CK2α) as a poor prognosticator in human colorectal cancer. PLoS ONE 2011, 6, e17193. [Google Scholar] [CrossRef]
- Giusiano, S.; Cochet, C.; Filhol, O.; Duchemin-Pelletier, E.; Secq, V.; Bonnier, P.; Carcopino, X.; Boubli, L.; Birnbaum, D.; Garcia, S.; et al. Protein kinase CK2α subunit over-expression correlates with metastatic risk in breast carcinomas: Quantitative immunohistochemistry in tissue microarrays. Eur. J. Cancer 2011, 47, 792–801. [Google Scholar] [CrossRef]
- Ruzzene, M.; Pinna, L.A. Addiction to protein kinase CK2: A common denominator of diverse cancer cells? Biochim. Biophys. Acta 2010, 1804, 499–504. [Google Scholar] [CrossRef]
- Broséus, J.; Chen, G.; Hergalant, S.; Ramstein, G.; Mounier, N.; Guéant, J.L.; Feugier, P.; Gisselbrecht, C.; Thieblemont, C.; Houlgatte, R. Relapsed diffuse large B-cell lymphoma present different genomic profiles between early and late relapses. Oncotarget 2016, 7, 83987–84002. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lu, Q.; Wang, Z.; Cai, P.; Zeng, Z.; Zhang, L.; Wang, M.; Ma, L.; Ruan, C.; Chen, S. Identification of a Novel CSNK2A1-PDGFRB Fusion Gene in a Patient with Myeloid Neoplasm with Eosinophilia. Cancer Res. Treat. 2021, 53, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Mandato, E.; Manni, S.; Zaffino, F.; Semenzato, G.; Piazza, F. Targeting CK2-driven non-oncogene addiction in B-cell tumors. Oncogene 2016, 35, 6045–6052. [Google Scholar] [CrossRef]
- Zhang, H.; Kong, Q.; Wang, J.; Jiang, Y.; Hua, H. Complex roles of cAMP-PKA-CREB signaling in cancer. Exp. Hematol. Oncol. 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Boldyreff, B.; Sarno, S.; Cesaro, L.; Issinger, O.G.; Pinna, L.A. CK2: A protein kinase in need of control. Pharmacol. Ther. 1999, 82, 303–313. [Google Scholar] [CrossRef]
- Torres, J.; Pulido, R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001, 276, 993–998. [Google Scholar] [CrossRef]
- Quotti Tubi, L.; Canovas Nunes, S.; Brancalion, A.; Doriguzzi Breatta, E.; Manni, S.; Mandato, E.; Zaffino, F.; Macaccaro, P.; Carrino, M.; Gianesin, K.; et al. Protein kinase CK2 regulates AKT, NF-κB and STAT3 activation, stem cell viability and proliferation in acute myeloid leukemia. Leukemia 2017, 31, 292–300. [Google Scholar] [CrossRef]
- Battistutta, R. Protein kinase CK2 in health and disease: Structural bases of protein kinase CK2 inhibition. Cell. Mol. Life Sci. 2009, 66, 1868–1889. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Ruiz de Azua, I.; Barella, L.F.; Sakamoto, W.; Zhu, L.; Cui, Y.; Lu, H.; Rebholz, H.; Matschinsky, F.M.; Doliba, N.M.; et al. CK2 acts as a potent negative regulator of receptor-mediated insulin release in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2015, 112, E6818–E6824. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.E.; Kwon, S.; Seok, S.; Xiao, Z.; Lee, K.W.; Kang, Y.; Li, X.; Shinoda, K.; Kajimura, S.; Kemper, B.; et al. Obesity-Linked Phosphorylation of SIRT1 by Casein Kinase 2 Inhibits Its Nuclear Localization and Promotes Fatty Liver. Mol. Cell. Biol. 2017, 37, e00006-17. [Google Scholar] [CrossRef] [PubMed]
- Hauck, L.; Harms, C.; An, J.; Rohne, J.; Gertz, K.; Dietz, R.; Endres, M.; von Harsdorf, R. Protein kinase CK2 links extracellular growth factor signaling with the control of p27(Kip1) stability in the heart. Nat. Med. 2008, 14, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Eom, G.H.; Cho, Y.K.; Ko, J.H.; Shin, S.; Choe, N.; Kim, Y.; Joung, H.; Kim, H.S.; Nam, K.I.; Kee, H.J.; et al. Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation 2011, 123, 2392–2403. [Google Scholar] [CrossRef] [PubMed]
- Stachowski, M.J.; Holewinski, R.J.; Grote, E.; Venkatraman, V.; Van Eyk, J.E.; Kirk, J.A. Phospho-Proteomic Analysis of Cardiac Dyssynchrony and Resynchronization Therapy. Proteomics 2018, 18, e1800079. [Google Scholar] [CrossRef]
- Cortés, M.; Malave, L.; Castello, J.; Flajolet, M.; Cenci, M.A.; Friedman, E.; Rebholz, H. CK2 Oppositely Modulates l-DOPA-Induced Dyskinesia via Striatal Projection Neurons Expressing D1 or D2 Receptors. J. Neurosci. 2017, 37, 11930–11946. [Google Scholar] [CrossRef]
- Perez, D.I.; Gil, C.; Martinez, A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med. Res. Rev. 2011, 31, 924–954. [Google Scholar] [CrossRef]
- Zhang, Q.; Xia, Y.; Wang, Y.; Shentu, Y.; Zeng, K.; Mahaman, Y.A.R.; Huang, F.; Wu, M.; Ke, D.; Wang, Q.; et al. CK2 Phosphorylating I. Front. Mol. Neurosci. 2018, 11, 146. [Google Scholar] [CrossRef]
- Ryu, M.Y.; Kim, D.W.; Arima, K.; Mouradian, M.M.; Kim, S.U.; Lee, G. Localization of CKII beta subunits in Lewy bodies of Parkinson’s disease. J. Neurol. Sci. 2008, 266, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Rebholz, H.; Zhou, M.; Nairn, A.C.; Greengard, P.; Flajolet, M. Selective knockout of the casein kinase 2 in d1 medium spiny neurons controls dopaminergic function. Biol. Psychiatry 2013, 74, 113–121. [Google Scholar] [CrossRef]
- Kajiwara, Y.; Buxbaum, J.D.; Grice, D.E. SLITRK1 binds 14-3-3 and regulates neurite outgrowth in a phosphorylation-dependent manner. Biol. Psychiatry 2009, 66, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712. [Google Scholar] [CrossRef] [PubMed]
- Secci, E.; Luchinat, E.; Banci, L. The Casein Kinase 2-Dependent Phosphorylation of NS5A Domain 3 from Hepatitis C Virus Followed by Time-Resolved NMR Spectroscopy. ChemBioChem 2016, 17, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhang, Z.H.; Huang, M.; Heise, T.; Zhang, J.; Liu, G.L. Phosphorylation of human La protein at Ser366 by casein kinase II contributes to hepatitis B virus replication and expression in vitro. J. Viral Hepat. 2013, 20, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Rane, N.S.; Yonkovich, J.L.; Hegde, R.S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004, 23, 4550–4559. [Google Scholar] [CrossRef]
- Zamponi, E.; Buratti, F.; Cataldi, G.; Caicedo, H.H.; Song, Y.; Jungbauer, L.M.; LaDu, M.J.; Bisbal, M.; Lorenzo, A.; Ma, J.; et al. Prion protein inhibits fast axonal transport through a mechanism involving casein kinase 2. PLoS ONE 2017, 12, e0188340. [Google Scholar] [CrossRef]
- Rane, N.S.; Kang, S.W.; Chakrabarti, O.; Feigenbaum, L.; Hegde, R.S. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev. Cell 2008, 15, 359–370. [Google Scholar] [CrossRef]
- Larson, S.R.; Bortell, N.; Illies, A.; Crisler, W.J.; Matsuda, J.L.; Lenz, L.L. Myeloid Cell CK2 Regulates Inflammation and Resistance to Bacterial Infection. Front. Immunol. 2020, 11, 590266. [Google Scholar] [CrossRef]
- Dong, G.; Yang, Y.; Zhang, H.; Yu, W.; He, H.; Dai, F.; Ma, C.; Wang, Y.; Zhu, F.; Xiong, H.; et al. Protein Kinase CK2 Maintains Reciprocal Balance Between Th17 and Treg Cells in the Pathogenesis of UC. Inflamm. Bowel Dis. 2022, 28, 830–842. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, J.H.; Park, J.W.; Kim, D.Y.; Hahm, J.H.; Nam, H.G.; Bae, Y.S. Downregulation of protein kinase CK2 activity induces age-related biomarkers in C. elegans. Oncotarget 2017, 8, 36950–36963. [Google Scholar] [CrossRef]
- Ryu, S.W.; Woo, J.H.; Kim, Y.H.; Lee, Y.S.; Park, J.W.; Bae, Y.S. Downregulation of protein kinase CKII is associated with cellular senescence. FEBS Lett. 2006, 580, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Kim, J.J.; Bae, Y.S. CK2 downregulation induces senescence-associated heterochromatic foci formation through activating SUV39h1 and inactivating G9a. Biochem. Biophys. Res. Commun. 2018, 505, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Bae, Y.S. Dephosphorylation of p53 Ser 392 Enhances Trimethylation of Histone H3 Lys 9 via SUV39h1 Stabilization in CK2 Downregulation-Mediated Senescence. Mol. Cells 2019, 42, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Bae, Y.S. Downregulation of JMJD2a and LSD1 is involved in CK2 inhibition-mediated cellular senescence through the p53-SUV39h1 pathway. BMB Rep. 2022, 55, 92–97. [Google Scholar] [CrossRef]
- Ferguson, A.D.; Sheth, P.R.; Basso, A.D.; Paliwal, S.; Gray, K.; Fischmann, T.O.; Le, H.V. Structural basis of CX-4945 binding to human protein kinase CK2. FEBS Lett. 2011, 585, 104–110. [Google Scholar] [CrossRef]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol. Cell. Biochem. 2011, 356, 37–43. [Google Scholar] [CrossRef]
- Chon, H.J.; Bae, K.J.; Lee, Y.; Kim, J. The casein kinase 2 inhibitor, CX-4945, as an anti-cancer drug in treatment of human hematological malignancies. Front. Pharmacol. 2015, 6, 70. [Google Scholar] [CrossRef]
- Lee, J.Y.; Yun, J.S.; Kim, W.K.; Chun, H.S.; Jin, H.; Cho, S.; Chang, J.H. Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945. BiomMed Res. Int. 2019, 2019, 6125068. [Google Scholar] [CrossRef]
- Son, Y.H.; Moon, S.H.; Kim, J. The protein kinase 2 inhibitor CX-4945 regulates osteoclast and osteoblast differentiation in vitro. Mol. Cells 2013, 36, 417–423. [Google Scholar] [CrossRef]
- Richter, A.; Sender, S.; Lenz, A.; Schwarz, R.; Hinz, B.; Knuebel, G.; Sekora, A.; Murua Escobar, H.; Junghanss, C.; Roolf, C. Influence of Casein kinase II inhibitor CX-4945 on BCL6-mediated apoptotic signaling in B-ALL in vitro and in vivo. BMC Cancer 2020, 20, 184. [Google Scholar] [CrossRef]
- Kim, H.M.; Jeong, I.; Kim, H.J.; Kang, S.K.; Kwon, W.S.; Kim, T.S.; Park, K.H.; Jung, M.; Soong, J.; Lin, S.C.; et al. Casein Kinase 2 Inhibitor, CX-4945, as a Potential Targeted Anticancer Agent in Gastric Cancer. Anticancer Res. 2018, 38, 6171–6180. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.R.; Lúcio, P.; Melão, A.; Antunes, I.; Cardoso, B.A.; Stansfield, R.; Bertilaccio, M.T.; Ghia, P.; Drygin, D.; Silva, M.G.; et al. Activity of the clinical-stage CK2-specific inhibitor CX-4945 against chronic lymphocytic leukemia. Leukemia 2014, 28, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Perea, S.E.; Baladrón, I.; Valenzuela, C.; Perera, Y. CIGB-300: A peptide-based drug that impairs the Protein Kinase CK2-mediated phosphorylation. Semin. Oncol. 2018, 45, 58–67. [Google Scholar] [CrossRef]
- Benavent Acero, F.; Capobianco, C.S.; Garona, J.; Cirigliano, S.M.; Perera, Y.; Urtreger, A.J.; Perea, S.E.; Alonso, D.F.; Farina, H.G. CIGB-300, an anti-CK2 peptide, inhibits angiogenesis, tumor cell invasion and metastasis in lung cancer models. Lung Cancer 2017, 107, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, M.F.; Capobianco, C.S.; Sidabra, J.E.; Garona, J.; Perera, Y.; Perea, S.E.; Alonso, D.F.; Farina, H.G. Preclinical efficacy of CIGB-300, an anti-CK2 peptide, on breast cancer metastasic colonization. Sci. Rep. 2020, 10, 14689. [Google Scholar] [CrossRef]
- Perea, S.E.; Reyes, O.; Baladron, I.; Perera, Y.; Farina, H.; Gil, J.; Rodriguez, A.; Bacardi, D.; Marcelo, J.L.; Cosme, K.; et al. CIGB-300, a novel proapoptotic peptide that impairs the CK2 phosphorylation and exhibits anticancer properties both in vitro and in vivo. Mol. Cell. Biochem. 2008, 316, 163–167. [Google Scholar] [CrossRef]
- Perera, Y.; Ramos, Y.; Padrón, G.; Caballero, E.; Guirola, O.; Caligiuri, L.G.; Lorenzo, N.; Gottardo, F.; Farina, H.G.; Filhol, O.; et al. CIGB-300 anticancer peptide regulates the protein kinase CK2-dependent phosphoproteome. Mol. Cell. Biochem. 2020, 470, 63–75. [Google Scholar] [CrossRef]
- Lian, H.; Su, M.; Zhu, Y.; Zhou, Y.; Soomro, S.H.; Fu, H. Protein Kinase CK2, a Potential Therapeutic Target in Carcinoma Management. Asian Pac. J. Cancer Prev. 2019, 20, 23–32. [Google Scholar] [CrossRef]
- Farina, H.G.; Benavent Acero, F.; Perera, Y.; Rodríguez, A.; Perea, S.E.; Castro, B.A.; Gomez, R.; Alonso, D.F.; Gomez, D.E. CIGB-300, a proapoptotic peptide, inhibits angiogenesis in vitro and in vivo. Exp. Cell Res. 2011, 317, 1677–1688. [Google Scholar] [CrossRef]
- Iegre, J.; Atkinson, E.L.; Brear, P.D.; Cooper, B.M.; Hyvönen, M.; Spring, D.R. Chemical probes targeting the kinase CK2: A journey outside the catalytic box. Org. Biomol. Chem. 2021, 19, 4380–4396. [Google Scholar] [CrossRef]
- Perea, S.E.; Reyes, O.; Puchades, Y.; Mendoza, O.; Vispo, N.S.; Torrens, I.; Santos, A.; Silva, R.; Acevedo, B.; López, E.; et al. Antitumor effect of a novel proapoptotic peptide that impairs the phosphorylation by the protein kinase 2 (casein kinase 2). Cancer Res. 2004, 64, 7127–7129. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ulloa, A.; Ramos, Y.; Gil, J.; Perera, Y.; Castellanos-Serra, L.; García, Y.; Betancourt, L.; Besada, V.; González, L.J.; Fernández-de-Cossio, J.; et al. Proteomic profile regulated by the anticancer peptide CIGB-300 in non-small cell lung cancer (NSCLC) cells. J. Proteome Res. 2010, 9, 5473–5483. [Google Scholar] [CrossRef] [PubMed]
- Solares, A.M.; Santana, A.; Baladrón, I.; Valenzuela, C.; González, C.A.; Díaz, A.; Castillo, D.; Ramos, T.; Gómez, R.; Alonso, D.F.; et al. Safety and preliminary efficacy data of a novel casein kinase 2 (CK2) peptide inhibitor administered intralesionally at four dose levels in patients with cervical malignancies. BMC Cancer 2009, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.R.; Perera, Y.; Lúcio, P.; Silva, M.G.; Perea, S.E.; Barata, J.T. Targeting chronic lymphocytic leukemia using CIGB-300, a clinical-stage CK2-specific cell-permeable peptide inhibitor. Oncotarget 2014, 5, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Cirigliano, S.M.; Díaz Bessone, M.I.; Berardi, D.E.; Flumian, C.; Bal de Kier Joffé, E.D.; Perea, S.E.; Farina, H.G.; Todaro, L.B.; Urtreger, A.J. The synthetic peptide CIGB-300 modulates CK2-dependent signaling pathways affecting the survival and chemoresistance of non-small cell lung cancer cell lines. Cancer Cell Int. 2017, 17, 42. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).