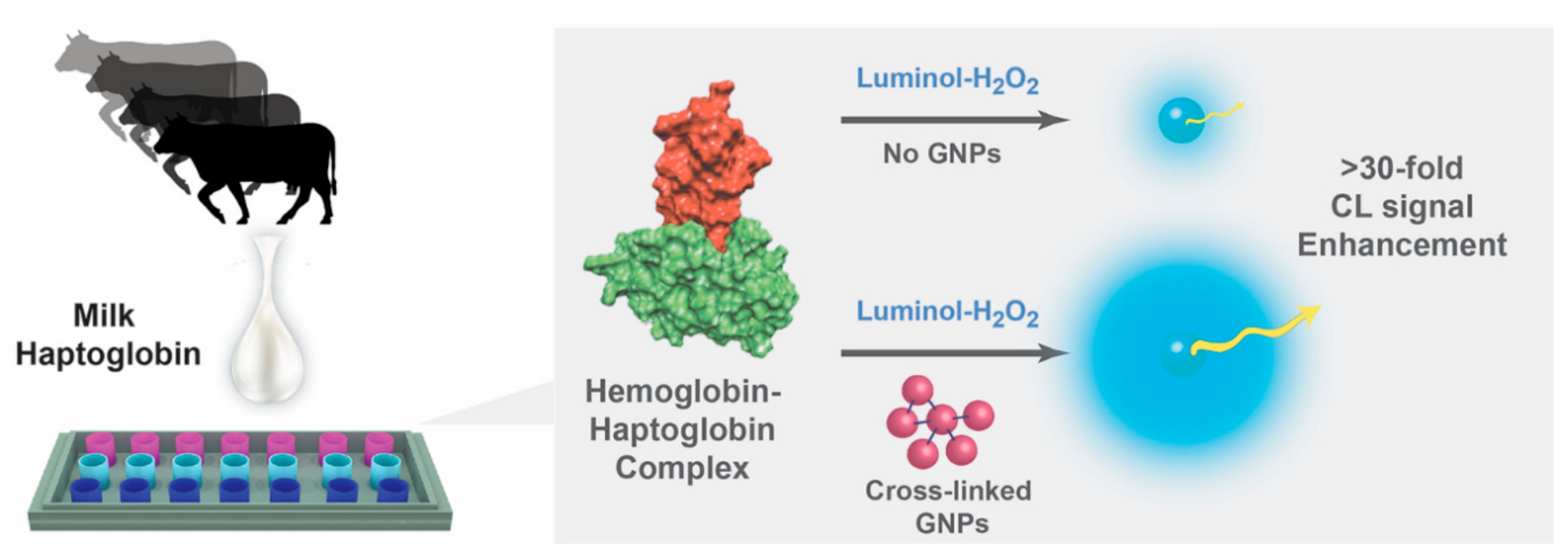
Gold Nanoparticle Size-Dependent Enhanced Chemiluminescence for Ultra-Sensitive Haptoglobin Biomarker Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis of Different Sizes of GNPs
2.3. CL Assay
2.4. Milk Sample Collection and Preparation
2.5. CL Assay Calibration
2.6. Haptoglobin Quantification
2.7. Instrumentation
2.8. Statistical Analysis
3. Results and Discussion
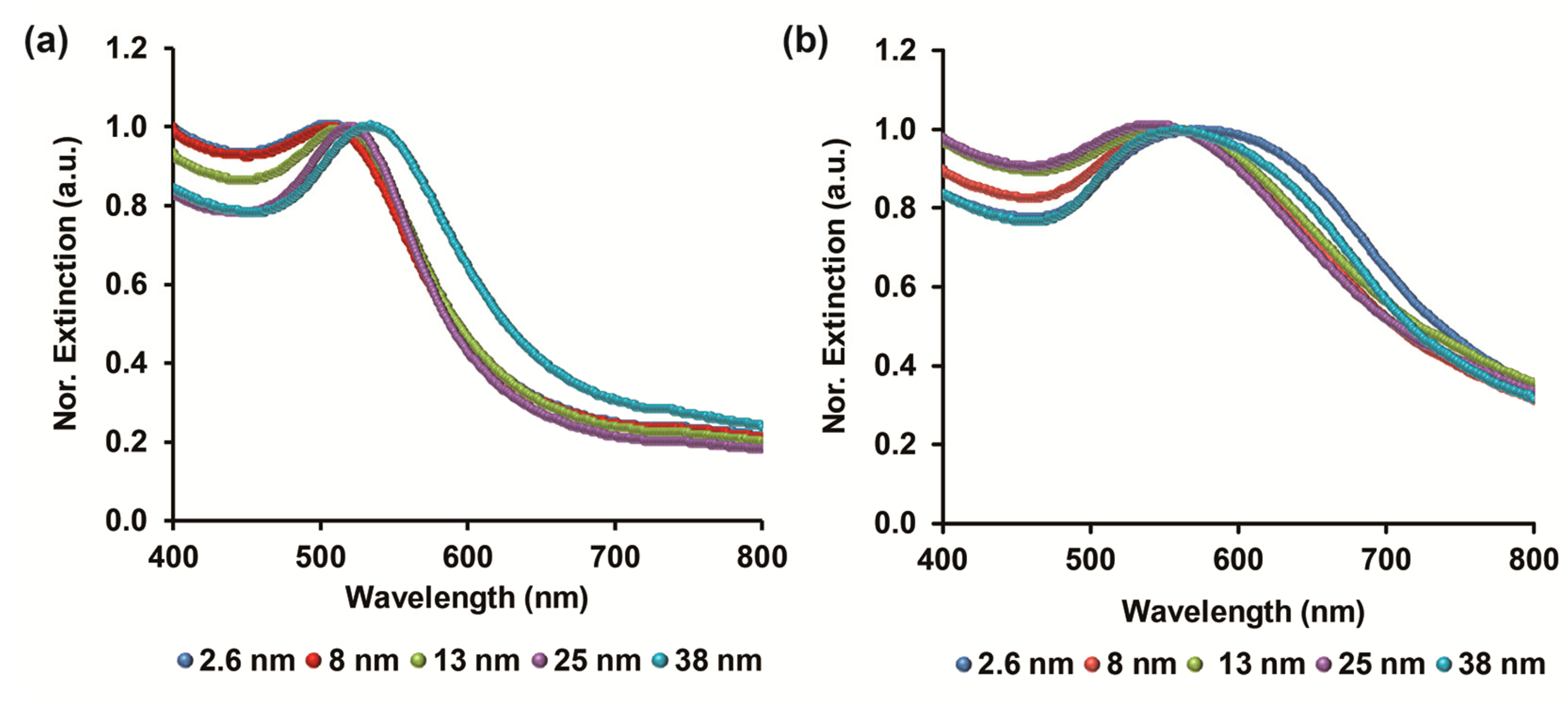
3.1. GNP Characterization
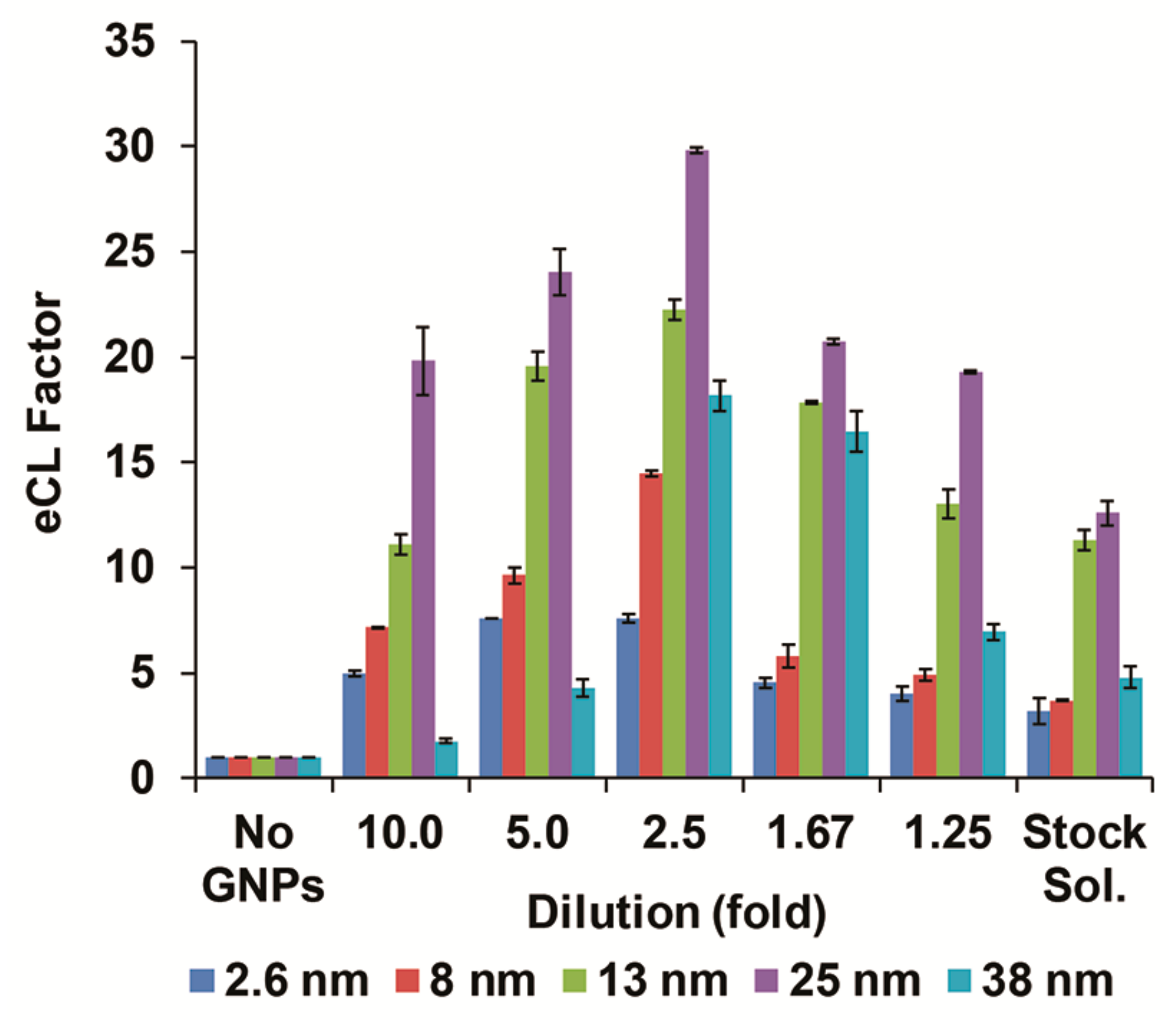
3.2. Enhanced CL Signal Optimization
3.3. Haptoglobin Quantification Based on eCL Factor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tiwari, A.; Dhoble, S.J. Recent advances and developments on integrating nanotechnology with chemiluminescence assays. Talanta 2018, 180, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, R.; Yang, X.; Qi, H.; Zhang, C. Recent advances in electrogenerated chemiluminescence biosensing methods for pharmaceuticals. J. Pharm. Anal. 2019, 9, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Shahvar, A.; Saraji, M.; Shamsaei, D. Smartphone-based chemiluminescence sensing for TLC imaging. Sens. Actuators B 2018, 255, 891–894. [Google Scholar] [CrossRef]
- Bui, M.P.N.; Brockgreitens, J.; Abbas, A. Gold Nanoplate-Enhanced Chemiluminescence and Macromolecular Shielding for Rapid Microbial Diagnostics. Adv. Healthcare Mater. 2018, 7, 1701506–1701516. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.-S.; Lee, J.H.; Park, J.-M.; Choi, H.N.; Lee, W.-Y. Luminol chemiluminescence biosensor for glycated hemoglobin (HbA1c) in human blood samples. Biosens. Bioelectron. 2016, 75, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Kang, S.H. Chemiluminescence detection of label-free C-reactive protein based on catalytic activity of gold nanoparticles. Talanta 2011, 84, 752–758. [Google Scholar] [CrossRef]
- Zangheri, M.; di Nardo, F.; Anfossi, L.; Giovannoli, C.; Baggiani, C.; Roda, A.; Mirasoli, M. A multiplex chemiluminescent biosensor for type B-fumonisins and aflatoxin B1 quantitative detection in maize flour. Analyst 2015, 140, 358–365. [Google Scholar] [CrossRef]
- Ryan, L.S.; Lippert, A.R. Ultrasensitive chemiluminescent detection of cathepsin B: Insights into the new frontier of chemiluminescent imaging. Angew. Chem. Int. Ed. 2018, 57, 622–624. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, X.; Ren, J. Recent advances in chemiluminescence detection coupled with capillary electrophoresis and microchip capillary electrophoresis. Electrophoresis 2016, 37, 2–18. [Google Scholar] [CrossRef]
- Hao, N.; Wang, K. Recent development of electrochemiluminescence sensors for food analysis. Anal. Bioanal. Chem. 2016, 408, 7035–7048. [Google Scholar] [CrossRef]
- Timofeeva, I.I.; Vakh, C.S.; Bulatov, A.V.; Worsfold, P.J. Flow analysis with chemiluminescence detection: Recent advances and applications. Talanta 2018, 179, 246–270. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Li, B. Enhanced effect of aggregated gold nanoparticles on luminol chemiluminescence system and its analytical application. Spectrochim. Acta Part A 2013, 111, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-F.; Cui, H.; Lai, C.-Z.; Liu, L.-J. Gold nanoparticle-catalyzed luminol chemiluminescence and its analytical applications. Anal. Chem. 2005, 77, 3324–3329. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Zhu, Z.; He, D.; Jin, L.; Zheng, H.; Hu, S. Simultaneous Determination of Size and Quantification of Gold Nanoparticles by Direct Coupling Thin layer Chromatography with Catalyzed Luminol Chemiluminescence. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Feltmeyer, A.; Saiapina, O.; Juzwik, J.; Arenz, B.; Abbas, A. Rapid and PCR-free DNA Detection by Nanoaggregation-Enhanced Chemiluminescence. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Nirala, N.R.; Pinker, N.; Desitti, C.; Shtenberg, G. Milk haptoglobin detection based on enhanced chemiluminescence of gold nanoparticles. Talanta 2019, 197, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhang, Z.-F.; Shi, M.-J. Chemiluminescent reactions induced by gold nanoparticles. J. Phys. Chem. B 2005, 109, 3099–3103. [Google Scholar] [CrossRef]
- Wang, J.; Mao, S.; Li, H.-F.; Lin, J.-M. Multi-DNAzymes-functionalized gold nanoparticles for ultrasensitive chemiluminescence detection of thrombin on microchip. Anal. Chim. Acta 2018, 1027, 76–82. [Google Scholar] [CrossRef]
- Zhong, Y.; Wu, X.; Li, J.; Lan, Q.; Jing, Q.; Min, L.; Ren, C.; Hu, X.; Lambert, A.; Cheng, Q.; et al. Multiplex immunoassay of chicken cytokines via highly-sensitive chemiluminescent imaging array. Anal. Chim. Acta 2019, 1049, 213–218. [Google Scholar] [CrossRef]
- Lyu, Z.-M.; Zhou, X.-L.; Wang, X.-N.; Li, P.; Xu, L.; Liu, E.H. Miniaturized electrochemiluminescent biochip prepared on gold nanoparticles-loaded mesoporous silica film for visual detection of hydrogen peroxide released from living cells. Sens. Actuators B 2019, 284, 437–443. [Google Scholar] [CrossRef]
- Kalmus, P.; Simojoki, H.; Pyörälä, S.; Taponen, S.; Holopainen, J.; Orro, T. Milk haptoglobin, milk amyloid A, and N-acetyl-β-d-glucosaminidase activity in bovines with naturally occurring clinical mastitis diagnosed with a quantitative PCR test. J. Dairy Sci. 2013, 96, 3662–3670. [Google Scholar] [CrossRef] [PubMed]
- Viguier, C.; Arora, S.; Gilmartin, N.; Welbeck, K.; O’Kennedy, R. Mastitis detection: Current trends and future perspectives. Trends Biotechnol. 2009, 27, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Ahmed, S.R.; Neethirajan, S. A nanocomposite-based biosensor for bovine haptoglobin on a 3D paper-based analytical device. Sens. Actuators B 2018, 265, 242–248. [Google Scholar] [CrossRef]
- Hiss, S.; Mielenz, M.; Bruckmaier, R.; Sauerwein, H. Haptoglobin concentrations in blood and milk after endotoxin challenge and quantification of mammary Hp mRNA expression. J. Dairy Sci. 2004, 87, 3778–3784. [Google Scholar] [CrossRef]
- Brown, K.R.; Fox, A.P.; Natan, M.J. Morphology-dependent electrochemistry of cytochrome c at Au colloid-modified SnO2 electrodes. J. Am. Chem. Soc. 1996, 118, 1154–1157. [Google Scholar] [CrossRef]
- Guarise, C.; Pasquato, L.; Scrimin, P. Reversible aggregation/deaggregation of gold nanoparticles induced by a cleavable dithiol linker. Langmuir 2005, 21, 5537–5541. [Google Scholar] [CrossRef] [PubMed]
- Chegel, V.; Rachkov, O.; Lopatynskyi, A.; Ishihara, S.; Yanchuk, I.; Nemoto, Y.; Hill, J.P.; Ariga, K. Gold Nanoparticles Aggregation: Drastic Effect of Cooperative Functionalities in a Single Molecular Conjugate. J. Phys. Chem. C 2012, 116, 2683–2690. [Google Scholar] [CrossRef]
- Daniel, M.-C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef]
- Kobayashi, M.; Juillerat, F.; Galletto, P.; Bowen, P.; Borkovec, M. Aggregation and Charging of Colloidal Silica Particles: Effect of Particle Size. Langmuir 2005, 21, 5761–5769. [Google Scholar] [CrossRef]
- Link, S.; El-Sayed, M.A. Spectral Properties and Relaxation Dynamics of Surface Plasmon Electronic Oscillations in Gold and Silver Nanodots and Nanorods. J. Phys. Chem. B 1999, 103, 8410–8426. [Google Scholar] [CrossRef]
- Duan, C.; Cui, H.; Zhang, Z.; Liu, B.; Guo, J.; Wang, W. Size-dependent inhibition and enhancement by gold nanoparticles of luminol− ferricyanide chemiluminescence. J. Phys. Chem. C 2007, 111, 4561–4566. [Google Scholar] [CrossRef]
- Hu, L.; Liao, H.; Feng, L.; Wang, M.; Fu, W. Accelerating the peroxidase-like activity of gold nanoclusters at neutral pH for colorimetric detection of heparin and heparinase activity. Anal. Chem. 2018, 90, 6247–6252. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Lee, K.; Gong, M.-S.; Joo, S.-W. Control of gold nanoparticle aggregates by manipulation of interparticle interaction. Langmuir 2005, 21, 9524–9528. [Google Scholar] [CrossRef] [PubMed]
- Makinen, M.W.; Milstien, J.B.; Kon, H. Specificity of interaction of haptoglobin with mammalian hemoglobin. Biochemistry 1972, 11, 3851–3860. [Google Scholar] [CrossRef] [PubMed]
- Pavlíček, Z.; Jaenicke, R. On the Mechanism of Hemoglobin-Haptoglobin Complex Formation. Eur. J. Biochem. 1971, 18, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, S.M.; Ghassabian, S. Linearity of Calibration Curves for Analytical Methods: A Review of Criteria for Assessment of Method Reliability. In Calibration and Validation of Analytical Methods: A Sampling of Current Approaches; IntechOpen Limited: London, UK, 2018; pp. 109–127. [Google Scholar]
- Krismastuti, F.S.; Pace, S.; Voelcker, N.H. Porous silicon resonant microcavity biosensor for matrix metalloproteinase detection. Adv. Funct. Mater. 2014, 24, 3639–3650. [Google Scholar] [CrossRef]
- Wang, Y.-R.; Yang, Y.-H.; Lu, C.-Y.; Chen, S.-H. Utilization of magnetic nanobeads for analyzing haptoglobin in human plasma as a marker of Alzheimer’s disease by capillary electrophoretic immunoassay with laser-induced fluorescence detection. Anal. Chim. Acta 2015, 865, 76–82. [Google Scholar] [CrossRef]
- Abadieh, R.; Safi, S.; Mohsenifar, A.; Bayat, M. Designation of a fluorescence-based nanobiosensor for detection of bovin haptopglobin. Bull. Georg. Natl. Acad. Sci. 2015, 9, 222–228. [Google Scholar]
- Åkerstedt, M.; Björck, L.; Waller, K.P.; Sternesjö, Å. Biosensor assay for determination of haptoglobin in bovine milk. J. Dairy Res. 2006, 73, 299–305. [Google Scholar] [CrossRef]
- Huang, G.; Ouyang, J.; Delanghe, J.R.; Baeyens, W.R.G.; Dai, Z. Chemiluminescent Image Detection of Haptoglobin Phenotyping after Polyacrylamide Gel Electrophoresis. Anal. Chem. 2004, 76, 2997–3004. [Google Scholar] [CrossRef]
- Nakagawa, H.; Yamamoto, O.; Oikawa, S.; Higuchi, H.; Watanabe, A.; Katoh, N. Detection of serum haptoglobin by enzyme-linked immunosorbent assay in cows with fatty liver. Res. Vet. Sci. 1997, 62, 137–141. [Google Scholar] [CrossRef]
- Rizwan, M.; Keasberry, N.A.; Ahmed, M.U. Efficient double electrochemiluminescence quenching based label-free highly sensitive detection of haptoglobin on a novel nanocomposite modified carbon nanofibers interface. Sens. Biosensing Res. 2019. [Google Scholar] [CrossRef]
- Thomas, F.C.; Geraghty, T.; Simões, P.B.A.; Mshelbwala, F.M.; Haining, H.; Eckersall, P.D. A pilot study of acute phase proteins as indicators of bovine mastitis caused by different pathogens. Res. Vet. Sci. 2018, 119, 176–181. [Google Scholar] [CrossRef] [PubMed]


| Sample | SCC | Pathogenic Bacteria |
|---|---|---|
| H | 60,000 | - |
| M1 | 300,000 | E. coli |
| M2 | 636,000 | E. coli |
| M3 | >1,000,000 | E. coli |
| M4 | 337,000 | CNS |
| M5 | 821,000 | CNS |
| M6 | >1,000,000 | CNS |
| Sample | Haptoglobin with Cross-Linked GNPs (µg mL−1) | Haptoglobin ELISA (µg mL−1) |
|---|---|---|
| H | 0.1 ± 0.1 | 0.1 ± 0.1 |
| M1 | 1.4 ± 1.2 | 1.2 ± 0.01 |
| M2 | 3.6 ± 1.4 | 3.9 ± 1.3 |
| M3 | 24.0 ± 4.5 | 21.0 ± 0.8 |
| M4 | 1.6 ± 0.9 | 1.7 ± 0.3 |
| M5 | 5.2 ± 1.7 | 4.8 ± 0.2 |
| M6 | 19.5 ± 3.0 | 16.6 ± 3.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nirala, N.R.; Shtenberg, G. Gold Nanoparticle Size-Dependent Enhanced Chemiluminescence for Ultra-Sensitive Haptoglobin Biomarker Detection. Biomolecules 2019, 9, 372. https://doi.org/10.3390/biom9080372
Nirala NR, Shtenberg G. Gold Nanoparticle Size-Dependent Enhanced Chemiluminescence for Ultra-Sensitive Haptoglobin Biomarker Detection. Biomolecules. 2019; 9(8):372. https://doi.org/10.3390/biom9080372
Chicago/Turabian StyleNirala, Narsingh R., and Giorgi Shtenberg. 2019. "Gold Nanoparticle Size-Dependent Enhanced Chemiluminescence for Ultra-Sensitive Haptoglobin Biomarker Detection" Biomolecules 9, no. 8: 372. https://doi.org/10.3390/biom9080372
APA StyleNirala, N. R., & Shtenberg, G. (2019). Gold Nanoparticle Size-Dependent Enhanced Chemiluminescence for Ultra-Sensitive Haptoglobin Biomarker Detection. Biomolecules, 9(8), 372. https://doi.org/10.3390/biom9080372






