Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Illumina Microarray Data
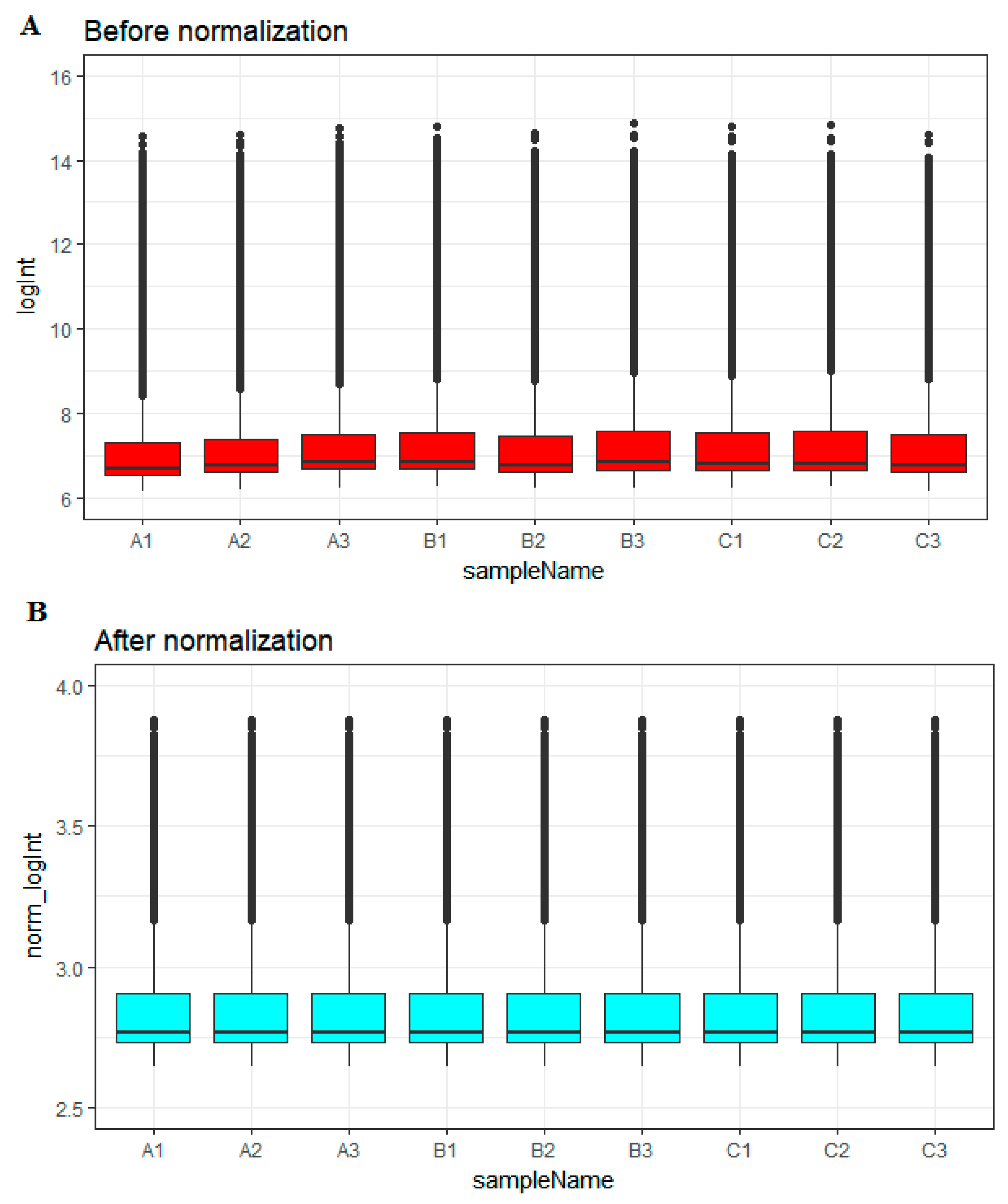
2.2. Data Preprocessing and Differential Expression Analysis
2.3. Pathway Enrichment Analysis
2.4. GO Term Enrichment Analysis
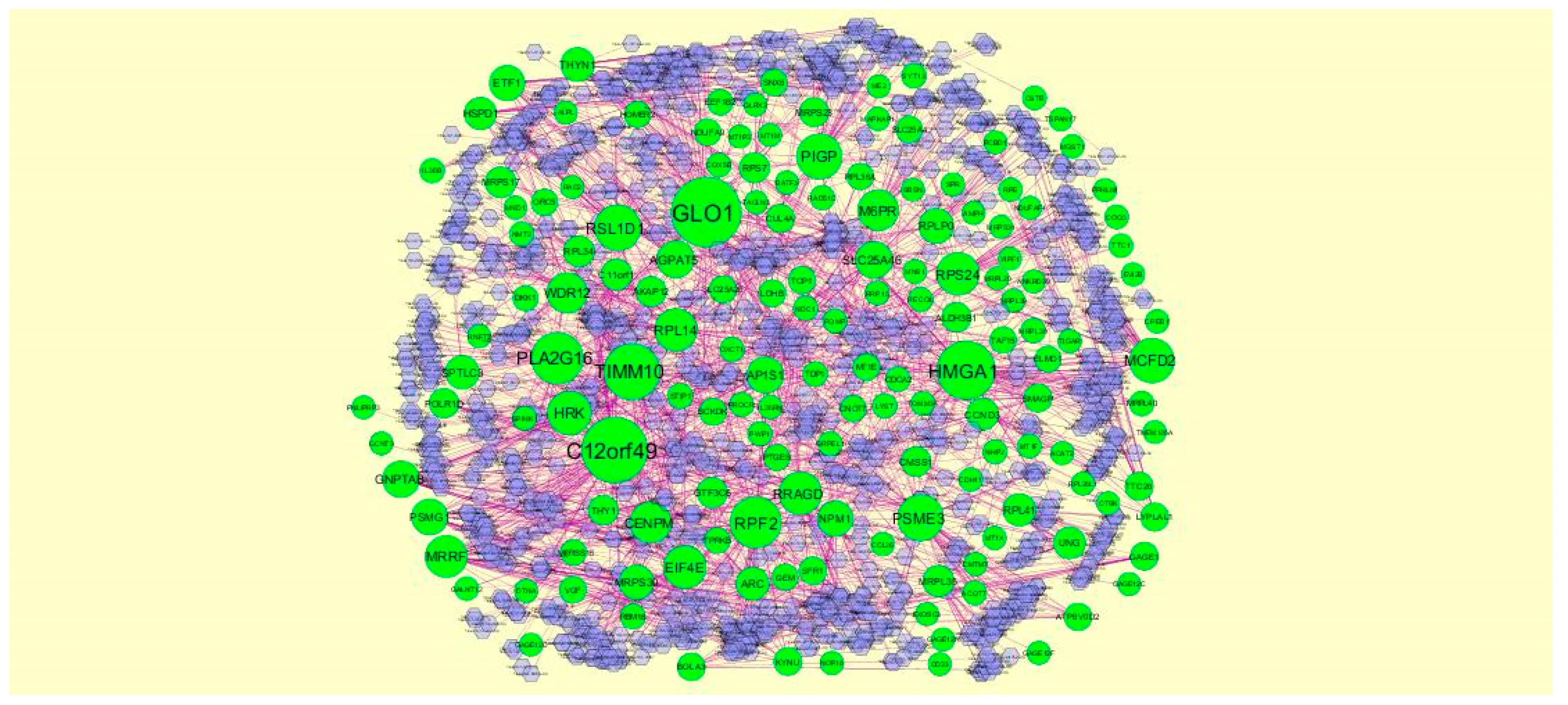
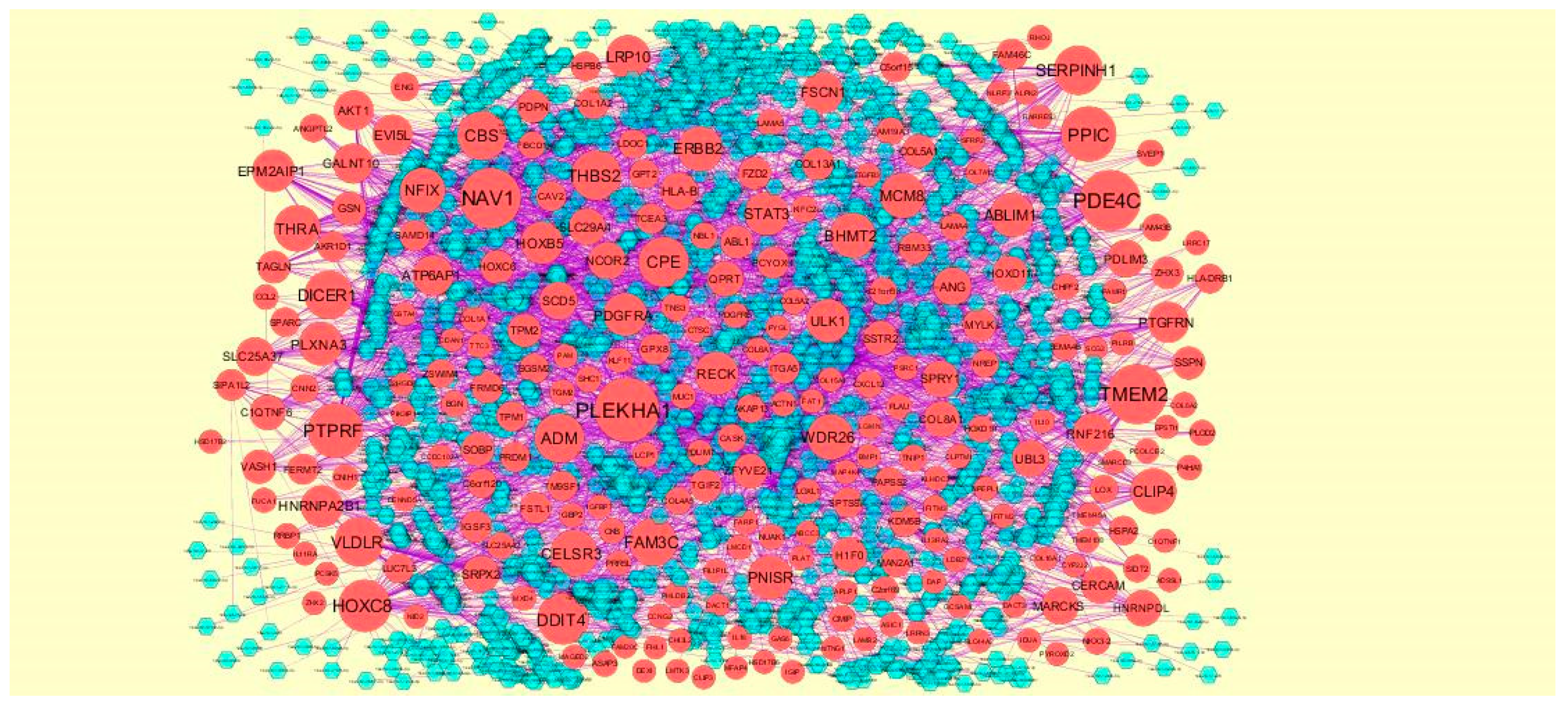
2.5. PPI Network Construction
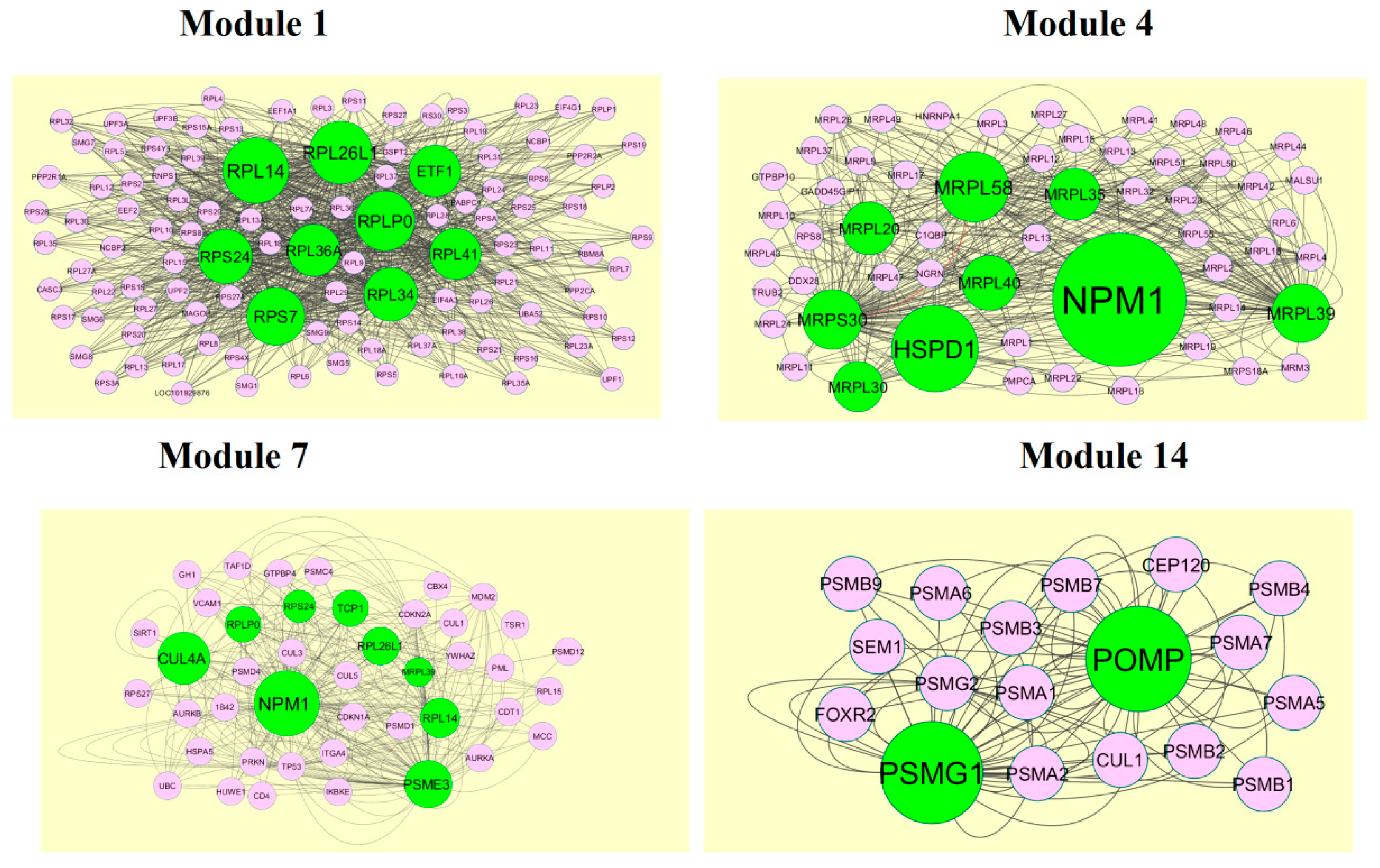
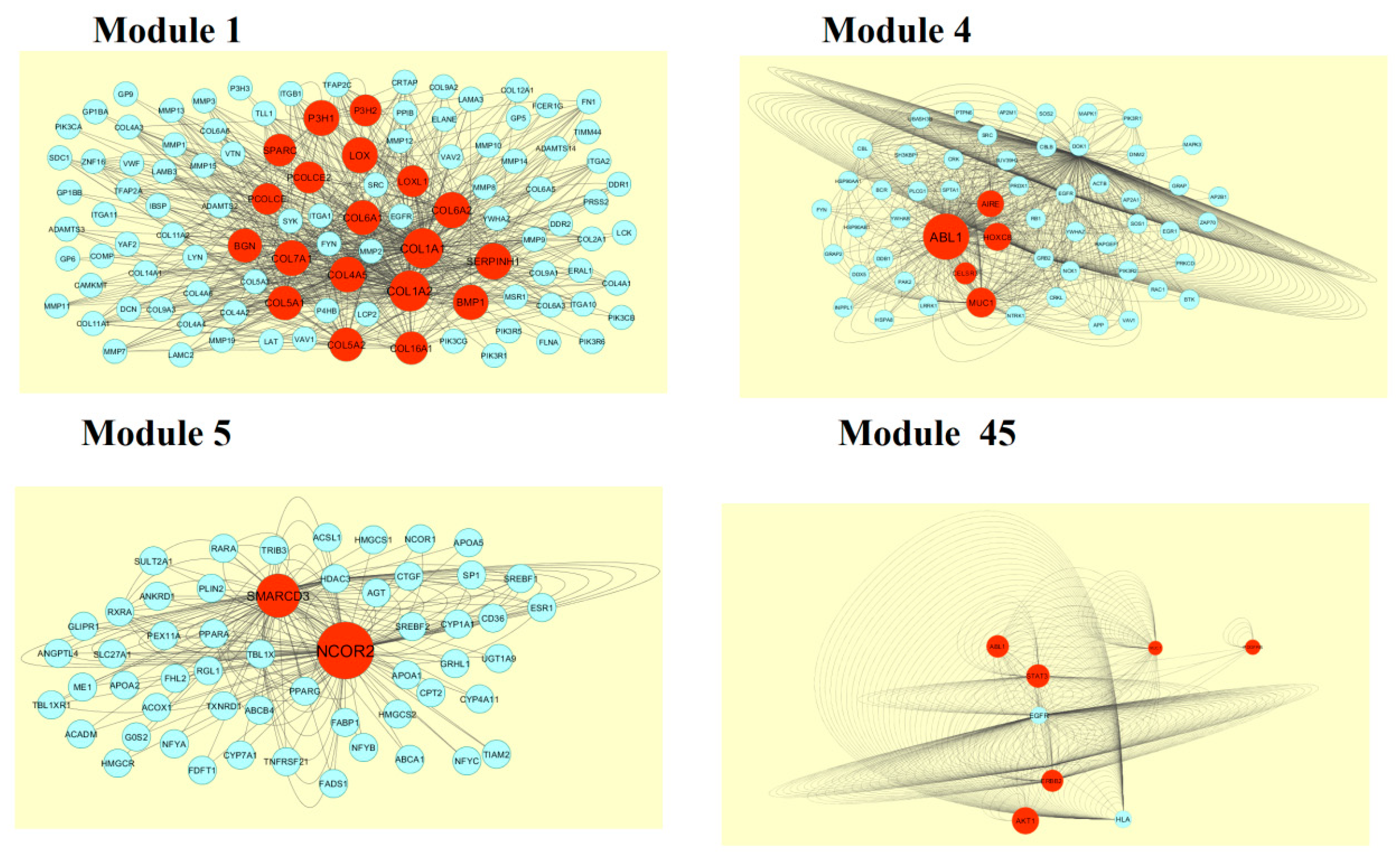
2.6. Module Analysis
2.7. Construction of the Target Gene-miRNA Network
2.8. Construction of the Target Gene-TF-Network
2.9. Survival Analysis of Hub Genes
2.10. The mRNA Expression Levels of Hub Genes in GBM
2.11. Mutation Analysis
3. Results
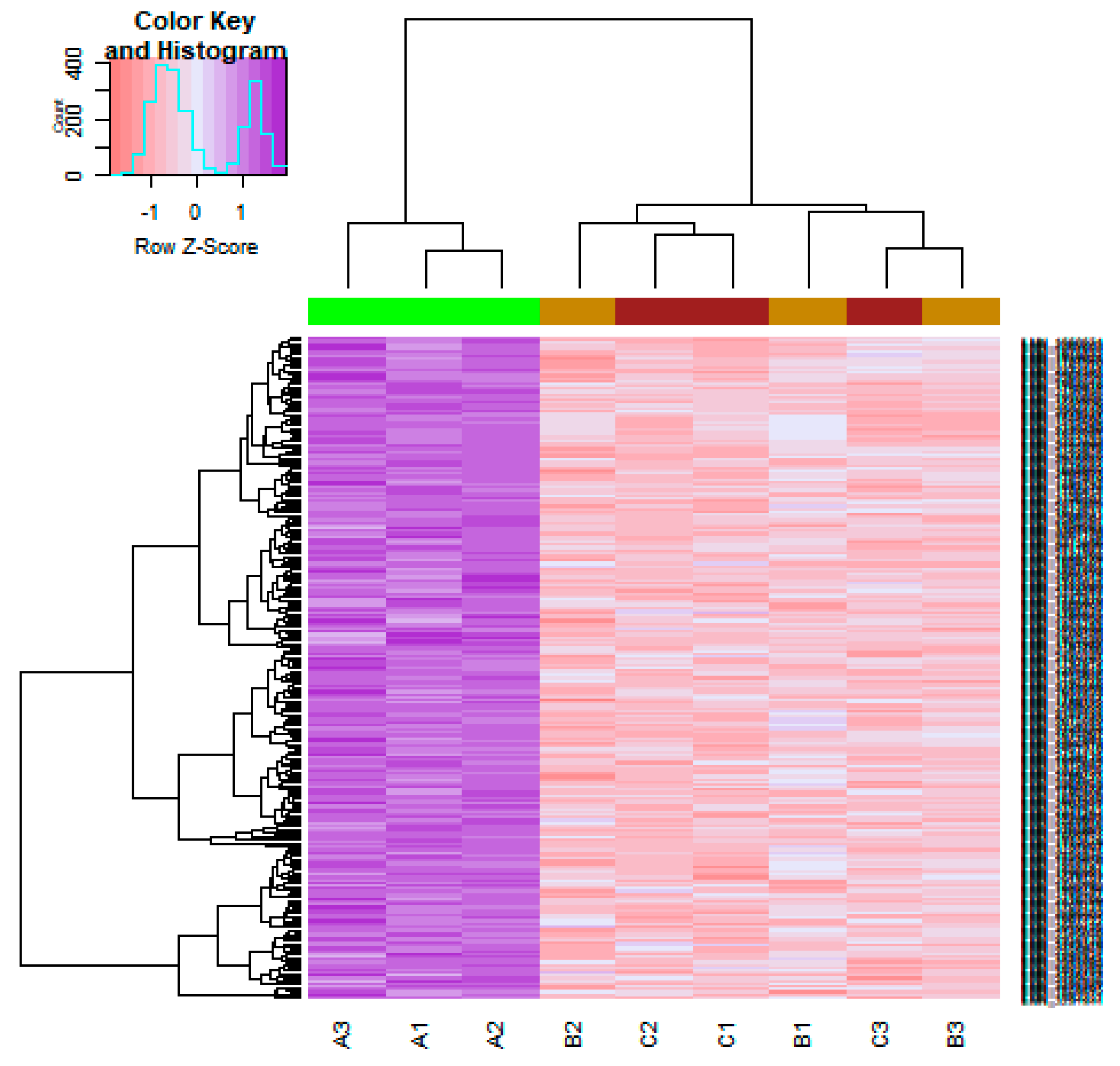
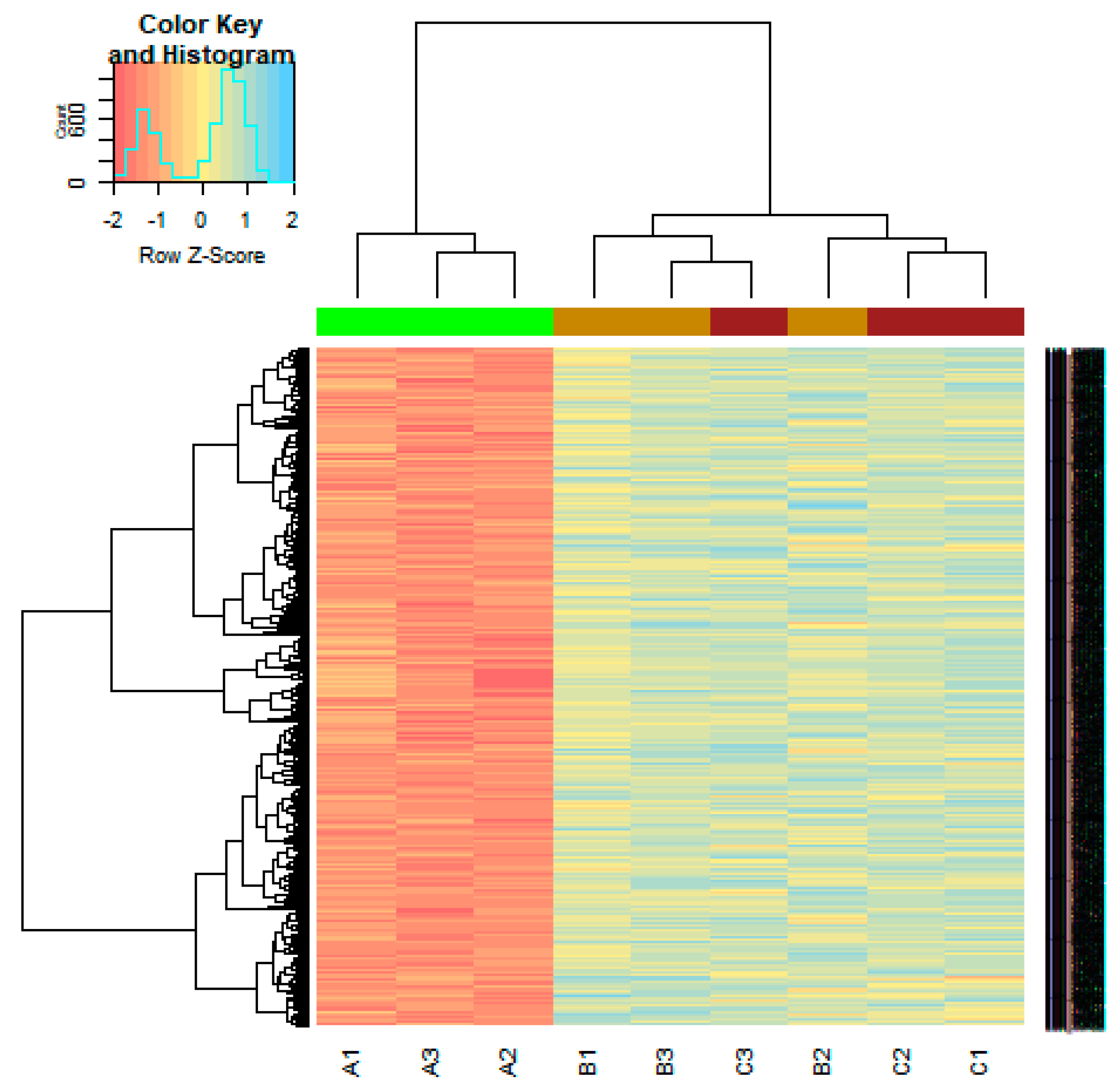
3.1. Identification of DEGs
3.2. Pathway Enrichment Analysis
3.3. GO Term Enrichment Analysis
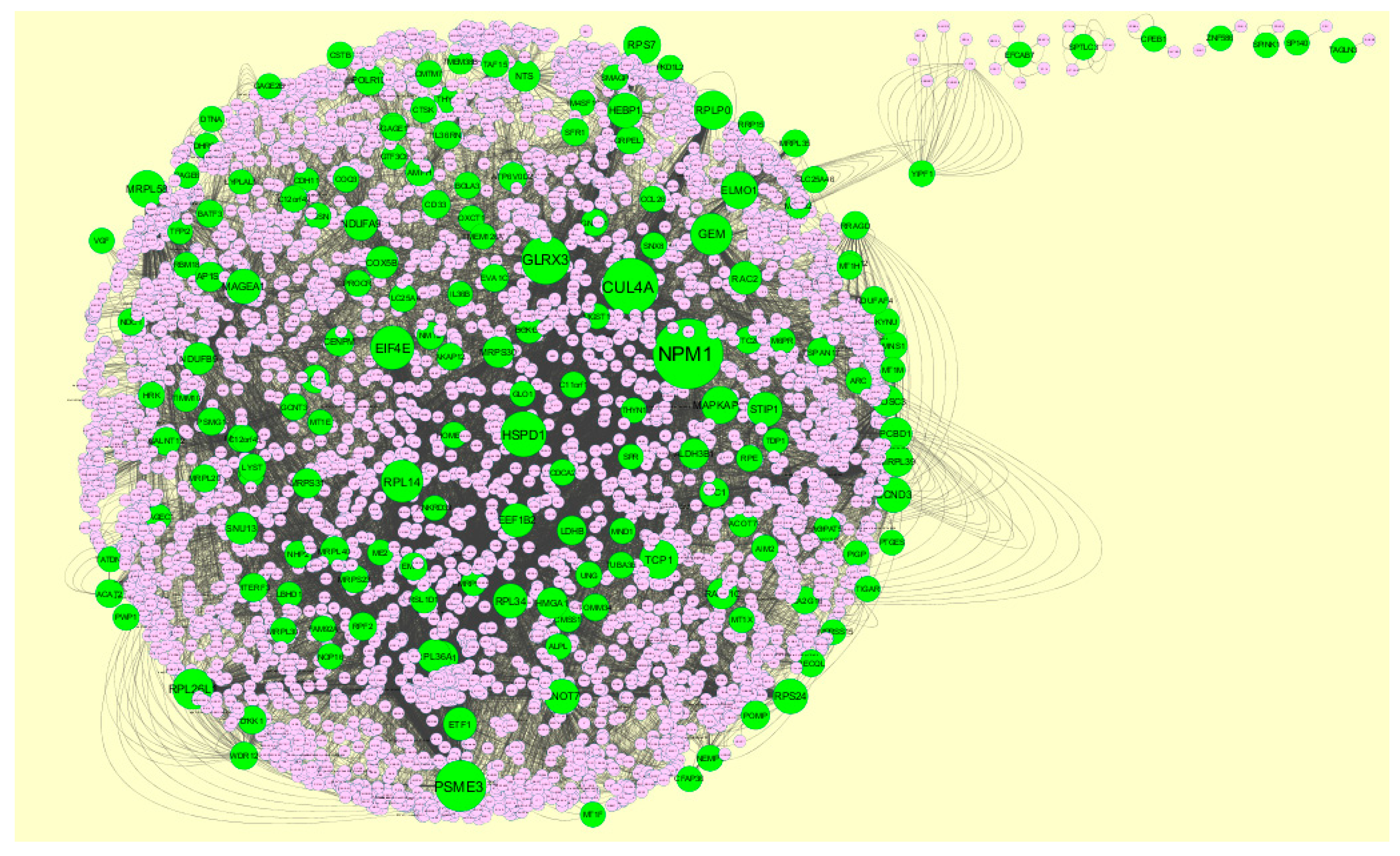
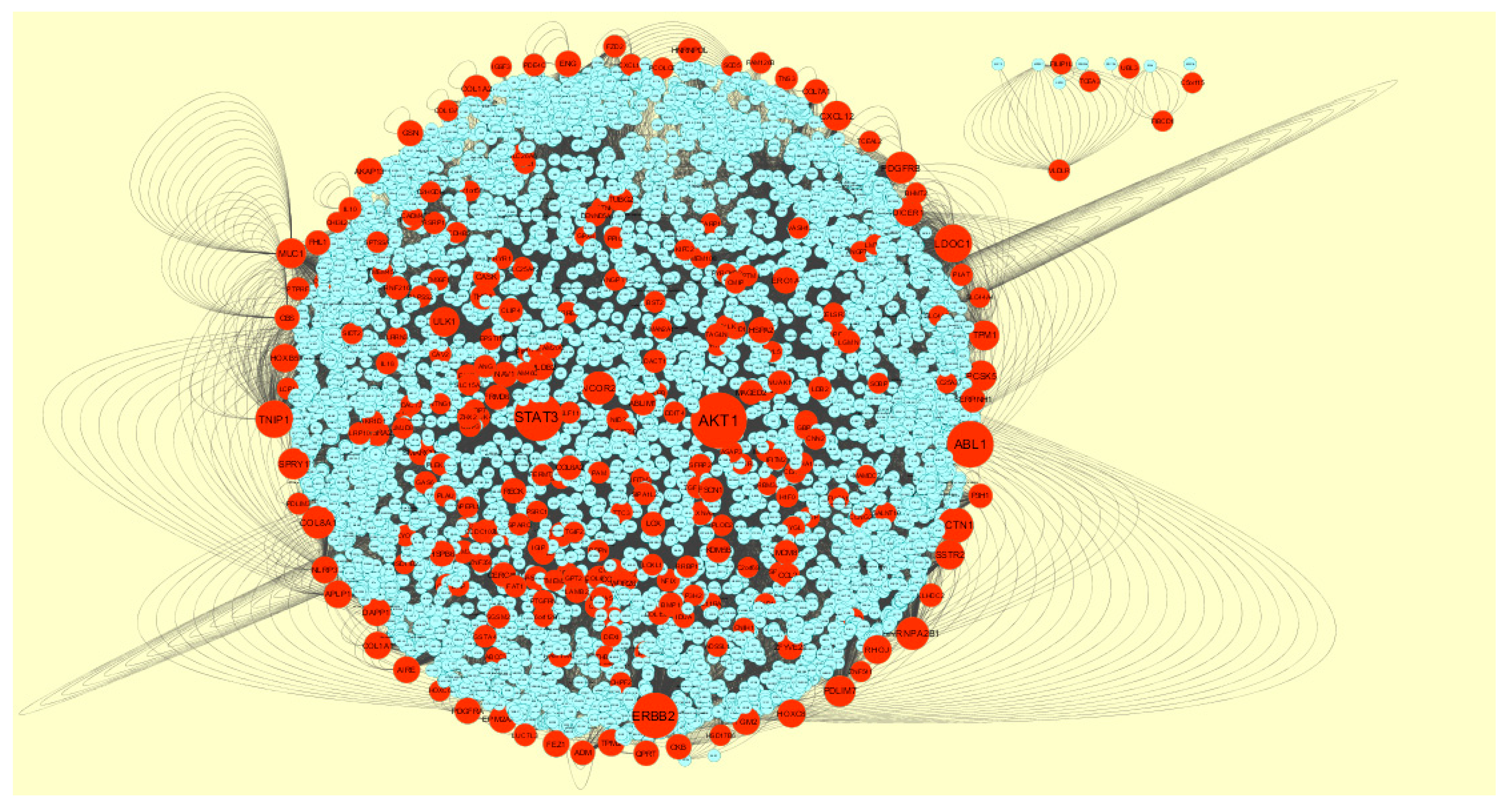
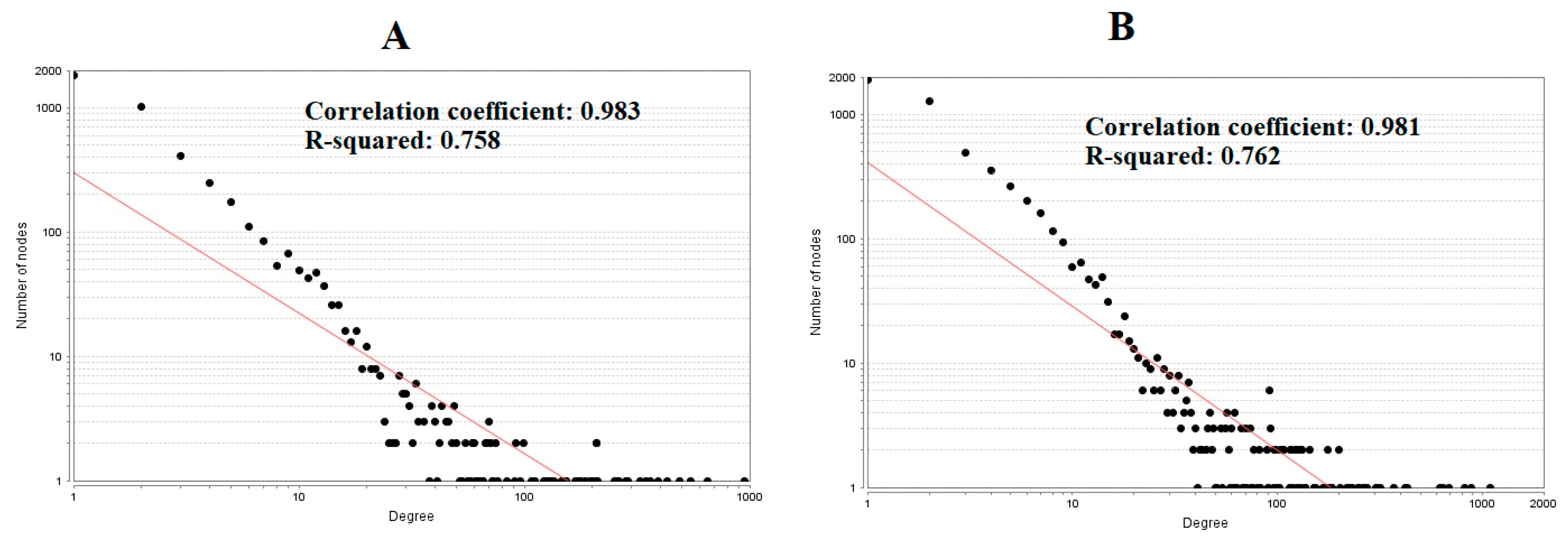
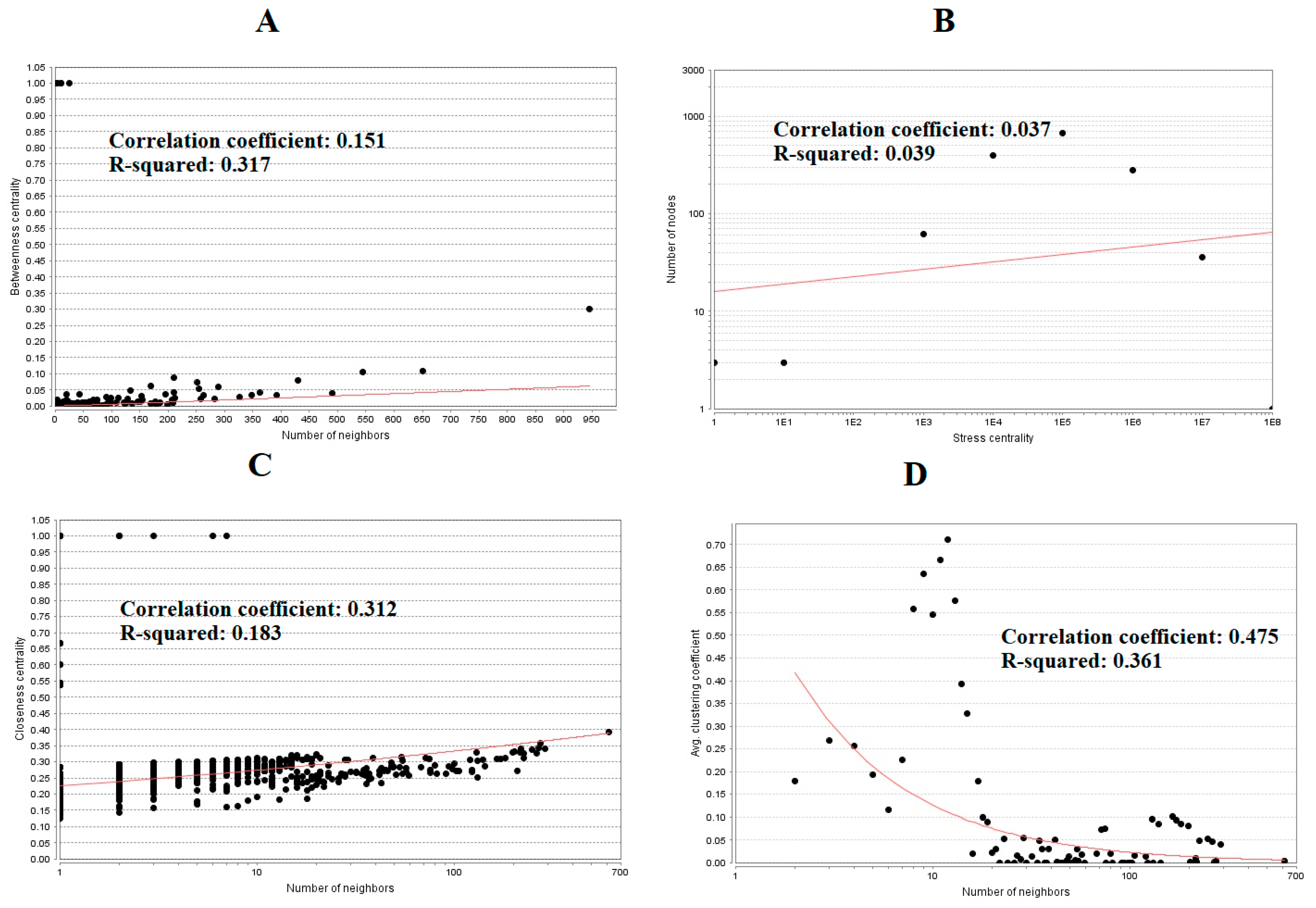
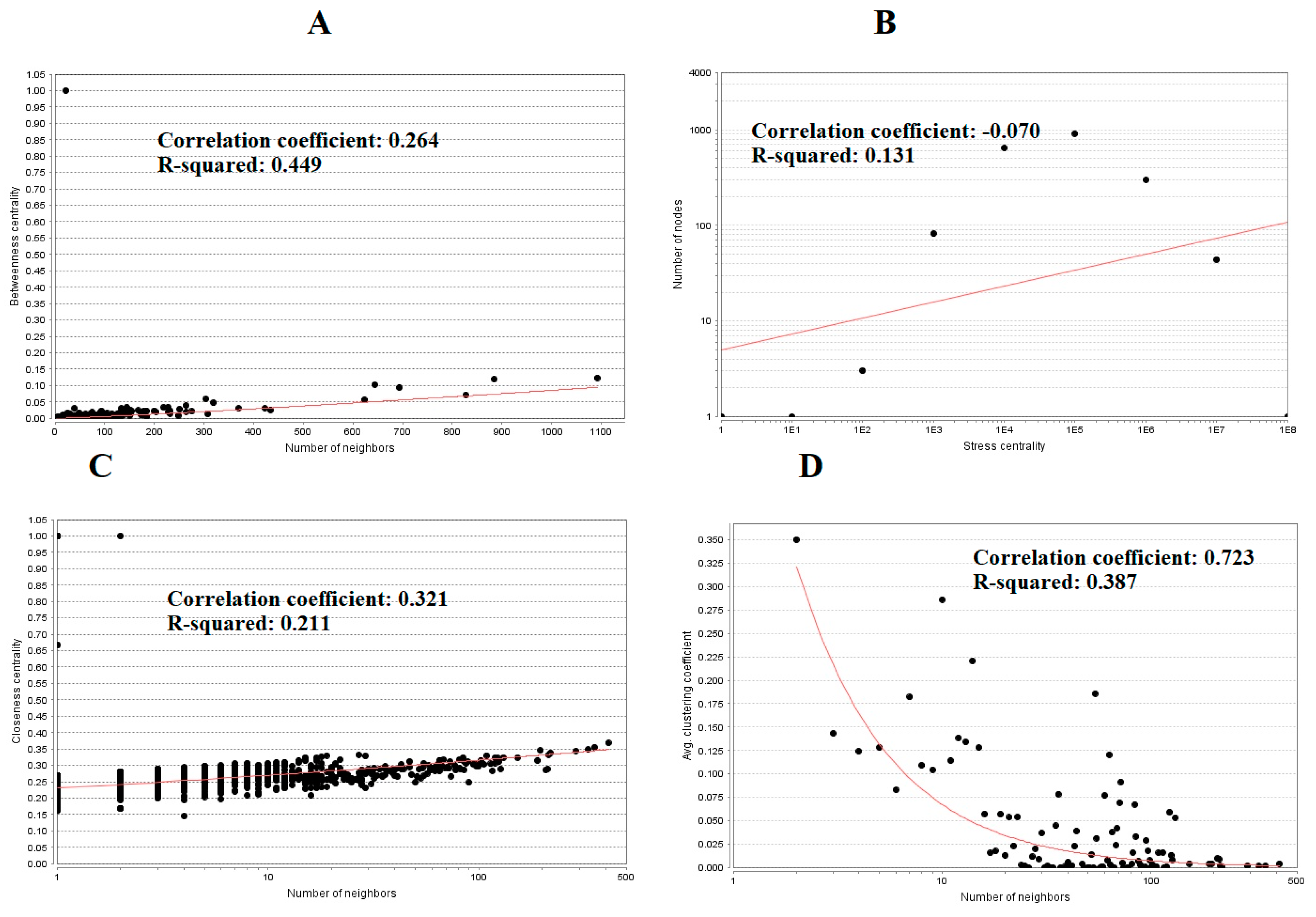
3.4. Construction PPI Network and Topological Analysis
3.5. Module Analysis
3.6. Construction of the Target Gene-miRNA Network
3.7. Construction of the Target Gene-TF Network
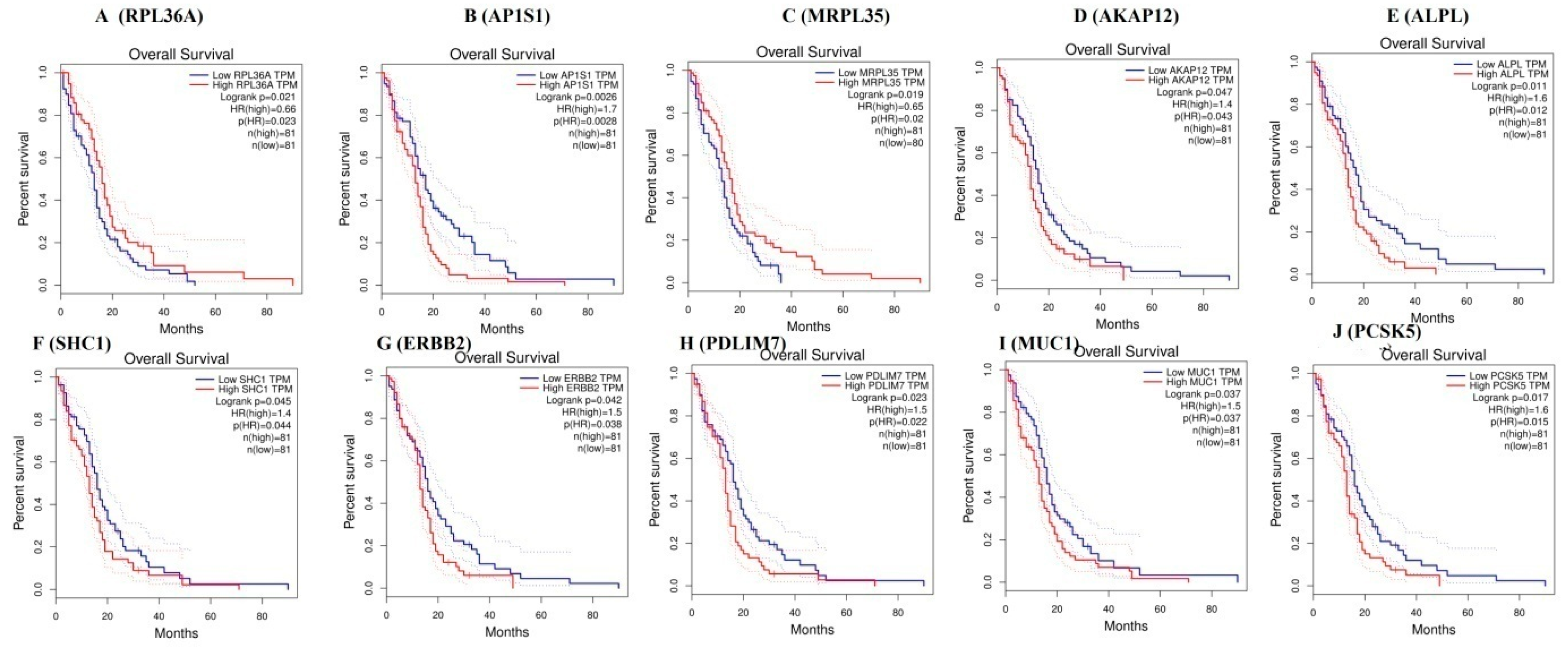
3.8. Survival Curve Analysis of Hub Genes
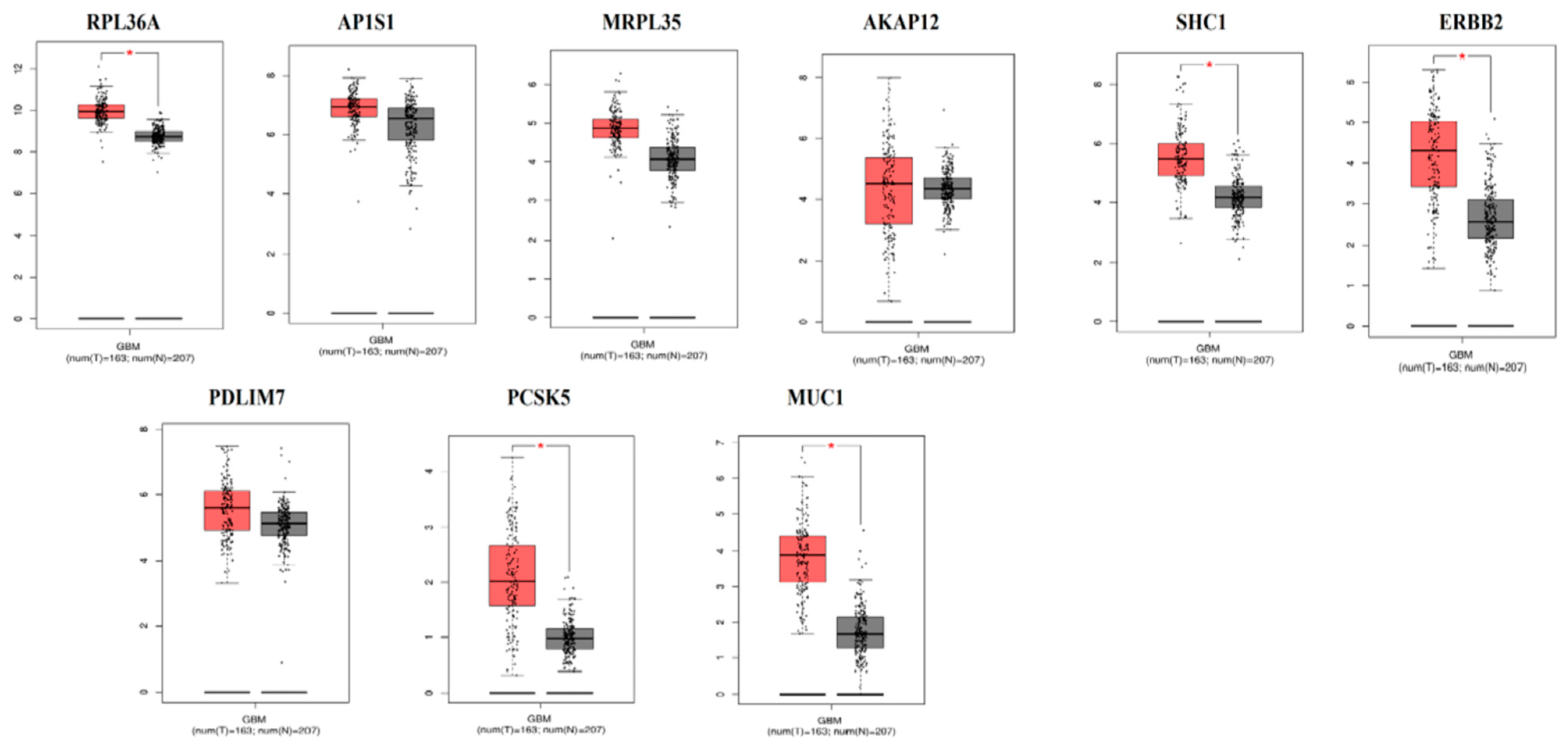
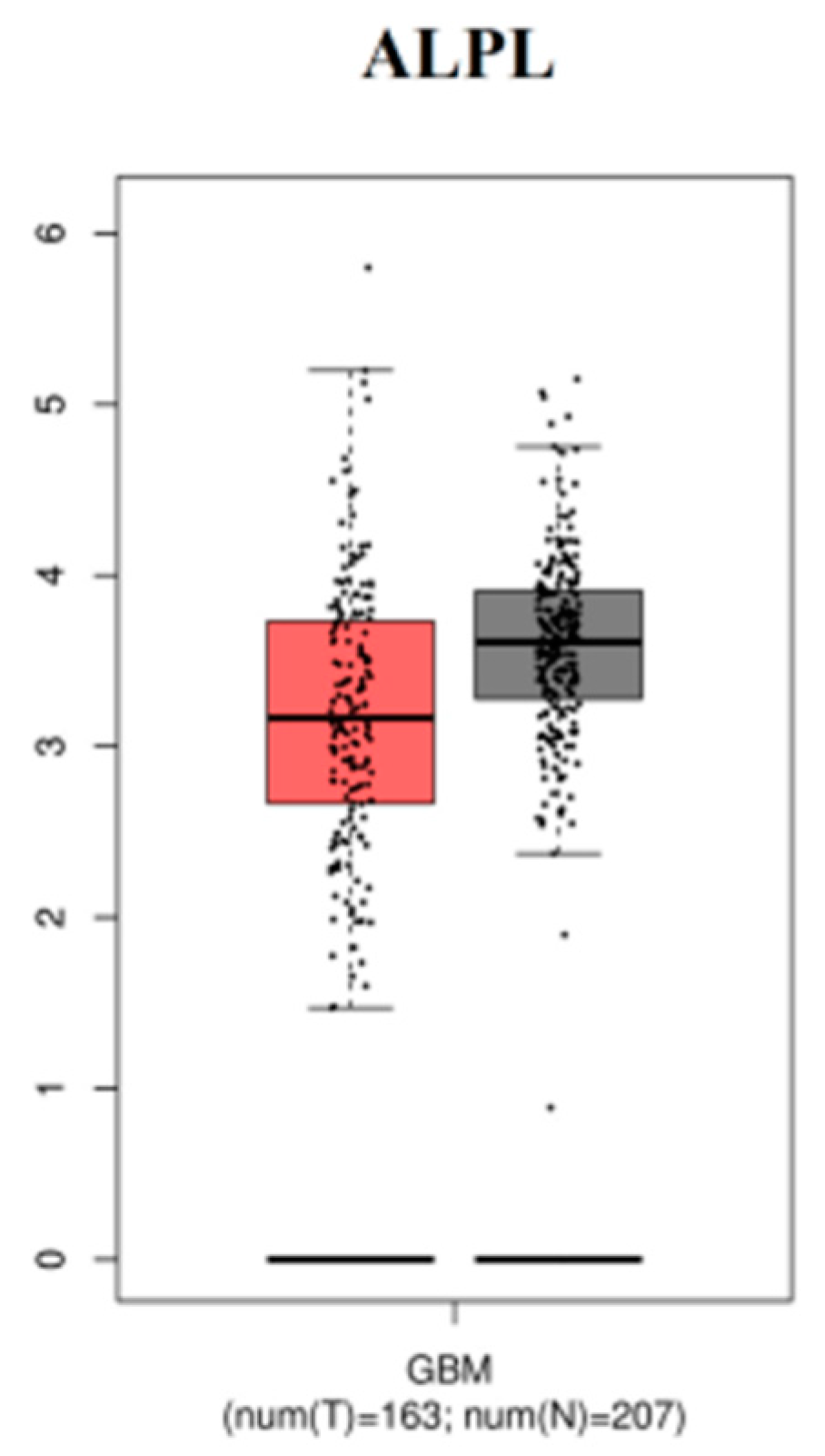
3.9. The mRNA Expression Levels of Hub Genes in GBM
3.10. Mutation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Carson-Walter, E.B.; Cooper, A.; Winans, B.N.; Johnson, M.D.; Walter, K.A. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J. Neurooncol. 2010, 99, 13–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lötsch, D.; Steiner, E.; Holzmann, K.; Spiegl-Kreinecker, S.; Pirker, C.; Hlavaty, J.; Petznek, H.; Hegedus, B.; Garay, T.; Mohr, T.; et al. Major vault protein supports glioblastoma survival and migration by upregulating the EGFR/PI3K signalling axis. Oncotarget 2013, 4, 1904–1918. [Google Scholar] [CrossRef] [PubMed]
- Soda, Y.; Myskiw, C.; Rommel, A.; Verma, I.M. Mechanisms of neovascularization and resistance to anti-angiogenic therapies in glioblastoma multiforme. J. Mol. Med. (Berl.) 2013, 91, 439–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochberg, F.; Toniolo, P.; Cole, P.; Salcman, M. Nonoccupational risk indicators of glioblastoma in adults. J. Neurooncol. 1990, 8, 55–60. [Google Scholar] [CrossRef]
- Srinivas, P.R.; Kramer, B.S.; Srivastava, S. Trends in biomarker research for cancer detection. Lancet. Oncol. 2001, 2, 698–704. [Google Scholar] [CrossRef]
- Mehrian-Shai, R.; Chen, C.D.; Shi, T.; Horvath, S.; Nelson, S.F.; Reichardt, J.K.; Sawyers, C.L. Insulin growth factor-binding protein 2 is a candidate biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 5563–5568. [Google Scholar] [CrossRef] [Green Version]
- Graff, J.R.; McNulty, A.M.; Hanna, K.R.; Konicek, B.W.; Lynch, R.L.; Bailey, S.N.; Banks, C.; Capen, A.; Goode, R.; Lewis, J.E.; et al. The protein kinase Cbeta-selective inhibitor, Enzastaurin (LY317615.HCl), suppresses signaling through the AKT pathway, induces apoptosis, and suppresses growth of human colon cancer and glioblastoma xenografts. Cancer Res. 2005, 65, 7462–7469. [Google Scholar] [CrossRef]
- Hiratsuka, M.; Inoue, T.; Toda, T.; Kimura, N.; Shirayoshi, Y.; Kamitani, H.; Watanabe, T.; Ohama, E.; Tahimic, C.G.; Kurimasa, A.; et al. Proteomics-based identification of differentially expressed genes in human gliomas: Down-regulation of SIRT2 gene. Biochem. Biophys. Res. Commun. 2003, 309, 558–566. [Google Scholar] [CrossRef]
- Groot, J.F.; Piao, Y.; Tran, H.; Gilbert, M.; Wu, H.K.; Liu, J.; Bekele, B.N.; Cloughesy, T.; Mehta, M.; Robins, H.I.; et al. Myeloid biomarkers associated with glioblastoma response to anti-VEGF therapy with aflibercept. Clin. Cancer Res. 2011, 17, 4872–4881. [Google Scholar] [CrossRef]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef]
- Moroni, M.; Veronese, S.; Benvenuti, S.; Marrapese, G.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Gambacorta, M.; Siena, S.; Bardelli, A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 2005, 6, 279–286. [Google Scholar] [CrossRef]
- Kalyankrishna, S.; Grandis, J.R. Epidermal growth factor receptor biology in head and neck cancer. J. Clin. Oncol. 2006, 24, 2666–2672. [Google Scholar] [CrossRef]
- Sainsbury, J.R.; Farndon, J.R.; Needham, G.K.; Malcolm, A.J.; Harris, A.L. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet 1987, 1, 1398–1402. [Google Scholar]
- Rossi, M.R.; La Duca, J.; Matsui, S.; Nowak, N.J.; Hawthorn, L.; Cowell, J.K. Novel amplicons on the short arm of chromosome 7 identified using high resolution array CGH contain over expressed genes in addition to EGFR in glioblastoma multiforme. Genes Chromosomes Cancer 2005, 44, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Perego, C.; Vanoni, C.; Massari, S.; Raimondi, A.; Pola, S.; Cattaneo, M.G.; Francolini, M.; Vicentini, L.M.; Pietrini, G. Invasive behaviour of glioblastoma cell lines is associated with altered organisation of the cadherin-catenin adhesion system. J. Cell Sci. 2002, 115, 3331–3340. [Google Scholar]
- Maksimovic, J.; Gordon, L.; Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome. Biol. 2012, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Kibbe, W.A.; Lin, S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Kim, K.I.; Wiel, M.A. Effects of dependence in high-dimensional multiple testing problems. BMC Bioinform. 2008, 9, 114. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Aoki-Kinoshita, K.F.; Kanehisa, M. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 2007, 396, 71–91. [Google Scholar] [CrossRef]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, 674–679. [Google Scholar] [CrossRef]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, 691–697. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Dahlquist, K.D.; Salomonis, N.; Vranizan, K.; Lawlor, S.C.; Conklin, B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002, 31, 19–20. [Google Scholar] [CrossRef] [Green Version]
- Petri, V.; Jayaraman, P.; Tutaj, M.; Hayman, G.T.; Smith, J.R.; De Pons, J.; Laulederkind, S.J.; Lowry, T.F.; Nigam, R.; Wang, S.J.; et al. The pathway ontology - updates and applications. J. Biomed. Semantics. 2014, 5, 7. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, 377–386. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, 258–261. [Google Scholar] [CrossRef]
- Alfarano, C.; Andrade, C.E.; Anthony, K.; Bahroos, N.; Bajec, M.; Bantoft, K.; Betel, D.; Bobechko, B.; Boutilier, K.; Burgess, E.; et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005, 33, 418–424. [Google Scholar] [CrossRef]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009, 37, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K.; O’Donnell, L.; Oster, S.; Theesfeld, C.; Sellam, A.; et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017, 45, D369–D379. [Google Scholar] [CrossRef]
- Giurgiu, M.; Reinhard, J.; Brauner, B.; Dunger-Kaltenbach, I.; Fobo, G.; Frishman, G.; Montrone, C.; Ruepp, A. CORUM: The comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019, 47, D559–D563. [Google Scholar] [CrossRef] [PubMed]
- Salwinski, L.; Miller, C.S.; Smith, A.J.; Pettit, F.K.; Bowie, J.U.; Eisenberg, D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004, 32, D449–D451. [Google Scholar] [CrossRef]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012, 40, D841–D846. [Google Scholar] [CrossRef]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef]
- Güldener, U.; Münsterkötter, M.; Oesterheld, M.; Pagel, P.; Ruepp, A.; Mewes, H.W.; Stümpflen, V. MPact: The MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006, 34, D436–D441. [Google Scholar] [CrossRef]
- Pagel, P.; Kovac, S.; Oesterheld, M.; Brauner, B.; Dunger-Kaltenbach, I.; Frishman, G.; Montrone, C.; Mark, P.; Stümpflen, V.; Mewes, H.W.; et al. The MIPS mammalian protein-protein interaction database. Bioinformatics 2005, 21, 832–834. [Google Scholar] [CrossRef]
- Brown, K.R.; Jurisica, I. Online predicted human interaction database. Bioinformatics 2005, 21, 2076–2082. [Google Scholar] [CrossRef] [PubMed]
- Razick, S.; Magklaras, G.; Donaldson, I.M. iRefIndex: A consolidated protein interaction database with provenance. BMC Bioinform. 2008, 9, 405. [Google Scholar] [CrossRef] [PubMed]
- Pritykin, Y.; Singh, M. Simple topological features reflect dynamics and modularity in protein interaction networks. PLoS Comput. Biol. 2013, 9, e1003243. [Google Scholar] [CrossRef] [PubMed]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Ng, H.K.; Srihari, S.; Leong, H.W.; Nesvizhskii, A.I. Examination of the relationship between essential genes in PPI network and hub proteins in reverse nearest neighbor topology. BMC Bioinform. 2010, 11, 505. [Google Scholar] [CrossRef] [PubMed]
- Zaki, N.; Efimov, D.; Berengueres, J. Protein complex detection using interaction reliability assessment and weighted clustering coefficient. BMC Bioinform. 2013, 14, 163. [Google Scholar] [CrossRef]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019, 47, D1013–D1017. [Google Scholar] [CrossRef]
- Ruepp, A.; Kowarsch, A.; Schmidl, D.; Buggenthin, F.; Brauner, B.; Dunger, I.; Fobo, G.; Frishman, G.; Montrone, C.; Theis, F.J. PhenomiR: A knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 2010, 11, R6. [Google Scholar] [CrossRef]
- Liu, X.; Wang, S.; Meng, F.; Wang, J.; Zhang, Y.; Dai, E.; Yu, X.; Li, X.; Jiang, W. SM2miR: A database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics 2013, 29, 409–411. [Google Scholar] [CrossRef]
- Rukov, J.L.; Wilentzik, R.; Jaffe, I.; Vinther, J.; Shomron, N. Pharmaco-miR: Linking microRNAs and drug effects. Brief. Bioinform. 2014, 15, 648–659. [Google Scholar] [CrossRef]
- Dai, E.; Yu, X.; Zhang, Y.; Meng, F.; Wang, S.; Liu, X.; Liu, D.; Wang, J.; Li, X.; Jiang, W. EpimiR: A database of curated mutual regulation between miRNAs and epigenetic modifications. Database (Oxf.) 2014, 2014, 23. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, R.K.; Chen, B.; Butte, A.J. Relating genes to function: Identifying enriched transcription factors using the ENCODE ChIP-Seq significance tool. Bioinformatics 2013, 29, 1922–1924. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef]
- Yook, S.H.; Oltvai, Z.N.; Barabási, A.L. Functional and topological characterization of protein interaction networks. Proteomics 2004, 4, 928–942. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Koschützki, D.; Schreiber, F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene. Regul. Syst. Bio. 2008, 2, 193–201. [Google Scholar] [CrossRef]
- Barrat, A.; Barthélemy, M.; Pastor-Satorras, R.; Vespignani, A. The architecture of complex weighted networks. Proc. Natl. Acad. Sci. USA 2004, 101, 3747–3752. [Google Scholar] [CrossRef] [Green Version]
- Baer, C.; Claus, R.; Plass, C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013, 73, 473–477. [Google Scholar] [CrossRef]
- Yang, J.Y.; Hung, M.C. A new fork for clinical application: Targeting forkhead transcription factors in cancer. Clin. Cancer Res. 2009, 15, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Sugawa, N.; Ekstrand, A.J.; James, C.D.; Collins, V.P. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc. Natl. Acad. Sci. USA 1990, 87, 8602–8606. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.F.; Miller, D.C.; Koslow, M.; Newcomb, E.W. Pathways leading to glioblastoma multiforme: A molecular analysis of genetic alterations in 65 astrocytic tumors. J. Neurosurg. 1994, 81, 427–436. [Google Scholar] [CrossRef]
- Matsumoto, T.; Kawashima, Y.; Nagashio, R.; Kageyama, T.; Kodera, Y.; Jiang, S.X.; Okayasu, I.; Kameya, T.; Sato, Y. A new possible lung cancer marker: VGF detection from the conditioned medium of pulmonary large cell neuroendocrine carcinoma-derived cells using secretome analysis. Int. J. Biol. Markers 2009, 24, 282–285. [Google Scholar] [CrossRef]
- Flanagan, J.; Healey, S.; Young, J.; Whitehall, V.; Chenevix-Trench, G. Analysis of the transcription regulator, CNOT7, as a candidate chromosome 8 tumor suppressor gene in colorectal cancer. Int. J. Cancer 2003, 106, 505–509. [Google Scholar] [CrossRef]
- Kanda, M.; Shimizu, D.; Tanaka, H.; Tanaka, C.; Kobayashi, D.; Hayashi, M.; Takami, H.; Niwa, Y.; Yamada, S.; Fujii, T.; et al. Synaptotagmin XIII expression and peritoneal metastasis in gastric cancer. Br. J. Surg. 2018, 105, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Lu, Y.R. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015, 15, 36. [Google Scholar] [CrossRef]
- Calcagno, D.Q.; Takeno, S.S.; Gigek, C.O.; Leal, M.F.; Wisnieski, F.; Chen, E.S.; Araújo, T.M.; Lima, E.M.; Melaragno, M.I.; Demachki, S.; et al. Identification of IL11RA and MELK amplification in gastric cancer by comprehensive genomic profiling of gastric cancer cell lines. World J. Gastroenterol. 2016, 22, 9506–9514. [Google Scholar] [CrossRef]
- Cai, D.; Cao, J.; Li, Z.; Zheng, X.; Yao, Y.; Li, W.; Yuan, Z. Up-regulation of bone marrow stromal protein 2 (BST2) in breast cancer with bone metastasis. BMC Cancer 2009, 9, 102. [Google Scholar] [CrossRef]
- Sun, W.; Fujimoto, J.; Tamaya, T. Coexpression of Gas6/Axl in human ovarian cancers. Oncology 2004, 66, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.C.; Chen, L.H.; Hsieh, Y.C.; Yang, P.W.; Lai, L.C.; Chuang, E.Y.; Lee, J.M.; Tsai, M.H. miR-338-5p inhibits cell proliferation, colony formation, migration, and cisplatin resistance in esophageal squamous cancer cells by targeting FERMT2. Carcinogenesis 2018. [Google Scholar] [CrossRef]
- Wu, Y.; Yu, H.; Zheng, S.L.; Feng, B.; Kapron, A.L.; Na, R.; Boyle, J.L.; Shah, S.; Shi, Z.; Ewing, C.M.; et al. Germline mutations in PPFIBP2 are associated with lethal prostate cancer. Prostate 2018, 78, 1222–1228. [Google Scholar] [CrossRef]
- Nakamura, M.; Ishida, E.; Shimada, K.; Nakase, H.; Sakaki, T.; Konishi, N. Frequent HRK inactivation associated with low apoptotic index in secondary glioblastomas. Acta Neuropathol. 2005, 110, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Sica, G.; Lama, G.; Anile, C.; Geloso, M.C.; La Torre, G.; De Bonis, P.; Maira, G.; Lauriola, L.; Jhanwar-Uniyal, M.; Mangiola, A. Assessment of angiogenesis by CD105 and nestin expression in peritumor tissue of glioblastoma. Int. J. Oncol. 2011, 38, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Teller, A.; Jechorek, D.; Hartig, R.; Adolf, D.; Reißig, K.; Roessner, A.; Franke, S. Dysregulation of apoptotic signaling pathways by interaction of RPLP0 and cathepsin X/Z in gastric cancer. Pathol. Res. Pract. 2015, 211, 62–70. [Google Scholar] [CrossRef]
- Quigley, D.A.; Fiorito, E.; Nord, S.; Van Loo, P.; Alnæs, G.G.; Fleischer, T.; Tost, J.; Moen Vollan, H.K.; Tramm, T.; Overgaard, J.; et al. The 5p12 breast cancer susceptibility locus affects MRPS30 expression in estrogen-receptor positive tumors. Mol. Oncol. 2014, 8, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tong, D.; Liu, F.; Li, D.; Li, J.; Cheng, X.; Wang, Z. RPS7 inhibits colorectal cancer growth via decreasing HIF-1α-mediated glycolysis. Oncotarget 2016, 7, 5800–5814. [Google Scholar] [CrossRef]
- Yu, M.J.; Zhao, N.; Shen, H.; Wang, H. Long Noncoding RNA MRPL39 Inhibits Gastric Cancer Proliferation and Progression by Directly Targeting miR-130. Genet. Test. Mol. Biomarkers 2018, 22, 656–663. [Google Scholar] [CrossRef]
- Gao, Y.; Li, F.; Zhou, H.; Yang, Y.; Wu, R.; Chen, Y.; Li, W.; Li, Y.; Xu, X.; Ke, C.; et al. Down-regulation of MRPS23 inhibits rat breast cancer proliferation and metastasis. Oncotarget 2017, 8, 71772–71781. [Google Scholar] [CrossRef]
- Dey, P.; Baddour, J.; Muller, F.; Wu, C.C.; Wang, H.; Liao, W.T.; Lan, Z.; Chen, A.; Gutschner, T.; Kang, Y.; et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 2017, 542, 119–123. [Google Scholar] [CrossRef]
- Li, L.D.; Sun, H.F.; Liu, X.X.; Gao, S.P.; Jiang, H.L.; Hu, X.; Jin, W. Down-Regulation of NDUFB9 Promotes Breast Cancer Cell Proliferation, Metastasis by Mediating Mitochondrial Metabolism. PLoS ONE 2015, 10, e0144441. [Google Scholar] [CrossRef]
- Wang, Y.; Sui, J.; Li, X.; Cao, F.; He, J.; Yang, B.; Zhu, X.; Sun, Y.; Pu, Y.D. RPS24 knockdown inhibits colorectal cancer cell migration and proliferation in vitro. Gene 2015, 571, 286–291. [Google Scholar] [CrossRef]
- Shriver, S.P.; Shriver, M.D.; Tirpak, D.L.; Bloch, L.M.; Hunt, J.D.; Ferrell, R.E.; Siegfried, J.M. Trinucleotide repeat length variation in the human ribosomal protein L14 gene (RPL14): Localization to 3p21.3 and loss of heterozygosity in lung and oral cancers. Mutat. Res. 1998, 406, 9–23. [Google Scholar] [CrossRef]
- Ji, P.; Wang, L.; Liu, J.; Mao, P.; Li, R.; Jiang, H.; Lou, M.; Xu, M.; Yu, X. Knockdown of RPL34 inhibits the proliferation and migration of glioma cells through the inactivation of JAK/STAT3 signaling pathway. J. Cell Biochem. 2019, 120, 3259–3267. [Google Scholar] [CrossRef]
- Jandial, R.; Neman, J.; Lim, P.P.; Tamae, D.; Kowolik, C.M.; Wuenschell, G.E.; Shuck, S.C.; Ciminera, A.K.; De Jesus, L.R.; Ouyang, C.; et al. Inhibition of GLO1 in Glioblastoma Multiforme Increases DNA-AGEs, Stimulates RAGE Expression, and Inhibits Brain Tumor Growth in Orthotopic Mouse Models. Int. J. Mol. Sci. 2018, 19, 406. [Google Scholar] [CrossRef]
- Li, J.; Zhu, S.; Tong, J.; Hao, H.; Yang, J.; Liu, Z.; Wang, Y. Suppression of lactate dehydrogenase A compromises tumor progression by downregulation of the Warburg effect in glioblastoma. Neuroreport 2016, 27, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Xi, J. Identification of COX5B as a novel biomarker in high-grade glioma patients. Onco Targets Ther. 2017, 10, 5463–5470. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, S.T.; Feng, Y.L.; Wan, T.; Gu, H.F.; Xu, J.; Yuan, L.J.; Zhou, Y.; Yu, X.J.; Huang, L.; et al. The Bidirectional Regulation between MYL5 and HIF-1α Promotes Cervical Carcinoma Metastasis. Theranostics 2017, 7, 3768–3780. [Google Scholar] [CrossRef]
- Elsheikh, S.E.; Green, A.R.; Rakha, E.A.; Samaka, R.M.; Ammar, A.A.; Powe, D.; Reis-Filho, J.S.; Ellis, I.O. Caveolin 1 and Caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br. J. Cancer 2008, 99, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Liu, T.; Fang, O.; Leach, L.; Hu, X.; Luo, Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene 2014, 33, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wei, H.; Gao, Z.; Chen, G.; Liu, Y.; Gao, X.; Bai, G.; He, S.; Liu, T.; Xu, W.; et al. COL5A1 may contribute the metastasis of lung adenocarcinoma. Gene 2018, 665, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Kamikawaji, K.; Seki, N.; Watanabe, M.; Mataki, H.; Kumamoto, T.; Takagi, K.; Mizuno, K.; Inoue, H. Regulation of LOXL2 and SERPINH1 by antitumor microRNA-29a in lung cancer with idiopathic pulmonary fibrosis. J. Hum. Genet. 2016, 61, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Kita, Y.; Mimori, K.; Tanaka, F.; Matsumoto, T.; Haraguchi, N.; Ishikawa, K.; Matsuzaki, S.; Fukuyoshi, Y.; Inoue, H.; Natsugoe, S.; et al. Clinical significance of LAMB3 and COL7A1 mRNA in esophageal squamous cell carcinoma. Eur. J. Surg. Oncol. 2009, 35, 52–58. [Google Scholar] [CrossRef]
- Zhao, Y.; Jia, L.; Mao, X.; Xu, H.; Wang, B.; Liu, Y. siRNA-targeted COL8A1 inhibits proliferation, reduces invasion and enhances sensitivity to D-limonence treatment in hepatocarcinoma cells. IUBMB Life 2009, 61, 74–79. [Google Scholar] [CrossRef]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Toritsuka, M.; Ohnishi, S.; Shimada, K.; Furuya, H.; Khadka, V.S.; Deng, Y.; et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017, 8, 36099–36114. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Duan, Y.T.; Li, J.F.; Su, L.P.; Yan, M.; Zhu, Z.G.; Liu, B.Y.; Yang, Q.M. Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget 2014, 5, 1885–1896. [Google Scholar] [CrossRef]
- Jian, H.; Zhao, Y.; Liu, B.; Lu, S. SEMA4b inhibits MMP9 to prevent metastasis of non-small cell lung cancer. Tumour. Biol. 2014, 35, 11051–11056. [Google Scholar] [CrossRef] [PubMed]
- Lal, G.; Hashimi, S.; Smith, B.J.; Lynch, C.F.; Zhang, L.; Robinson, R.A.; Weigel, R.J. Extracellular matrix 1 (ECM1) expression is a novel prognostic marker for poor long-term survival in breast cancer: A Hospital-based Cohort Study in Iowa. Ann. Surg. Oncol. 2009, 16, 2280–2287. [Google Scholar] [CrossRef]
- Ghellal, A.; Li, C.; Hayes, M.; Byrne, G.; Bundred, N.; Kumar, S. Prognostic significance of TGF beta 1 and TGF beta 3 in human breast carcinoma. Anticancer Res. 2000, 20, 4413–4418. [Google Scholar] [PubMed]
- Endo, M.; Yamamoto, Y.; Nakano, M.; Masuda, T.; Odagiri, H.; Horiguchi, H.; Miyata, K.; Kadomatsu, T.; Motokawa, I.; Okada, S.; et al. Serum ANGPTL2 levels reflect clinical features of breast cancer patients: Implications for the pathogenesis of breast cancer metastasis. Int. J. Biol. Markers 2014, 29, e239–e245. [Google Scholar] [CrossRef]
- Pandey, P.; Kharbanda, S.; Kufe, D. Association of the DF3/MUC1 breast cancer antigen with Grb2 and the Sos/Ras exchange protein. Cancer Res. 1995, 55, 4000–4003. [Google Scholar]
- Cairns, J.; Ingle, J.N.; Wickerham, L.D.; Weinshilboum, R.; Liu, M.; Wang, L. SNPs near the cysteine proteinase cathepsin O gene (CTSO) determine tamoxifen sensitivity in ERα-positive breast cancer through regulation of BRCA1. PLoS Genet. 2017, 13, e1007031. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.J.; Liu, Y.; Zhou, X.L.; Wang, F.Z.; Zhang, X.D.; Ye, L.H. Myosin light chain kinase is responsible for high proliferative ability of breast cancer cells via anti-apoptosis involving p38 pathway. Acta Pharmacol. Sin. 2010, 31, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ding, Y.; Li, A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J. Surg. Oncol. 2016, 14, 297. [Google Scholar] [CrossRef]
- Hamada, S.; Masamune, A.; Miura, S.; Satoh, K.; Shimosegawa, T. MiR-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein SHC1 and pro-apoptotic regulator BAX. Cell Signal. 2014, 26, 179–185. [Google Scholar] [CrossRef]
- Wragg, J.W.; Finnity, J.P.; Anderson, J.A.; Ferguson, H.J.; Porfiri, E.; Bhatt, R.I.; Murray, P.G.; Heath, V.L.; Bicknell, R. MCAM and LAMA4 Are Highly Enriched in Tumor Blood Vessels of Renal Cell Carcinoma and Predict Patient Outcome. Cancer Res. 2016, 76, 2314–2326. [Google Scholar] [CrossRef]
- Bartolini, A.; Cardaci, S.; Lamba, S.; Oddo, D.; Marchiò, C.; Cassoni, P.; Amoreo, C.A.; Corti, G.; Testori, A.; Bussolino, F.; et al. BCAM and LAMA5 Mediate the Recognition between Tumor Cells and the Endothelium in the Metastatic Spreading of KRAS-Mutant Colorectal Cancer. Clin. Cancer Res. 2016, 22, 4923–4933. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Adachi, Y.; Nagayama, T. Expression of a secretory protein C1qTNF6, a C1qTNF family member, in hepatocellular carcinoma. Anal. Cell. Pathol. (Amst.) 2011, 34, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.T.; Liu, X.P.; Liu, T.Z.; Wang, X.H. The clinical significance of COL5A2 in patients with bladder cancer: A retrospective analysis of bladder cancer gene expression data. Med. (Baltim.) 2018, 97, e0091. [Google Scholar] [CrossRef]
- Glait-Santar, C.; Benayahu, D. Regulation of SVEP1 gene expression by 17β-estradiol and TNFα in pre-osteoblastic and mammary adenocarcinoma cells. J. Steroid Biochem. Mol. Biol. 2012, 130, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Renard, I.; Joniau, S.; Cleynenbreugel, B.; Collette, C.; Naômé, C.; Vlassenbroeck, I.; Nicolas, H.; Leval, J.; Straub, J.; Criekinge, W.; et al. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur. Urol. 2010, 58, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Allione, A.; Pardini, B.; Viberti, C.; Giribaldi, G.; Turini, S.; Di Gaetano, C.; Guarrera, S.; Cordero, F.; Oderda, M.; Allasia, M.; et al. MMP23B expression and protein levels in blood and urine are associated with bladder cancer. Carcinogenesis 2018, 39, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Smith, P.; Purdie, C.; Quinlan, P.; Baker, L.; Aman, P.; Thompson, A.M.; Crook, T. The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer. Br. J. Cancer 2009, 100, 1687–1696. [Google Scholar] [CrossRef]
- Lo, P.H.; Tanikawa, C.; Katagiri, T.; Nakamura, Y.; Matsuda, K. Identification of novel epigenetically inactivated gene PAMR1 in breast carcinoma. Oncol. Rep. 2015, 33, 267–273. [Google Scholar] [CrossRef]
- Khaket, T.P.; Singh, M.P.; Khan, I.; Bhardwaj, M.; Kang, S.C. Targeting of cathepsin C induces autophagic dysregulation that directs ER stress mediated cellular cytotoxicity in colorectal cancer cells. Cell Signal. 2018, 46, 92–102. [Google Scholar] [CrossRef]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genßler, S.; Schönfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ERBB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2015, 108. [Google Scholar] [CrossRef] [PubMed]
- Szerlip, N.J.; Pedraza, A.; Chakravarty, D.; Azim, M.; McGuire, J.; Fang, Y.; Ozawa, T.; Holland, E.C.; Huse, J.T.; Jhanwar, S.; et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. USA 2012, 109, 3041–3046. [Google Scholar] [CrossRef] [Green Version]
- Plate, K.H.; Breier, G.; Farrell, C.L.; Risau, W. Platelet-derived growth factor receptor-beta is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas. Lab. Investig. 1992, 67, 529–534. [Google Scholar]
- Feng, L.; Ma, J.; Ji, H.; Liu, Y.; Hu, W. miR-330-5p suppresses glioblastoma cell proliferation and invasiveness through targeting ITGA5. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, F.E.; Lamba, S.; Zanon, C.; Tilborg, A.A.; Leenstra, S.; Troost, D.; Hulsebos, T.; Vandertop, W.P.; Bardelli, A. Absence of AKT1 mutations in glioblastoma. PLoS ONE 2009, 4, e5638. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Y.; Wu, Y.; Gao, Y.; Li, Q.; Abdulrahman, A.A.; Liu, X.F.; Ji, G.Q.; Gao, J.; Li, L.; et al. Identification of COL1A1 as an invasion-related gene in malignant astrocytoma. Int. J. Oncol. 2018, 53, 2542–2554. [Google Scholar] [CrossRef]
- Fujita, A.; Sato, J.R.; Festa, F.; Gomes, L.R.; Oba-Shinjo, S.M.; Marie, S.K.; Ferreira, C.E.; Sogayar, M.C. Identification of COL6A1 as a differentially expressed gene in human astrocytomas. Genet. Mol. Res. 2008, 7, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Kazuno, M.; Tokunaga, T.; Oshika, Y.; Tanaka, Y.; Tsugane, R.; Kijima, H.; Yamazaki, H.; Ueyama, Y.; Nakamura, M. Thrombospondin-2 (TSP2) expression is inversely correlated with vascularity in glioma. Eur. J. Cancer 1999, 35, 502–506. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, S.; Wang, J.; Long, H.; Fang, L.; Wang, G.; Li, Z.; Que, T.; Liu, Y.; Li, Y.; et al. Hypoxia-induced PLOD2 promotes proliferation, migration and invasion via PI3K/Akt signaling in glioma. Oncotarget 2017, 8, 41947–41962. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Jin, G.; Mi, R.; Zhang, J.; Zhang, J.; Xu, H.; Cheng, S.; Zhang, Y.; Song, W.; Liu, F. Knockdown of P4HA1 inhibits neovascularization via targeting glioma stem cell-endothelial cell transdifferentiation and disrupting vascular basement membrane. Oncotarget 2017, 8, 35877–35889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, S.; Czub, S.; Greif, M.; Vince, G.H.; Süss, N.; Kerkau, S.; Rieckmann, P.; Roggendorf, W.; Roosen, K.; Tonn, J.C. Microglial/macrophage expression of interleukin 10 in human glioblastomas. Int. J. Cancer 1999, 82, 12–16. [Google Scholar] [CrossRef]
- Gagliano, N.; Moscheni, C.; Torri, C.; Magnani, I.; Bertelli, A.A.; Gioia, M. Effect of resveratrol on matrix metalloproteinase-2 (MMP-2) and Secreted Protein Acidic and Rich in Cysteine (SPARC) on human cultured glioblastoma cells. Biomed. Pharmacother. 2005, 59, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Kast, R.E. The role of interleukin-18 in glioblastoma pathology implies therapeutic potential of two old drugs-disulfiram and ritonavir. Chin. J. Cancer 2015, 34, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhao, J.; Zhang, L.; Zhao, J.; Zhuang, Y.; Liang, P. SRPX2 Enhances the Epithelial-Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Cells. Cell. Mol. Neurobiol. 2016, 36, 1067–1076. [Google Scholar] [CrossRef]
- Vakilian, A.; Khorramdelazad, H.; Heidari, P.; Sheikh Rezaei, Z.; Hassanshahi, G. CCL2/CCR2 signaling pathway in glioblastoma multiforme. Neurochem Int. 2017, 103, 1–7. [Google Scholar] [CrossRef]
- Yamashita, D.; Kondo, T.; Ohue, S.; Takahashi, H.; Ishikawa, M.; Matoba, R.; Suehiro, S.; Kohno, S.; Harada, H.; Tanaka, J.; et al. miR340 suppresses the stem-like cell function of glioma-initiating cells by targeting tissue plasminogen activator. Cancer Res. 2015, 75, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Mishima, K.; Mazar, A.P.; Gown, A.; Skelly, M.; Ji, X.D.; Wang, X.D.; Jones, T.R.; Cavenee, W.K.; Huang, H.J. A peptide derived from the non-receptor-binding region of urokinase plasminogen activator inhibits glioblastoma growth and angiogenesis in vivo in combination with cisplatin. Proc. Natl. Acad. Sci. USA 2000, 97, 8484–8489. [Google Scholar] [CrossRef]
- Sciaccaluga, M.; Fioretti, B.; Catacuzzeno, L.; Pagani, F.; Bertollini, C.; Rosito, M.; Catalano, M.; D’Alessandro, G.; Santoro, A.; Cantore, G.; et al. CXCL12-induced glioblastoma cell migration requires intermediate conductance Ca2+-activated K+ channel activity. Am. J. Physiol. Cell Physiol. 2010, 299, C175–C184. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak-Celińska, A.; Słocińska, M.; Barciszewska, A.M.; Nowak, S.; Baer-Dubowska, W. Wnt pathway antagonists, SFRP1, SFRP2, SOX17, and PPP2R2B, are methylated in gliomas and SFRP1 methylation predicts shorter survival. J. Appl. Genet. 2016, 57, 189–197. [Google Scholar] [CrossRef]
- Sie, M.; Wagemakers, M.; Molema, G.; Mooij, J.J.; Bont, E.S.; Dunnen, W.F. The angiopoietin 1/angiopoietin 2 balance as a prognostic marker in primary glioblastoma multiforme. J. Neurosurg. 2009, 110, 147–155. [Google Scholar] [CrossRef]
- Zeng, A.; Yin, J.; Wang, Z.; Zhang, C.; Li, R.; Zhang, Z.; Yan, W.; You, Y. miR-17-5p-CXCL14 axis related transcriptome profile and clinical outcome in diffuse gliomas. Oncoimmunology 2018, 7, e1510277. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Yang, Q.Y.; Sai, K.; Chen, F.R.; Pang, J.C.; Ng, H.K.; Kwan, A.L.; Chen, Z.P. TGM2 inhibition attenuates ID1 expression in CD44-high glioma-initiating cells. Neuro Oncol. 2013, 15, 1353–1365. [Google Scholar] [CrossRef] [Green Version]
- Nie, E.; Miao, F.; Jin, X.; Wu, W.; Zhou, X.; ZENG, A.; Yu, T.; Zhi, T.; Shi, Z.; Wang, Y.; et al. Fstl1/DIP2A/MGMT signaling pathway plays important roles in temozolomide resistance in glioblastoma. Oncogene 2018. [Google Scholar] [CrossRef]
- Santosh, V.; Arivazhagan, A.; Sreekanthreddy, P.; Srinivasan, H.; Thota, B.; Srividya, M.R.; Vrinda, M.; Sridevi, S.; Shailaja, B.C.; Samuel, C.; et al. Grade-specific expression of insulin-like growth factor-binding proteins-2,-3, and -5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Pen, A.; Moreno, M.J.; Durocher, Y.; Deb-Rinker, P.; Stanimirovic, D.B. Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene 2008, 27, 6834–6844. [Google Scholar] [CrossRef]
- Laczko, R.; Szauter, K.M.; Jansen, M.K.; Hollosi, P.; Muranyi, M.; Molnar, J.; Fong, K.S.; Hinek, A.; Csiszar, K. Active lysyl oxidase (LOX) correlates with focal adhesion kinase (FAK)/paxillin activation and migration in invasive astrocytes. Neuropathol. Appl. Neurobiol. 2007, 33, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, L.; Yin, L. Silencing LncRNA LOXL1-AS1 attenuates mesenchymal characteristics of glioblastoma via NF-κB pathway. Biochem. Biophys. Res. Commun. 2018, 500, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Atala, A. eIF4E Phosphorylation Promotes Tumorigenesis and is Associated with Prostate Cancer Progression. J. Urol. 2011, 185, 1533. [Google Scholar] [CrossRef]
- Holmberg Olausson, K.; Elsir, T.; Moazemi Goudarzi, K.; Nistér, M.; Lindström, M.S. NPM1 histone chaperone is upregulated in glioblastoma to promote cell survival and maintain nucleolar shape. Sci. Rep. 2015, 5, 16495. [Google Scholar] [CrossRef]
- Kochanek, D.M.; Wells, D.G. CPEB1 regulates the expression of MTDH/AEG-1 and glioblastoma cell migration. Mol. Cancer Res. 2013, 11, 149–160. [Google Scholar] [CrossRef]
- Ni, X.G.; Zhou, L.; Wang, G.Q.; Liu, S.M.; Bai, X.F.; Liu, F.; Peppelenbosch, M.P.; Zhao, P. The ubiquitin-proteasome pathway mediates gelsolin protein downregulation in pancreatic cancer. Mol. Med. 2008, 14, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Enokida, H.; Tatarano, S.; Kawahara, K.; Uchida, Y.; Nishiyama, K.; Fujimura, L.; Kikkawa, N.; Seki, N.; Nakagawa, M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br. J. Cancer 2010, 102, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Zhou, T.; Li, Y.; Yu, Z.; Sun, L. p53 target miR-29c-3p suppresses colon cancer cell invasion and migration through inhibition of PHLDB2. Biochem. Biophys. Res. Commun. 2017, 487, 90–95. [Google Scholar] [CrossRef]
- Chang, C.Y.; Lin, S.C.; Su, W.H.; Ho, C.M.; Jou, Y.S. Somatic LMCD1 mutations promoted cell migration and tumor metastasis in hepatocellular carcinoma. Oncogene 2012, 31, 2640–2652. [Google Scholar] [CrossRef]
- Raghu, H.; Lakka, S.S.; Gondi, C.S.; Mohanam, S.; Dinh, D.H.; Gujrati, M.; Rao, J.S. Suppression of uPA and uPAR attenuates angiogenin mediated angiogenesis in endothelial and glioblastoma cell lines. PLoS ONE 2010, 5, e12458. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, H.; Wang, K.; Zhang, C.; Wang, Y.; Yao, K.; Yang, P.; Han, L.; Kang, C.; Zhang, W.; et al. Multidimensional analysis of gene expression reveals TGFB1I1-induced EMT contributes to malignant progression of astrocytomas. Oncotarget 2014, 5, 12593–12606. [Google Scholar] [CrossRef]
- Ren, S.; Xu, C.; Cui, Z.; Yu, Y.; Xu, W.; Wang, F.; Lu, J.; Wei, M.; Lu, X.; Gao, X.; et al. Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. J. Mol. Med. (Berl.) 2012, 90, 1121–1132. [Google Scholar] [CrossRef]
- Li, B.; Chen, M.; Lu, M.; Xin-Xiang, J.; Meng-Xiong, P.; Jun-Wu, M. Glutaredoxin 3 promotes migration and invasion via the Notch signalling pathway in oral squamous cell carcinoma. Free Radic. Res. 2018, 52, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Roessler, M.; Rollinger, W.; Mantovani-Endl, L.; Hagmann, M.L.; Palme, S.; Berndt, P.; Engel, A.M.; Pfeffer, M.; Karl, J.; Bodenmüller, H.; et al. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell Proteomics 2006, 5, 2092–2101. [Google Scholar] [CrossRef]
- Li, G.; Li, M.; Liang, X.; Xiao, Z.; Zhang, P.; Shao, M.; Peng, F.; Chen, Y.; Li, Y.; Chen, Z. Identifying DCN and HSPD1 as Potential Biomarkers in Colon Cancer Using 2D-LC-MS/MS Combined with iTRAQ Technology. J. Cancer 2017, 8, 479–489. [Google Scholar] [CrossRef]
- Idbaih, A.; Paris, S.; Boisselier, B.; Marie, Y.; Sanson, M.; Thillet, J.; Hoang-Xuan, K.; Delattre, J.Y. Mutational analysis of RAC2 in gliomas. J. Neurooncol. 2008, 87, 365–366. [Google Scholar] [CrossRef]
- Gray, G.K.; McFarland, B.C.; Nozell, S.E.; Benveniste, E.N. NF-κB and STAT3 in glioblastoma: Therapeutic targets coming of age. Expert Rev. Neurother. 2014, 14, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Golan-Gerstl, R.; Cohen, M.; Shilo, A.; Suh, S.S.; Bakàcs, A.; Coppola, L.; Karni, R. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011, 71, 4464–4472. [Google Scholar] [CrossRef]
- Kiviniemi, A.; Gardberg, M.; Kivinen, K.; Posti, J.P.; Vuorinen, V.; Sipilä, J.; Rahi, M.; Sankinen, M.; Minn, H. Somatostatin receptor 2A in gliomas: Association with oligodendrogliomas and favourable outcome. Oncotarget 2017, 8, 49123–49132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desotelle, J.; Truong, M.; Ewald, J.; Weeratunga, P.; Yang, B.; Huang, W.; Jarrard, D. CpG island hypermethylation frequently silences FILIP1L isoform 2 expression in prostate cancer. J. Urol. 2013, 189, 329–335. [Google Scholar] [CrossRef]
- Cha, Y.; Kim, D.K.; Hyun, J.; Kim, S.J.; Park, K.S. TCEA3 binds to TGF-beta receptor I and induces Smad-independent, JNK-dependent apoptosis in ovarian cancer cells. Cell Signal. 2013, 25, 1245–1251. [Google Scholar] [CrossRef]
- Hallal, S.; Russell, B.P.; Wei, H.; Lee, M.Y.T.; Toon, C.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Kaufman, K.L. Extracellular Vesicles from Neurosurgical Aspirates Identifies Chaperonin Containing TCP1 Subunit 6A as a Potential Glioblastoma Biomarker with Prognostic Significance. Proteomics 2019, 19, e1800157. [Google Scholar] [CrossRef]
- Khor, G.H.; Froemming, G.R.; Zain, R.B.; Abraham, T.M.; Lin, T.K. Involvement of CELSR3 Hypermethylation in Primary Oral Squamous Cell Carcinoma. Asian Pac. J. Cancer Prev. 2016, 17, 219–223. [Google Scholar] [CrossRef]
- Sibin, M.K.; Harshitha, S.M.; Narasingarao, K.V.; Dhananjaya, I.B.; Dhaval, P.S.; Chetan, G.K. Effect of rs11614913 Polymorphism on Mature miR196a2 Expression and its Target Gene HOXC8 Expression in Human Glioma. J. Mol. Neurosci. 2017, 61, 144–151. [Google Scholar] [CrossRef]
- Alrfaei, B.M.; Vemuganti, R.; Kuo, J.S. microRNA-100 targets SMRT/NCOR2, reduces proliferation, and improves survival in glioblastoma animal models. PLoS ONE 2013, 8, e80865. [Google Scholar] [CrossRef]
- Ye, Y.; Tang, X.; Sun, Z.; Chen, S. Upregulated WDR26 serves as a scaffold to coordinate PI3K/AKT pathway-driven breast cancer cell growth, migration, and invasion. Oncotarget 2016, 7, 17854–17869. [Google Scholar] [CrossRef]
- Wang, X.; Wang, K.; Han, L.; Zhang, A.; Shi, Z.; Zhang, K.; Zhang, H.; Yang, S.; Pu, P.; Shen, C.; et al. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/β-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett. 2013, 331, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Heravi-Moussavi, A.; Anglesio, M.S.; Cheng, S.W.; Senz, J.; Yang, W.; Prentice, L.; Fejes, A.P.; Chow, C.; Tone, A.; Kalloger, S.E.; et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012, 366, 234–242. [Google Scholar] [CrossRef]
- Du, H.Q.; Wang, Y.; Jiang, Y.; Wang, C.H.; Zhou, T.; Liu, H.Y.; Xiao, H. Silencing of the TPM1 gene induces radioresistance of glioma U251 cells. Oncol. Rep. 2015, 33, 2807–2814. [Google Scholar] [CrossRef]
- Srivastava, C.; Irshad, K.; Dikshit, B.; Chattopadhyay, P.; Sarkar, C.; Gupta, D.K.; Sinha, S.; Chosdol, K. FAT1 modulates EMT and stemness genes expression in hypoxic glioblastoma. Int. J. Cancer 2018, 142, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Hoelzinger, D.B.; Mariani, L.; Weis, J.; Woyke, T.; Berens, T.J.; McDonough, W.S.; Sloan, A.; Coons, S.W.; Berens, M.E. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia 2005, 7, 7–16. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshabi, A.M.; Vastrad, B.; Shaikh, I.A.; Vastrad, C. Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis. Biomolecules 2019, 9, 201. https://doi.org/10.3390/biom9050201
Alshabi AM, Vastrad B, Shaikh IA, Vastrad C. Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis. Biomolecules. 2019; 9(5):201. https://doi.org/10.3390/biom9050201
Chicago/Turabian StyleAlshabi, Ali Mohamed, Basavaraj Vastrad, Ibrahim Ahmed Shaikh, and Chanabasayya Vastrad. 2019. "Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis" Biomolecules 9, no. 5: 201. https://doi.org/10.3390/biom9050201
APA StyleAlshabi, A. M., Vastrad, B., Shaikh, I. A., & Vastrad, C. (2019). Identification of Crucial Candidate Genes and Pathways in Glioblastoma Multiform by Bioinformatics Analysis. Biomolecules, 9(5), 201. https://doi.org/10.3390/biom9050201





