Computational Simulation of Adapter Length-Dependent LASSO Probe Capture Efficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Long-Adapter Single-Stranded Oligonucleotide Probe & Target Cloning
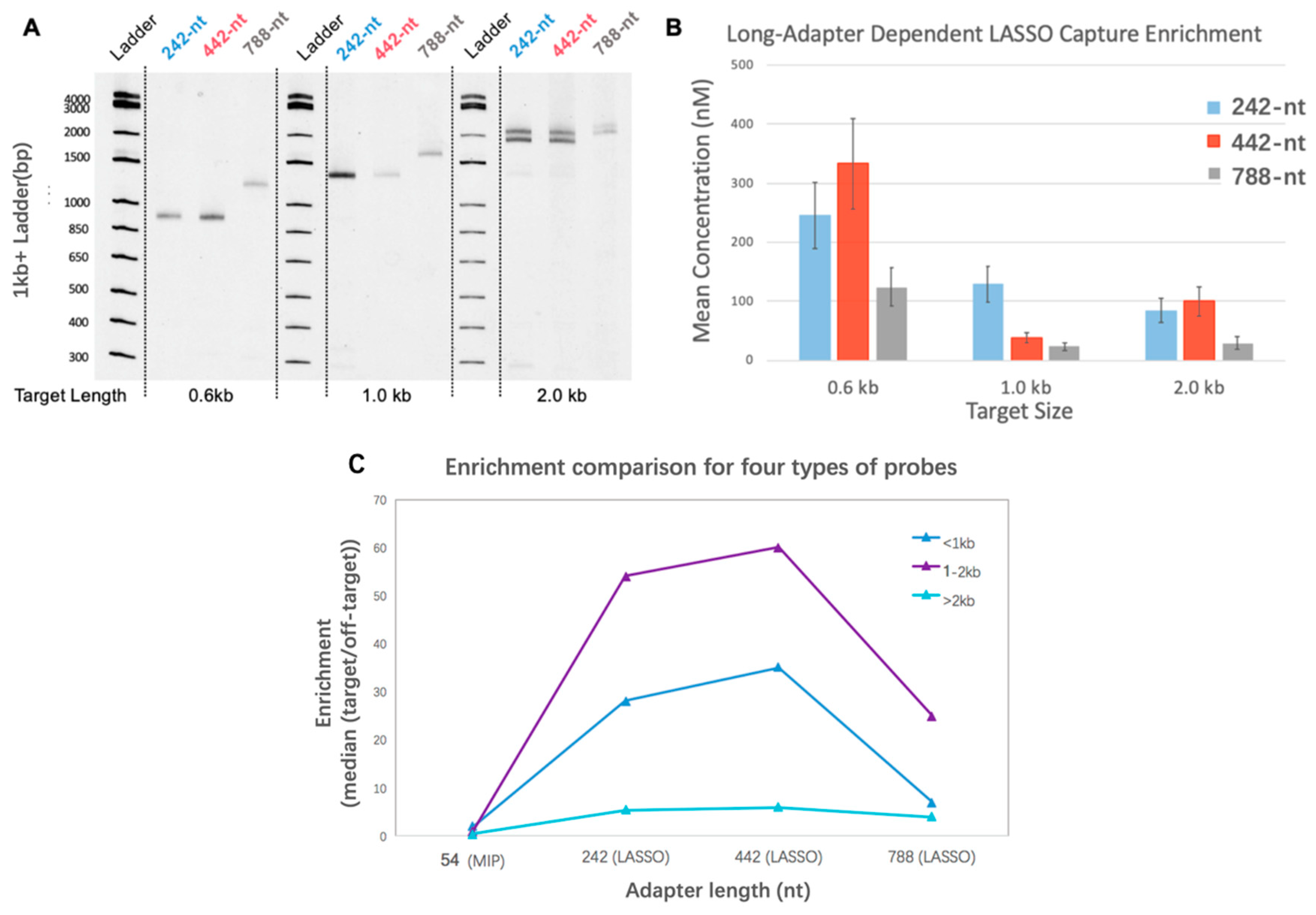
2.2. LASSO Capture Quantification for Single-Target and Library Experiments
2.3. Probe–Target Interaction Simulation
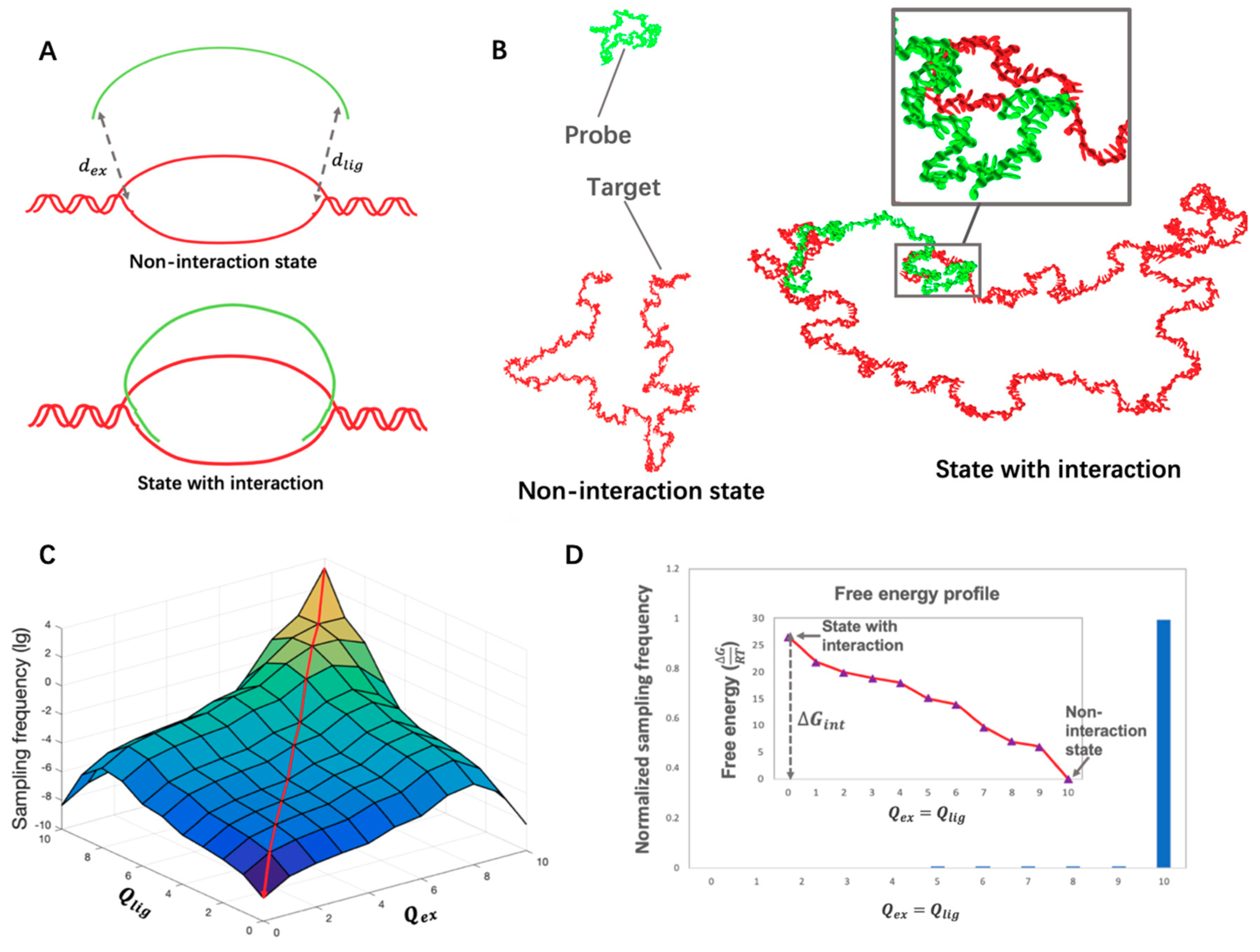
2.3.1. Order Parameter
2.3.2. Umbrella Sampling
2.3.3. Box Size and Salt Concentration
2.3.4. Procedure of Simulation
3. Results
3.1. Adapter Length-Dependent LASSO Probe Efficiency
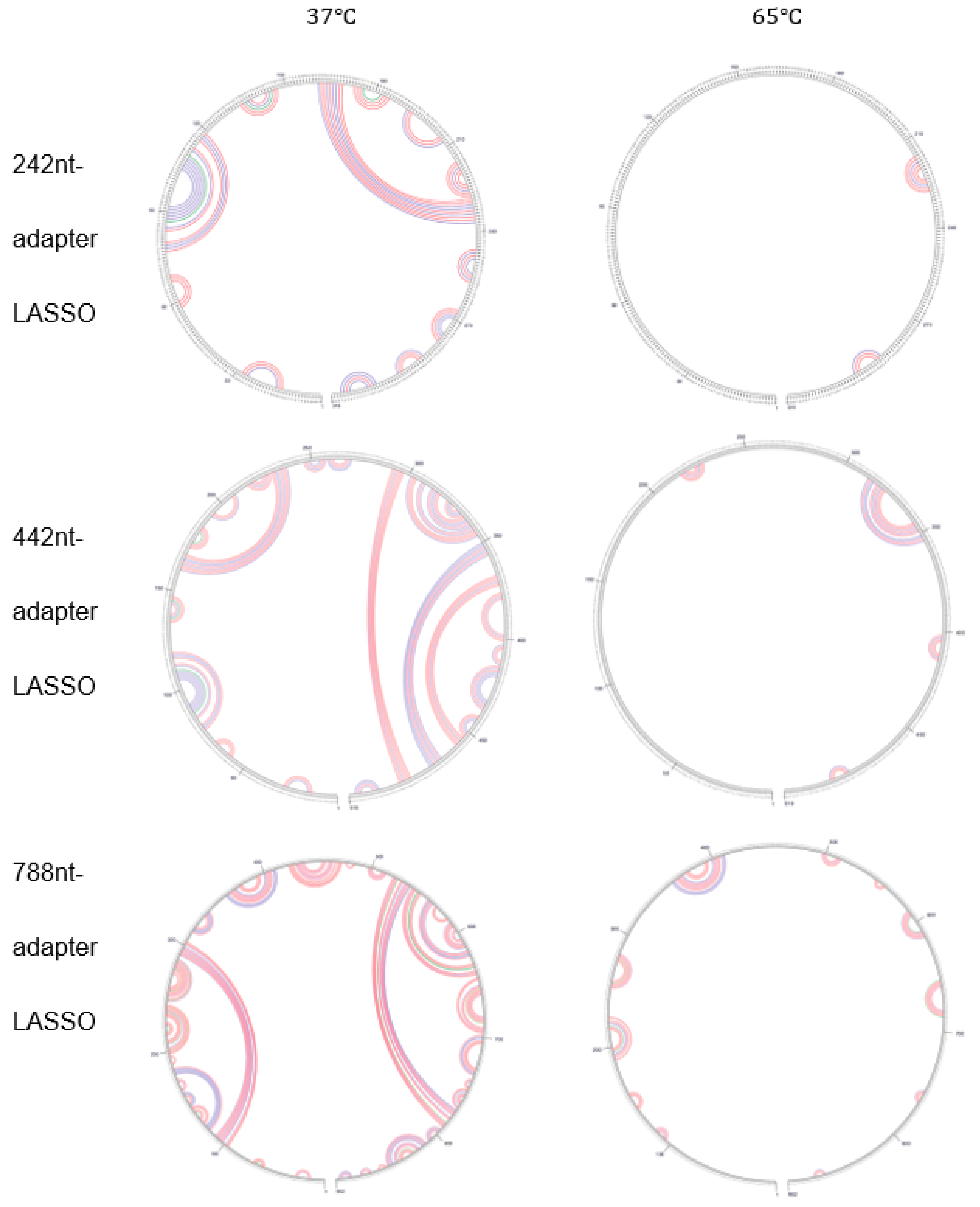
3.2. The Effect of Secondary Structure Formed by LASSO
3.3. Calculation of Free Energy of Probe–Target Interaction
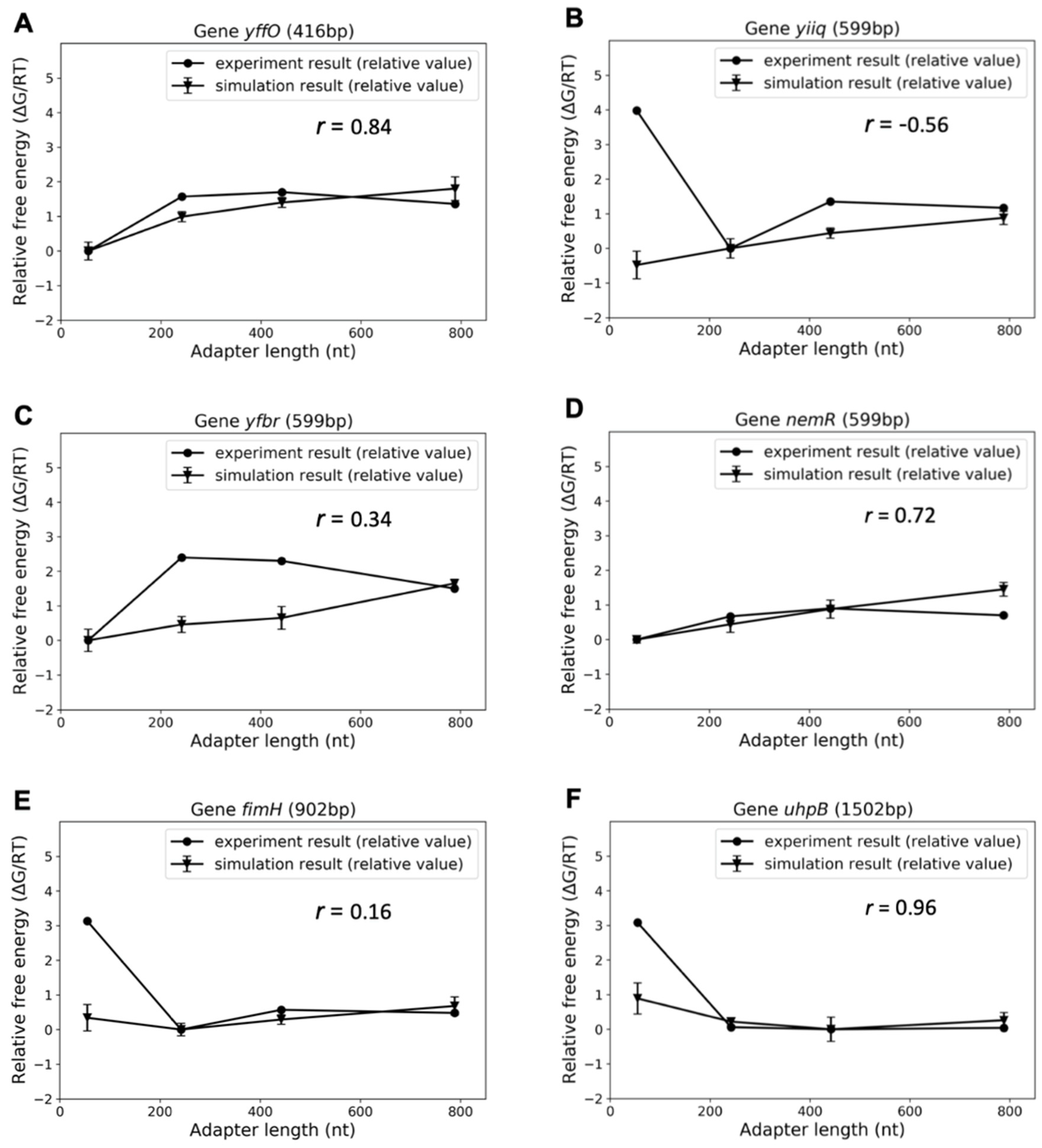
3.4. Comparison with Experiments
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tosi, L.; Sridhara, V.; Yang, Y.; Guan, D.; Shpilker, P.; Segata, N.; Larman, H.B.; Parekkadan, B. Long-adapter single-strand oligonucleotide probes for the massively multiplexed cloning of kilobase genome regions. Nat. Biomed. Eng. 2017, 1, 0092. [Google Scholar] [CrossRef]
- Landegren, U.; Schallmeiner, E.; Nilsson, M.; Fredriksson, S.; Baner, J.; Gullberg, M.; Jarvius, J.; Gustafsdottir, S.; Dahl, F.; Soderberg, O.; et al. Molecular tools for a molecular medicine: Analyzing genes, transcripts and proteins using padlock and proximity probes. J. Mol. Recognit. 2004, 17, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Hardenbol, P.; Banér, J.; Jain, M.; Nilsson, M.; Namsaraev, E.A.; Karlin-Neumann, G.A.; Davis, R.W. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003, 21, 673. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Wang, W.; Krishnakumar, S.; Palm, C.; Chi, A.K.; Enns, G.M.; Davis, R.W.; Speed, T.P.; Mindrinos, M.N.; Scharfe, C. High-quality DNA sequence capture of 524 disease candidate genes. Proc. Natl. Acad. Sci. USA 2011, 108, 6549–6554. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, J.B.; Pritchard, C.C.; Salipante, S.J.; O’Roak, B.J.; Shendure, J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013, 23, 843–854. [Google Scholar] [CrossRef]
- Ouldridge, T.E.; Louis, A.A.; Doye, J.P. DNA nanotweezers studied with a coarse-grained model of DNA. Phys. Rev. Lett. 2010, 104, 178101. [Google Scholar] [CrossRef]
- Harrison, R.M.; Romano, F.; Ouldridge, T.E.; Louis, A.A.; Doye, J.P. Coarse-grained modelling of strong DNA bending II: Cyclization. Arx. Prepr. Arx. 2015, 1506, 09008. [Google Scholar]
- Whitelam, S.; Geissler, P.L. Avoiding unphysical kinetic traps in Monte Carlo simulations of strongly attractive particles. J. Chem. Phys. 2007, 127, 154101. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmidet, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Ouldridge, T.E. Coarse-Grained Modelling of DNA and DNA Self-Assembly. Ph.D. Thesis, University of Oxford, Oxford, UK, 2011. [Google Scholar]
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- Grigoryev, S.A.; Arya, G.; Correll, S.; Woodcock, C.L.; Schlick, T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA 2009, 106, 13317–13322. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Flory, P.J.; Volkenstein, M. Statistical Mechanics of Chain Molecules; Wiley Online Library: Hoboken, NJ, USA, 1969. [Google Scholar]
- Ouldridge, T.E.; Louis, A.A.; Doye, J.P. Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. J. Chem. Phys. 2011, 134, 085101. [Google Scholar] [CrossRef] [PubMed]
- Ouldridge, T.E.; Sulc, P.; Romano, F.; Doye, J.P.; Louis, A.A. DNA hybridization kinetics: Zippering, internal displacement and sequence dependence. Nucleic Acids Res. 2013, 41, 8886–8895. [Google Scholar] [CrossRef] [PubMed]
- Sulc, P.; Romano, F.; Ouldridge, T.E.; Rovigatti, L.; Doye, J.P.; Louis, A.A. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012, 137, 135101. [Google Scholar] [CrossRef] [PubMed]
- Romano, F.; Hudson, A.; Doye, J.P.; Ouldridge, T.E.; Louis, A.A. The effect of topology on the structure and free energy landscape of DNA kissing complexes. J. Chem. Phys. 2012, 136, 215102. [Google Scholar] [CrossRef]
- Wang, Q.; Pettitt, B.M. Sequence Affects the Cyclization of DNA Minicircles. J. Phys. Chem. Lett. 2016, 7, 1042–1046. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Lee Rodgers, J.; Nicewander, W.A. Thirteen ways to look at the correlation coefficient. Am. Stat. 1988, 42, 59–66. [Google Scholar] [CrossRef]
- Tee, S.R.; Wang, Z. How well can DNA rupture DNA? Shearing and unzipping forces inside DNA nanostructures. ACS Omega 2018, 3, 292–301. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Shukor, S.; Li, S.; Tamayo, A.; Tosi, L.; Larman, B.; Nanda, V.; Olson, W.K.; Parekkadan, B. Computational Simulation of Adapter Length-Dependent LASSO Probe Capture Efficiency. Biomolecules 2019, 9, 199. https://doi.org/10.3390/biom9050199
Liu J, Shukor S, Li S, Tamayo A, Tosi L, Larman B, Nanda V, Olson WK, Parekkadan B. Computational Simulation of Adapter Length-Dependent LASSO Probe Capture Efficiency. Biomolecules. 2019; 9(5):199. https://doi.org/10.3390/biom9050199
Chicago/Turabian StyleLiu, Jingqian, Syukri Shukor, Shuxiang Li, Alfred Tamayo, Lorenzo Tosi, Benjamin Larman, Vikas Nanda, Wilma K. Olson, and Biju Parekkadan. 2019. "Computational Simulation of Adapter Length-Dependent LASSO Probe Capture Efficiency" Biomolecules 9, no. 5: 199. https://doi.org/10.3390/biom9050199
APA StyleLiu, J., Shukor, S., Li, S., Tamayo, A., Tosi, L., Larman, B., Nanda, V., Olson, W. K., & Parekkadan, B. (2019). Computational Simulation of Adapter Length-Dependent LASSO Probe Capture Efficiency. Biomolecules, 9(5), 199. https://doi.org/10.3390/biom9050199



