Mechanisms of Spindle Positioning: Lessons from Worms and Mammalian Cells
Abstract
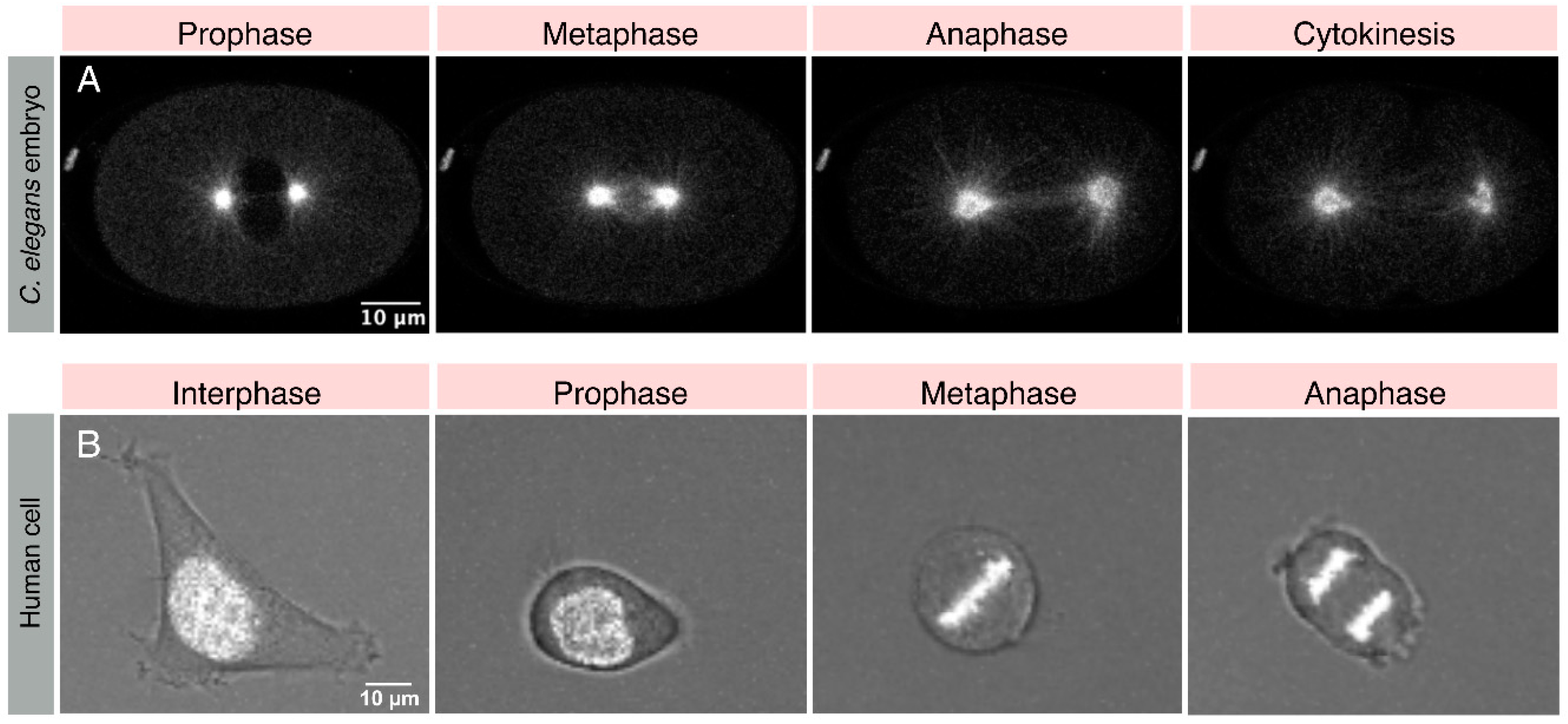
1. Introduction
2. Regulation of Spindle Positioning: Function of Key Players, Physical Environment, and Chemical Cues
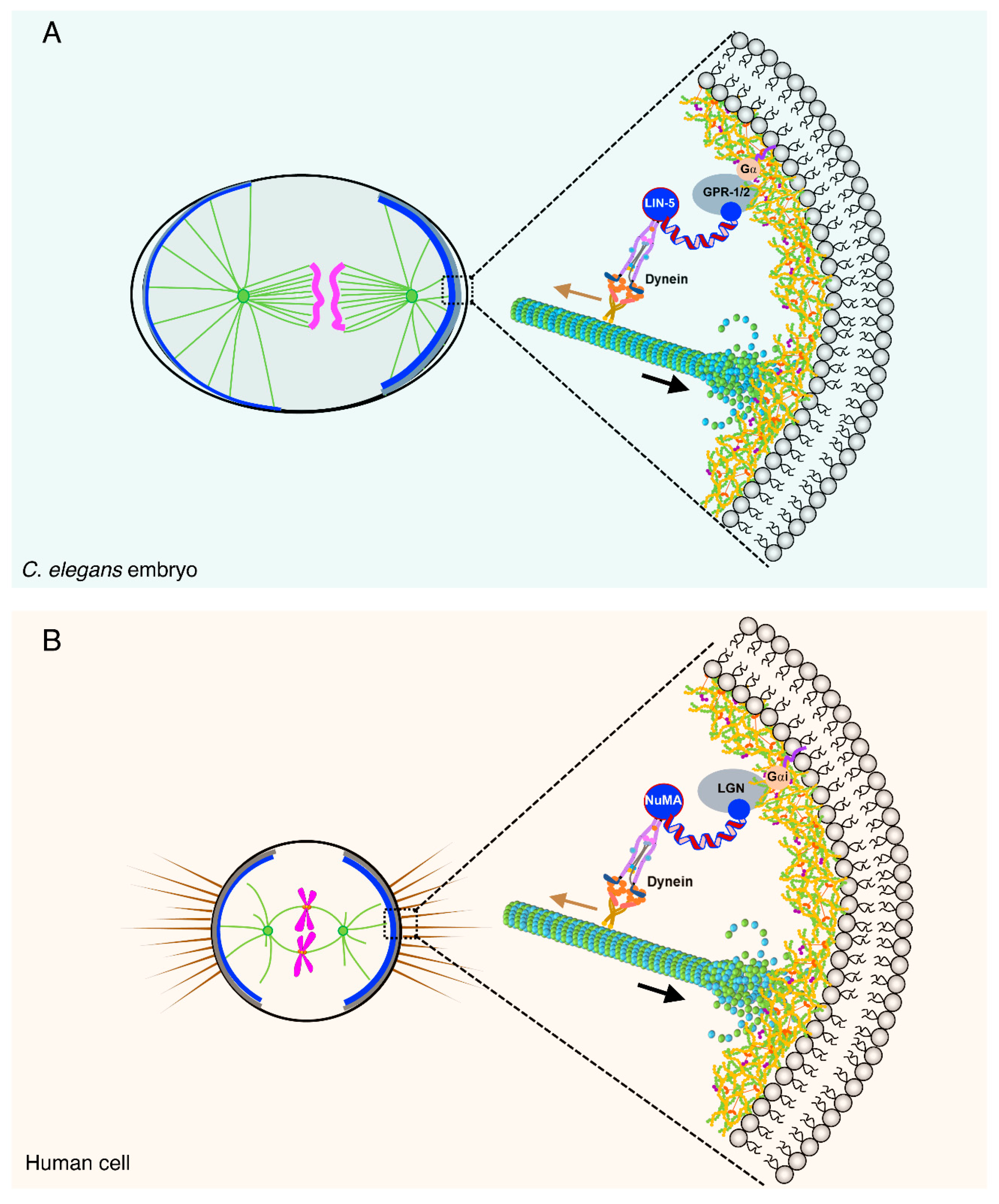
2.1. The Ternary Complex and Associated Proteins: The Dynein Capturing Machinery at the Cell Cortex
2.2. Linking Extrinsic Mechanical Forces to Spindle Positioning
2.3. Extrinsic Chemical Code for Guiding Positioning of the Mitotic Spindle
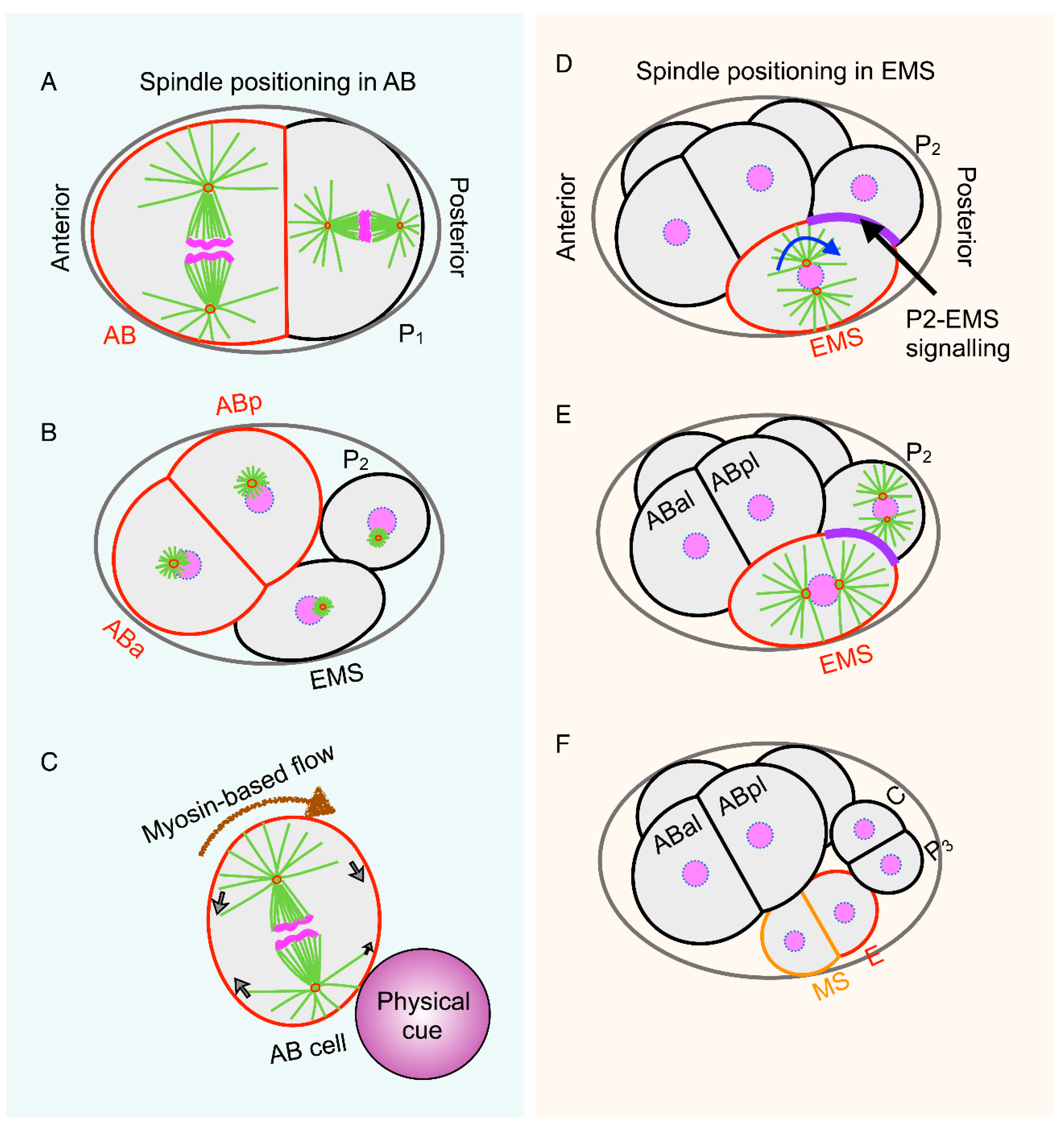
3. Spatiotemporal Control of Spindle Positioning
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Walczak, C.E.; Heald, R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008, 265, 111–158. [Google Scholar] [PubMed]
- Jongsma, M.L.; Berlin, I.; Neefjes, J. On the move: Organelle dynamics during mitosis. Trends Cell Biol. 2015, 25, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Knoblich, J.A. Mechanisms of asymmetric stem cell division. Cell 2008, 132, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Gonczy, P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008, 9, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Morin, X.; Bellaiche, Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 2011, 21, 102–119. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.E.; Fuchs, E. Oriented divisions, fate decisions. Curr. Opin. Cell Biol. 2013, 25, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, F.; Echard, A.; Morin, X. Regulation of mitotic spindle orientation: An integrated view. EMBO Rep. 2016, 17, 1106–1130. [Google Scholar] [CrossRef] [PubMed]
- Seldin, L.; Macara, I. Epithelial spindle orientation diversities and uncertainties: Recent developments and lingering questions. F1000Res 2017, 6, 984. [Google Scholar] [CrossRef] [PubMed]
- Bergstralh, D.T.; Dawney, N.S.; St Johnston, D. Spindle orientation: A question of complex positioning. Development 2017, 144, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, M.S.; Hiramoto, Y. Analysis of the Role of Astral Rays in Pronuclear Migration in Sand Dollar Eggs by the Colcemid-Uv Method. Dev. Growth Differ. 1986, 28, 143–156. [Google Scholar] [CrossRef]
- Kimura, K.; Kimura, A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc. Natl. Acad. Sci. USA 2011, 108, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Wuhr, M.; Tan, E.S.; Parker, S.K.; Detrich, H.W., 3rd; Mitchison, T.J. A model for cleavage plane determination in early amphibian and fish embryos. Curr. Biol. 2010, 20, 2040–2045. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Wuhr, M.; Nguyen, P.; Ishihara, K.; Groen, A.; Field, C.M. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken) 2012, 69, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Fink, J.; Carpi, N.; Betz, T.; Betard, A.; Chebah, M.; Azioune, A.; Bornens, M.; Sykes, C.; Fetler, L.; Cuvelier, D.; et al. External forces control mitotic spindle positioning. Nat. Cell Biol. 2011, 13, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Bagonis, M.; Danuser, G.; Pellman, D. Direct Microtubule-Binding by Myosin-10 Orients Centrosomes toward Retraction Fibers and Subcortical Actin Clouds. Dev. Cell 2015, 34, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Siller, K.H.; Doe, C.Q. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 2009, 11, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Knoblich, J.A. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 2010, 11, 849–860. [Google Scholar] [CrossRef] [PubMed]
- McCaffrey, L.M.; Macara, I.G. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011, 21, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.S.; Johnston, C.A. Molecular pathways regulating mitotic spindle orientation in animal cells. Development 2013, 140, 1843–1856. [Google Scholar] [CrossRef] [PubMed]
- Rose, L.; Gonczy, P. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook 2014, 1–43. [Google Scholar] [CrossRef] [PubMed]
- St Johnston, D. Establishing and transducing cell polarity: Common themes and variations. Curr. Opin. Cell Biol. 2018, 51, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Aist, J.R.; Berns, M.W. Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): New evidence from ultrastructural and laser microbeam experiments. J. Cell Biol. 1981, 91 Pt 1, 446–458. [Google Scholar] [CrossRef]
- Aist, J.R.; Liang, H.; Berns, M.W. Astral and spindle forces in PtK2 cells during anaphase B: A laser microbeam study. J. Cell Sci. 1993, 104 Pt 4, 1207–1216. [Google Scholar]
- Grill, S.W.; Gonczy, P.; Stelzer, E.H.; Hyman, A.A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 2001, 409, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.W.; Howard, J.; Schaffer, E.; Stelzer, E.H.; Hyman, A.A. The distribution of active force generators controls mitotic spindle position. Science 2003, 301, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Redemann, S.; Pecreaux, J.; Goehring, N.W.; Khairy, K.; Stelzer, E.H.; Hyman, A.A.; Howard, J. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE 2010, 5, e12301. [Google Scholar] [CrossRef] [PubMed]
- Gotta, M.; Ahringer, J. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell Biol. 2001, 3, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Colombo, K.; Grill, S.W.; Kimple, R.J.; Willard, F.S.; Siderovski, D.P.; Gonczy, P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science 2003, 300, 1957–1961. [Google Scholar] [CrossRef] [PubMed]
- Gotta, M.; Dong, Y.; Peterson, Y.K.; Lanier, S.M.; Ahringer, J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 2003, 13, 1029–1037. [Google Scholar] [CrossRef]
- Srinivasan, D.G.; Fisk, R.M.; Xu, H.; van den Heuvel, S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003, 17, 1225–1239. [Google Scholar] [CrossRef] [PubMed]
- Tsou, M.F.; Ku, W.; Hayashi, A.; Rose, L.S. PAR-dependent and geometry-dependent mechanisms of spindle positioning. J. Cell Biol. 2003, 160, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Macara, I.G. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 2004, 119, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Busso, C.; Gonczy, P. Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 2012, 199, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Gonczy, P. Mechanisms of spindle positioning: Cortical force generators in the limelight. Curr. Opin. Cell Biol. 2013, 25, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Rose, L.S. Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev. Biol. 2008, 315, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Fielmich, L.E.; Grigoriev, I.; Katrukha, E.A.; Akhmanova, A.; van den Heuvel, S. Two populations of cytoplasmic dynein contribute to spindle positioning in C. elegans embryos. J. Cell Biol. 2017, 216, 2777–2793. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, F.; Nishida, E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007, 26, 1487–1498. [Google Scholar] [CrossRef] [PubMed]
- Thery, M.; Racine, V.; Pepin, A.; Piel, M.; Chen, Y.; Sibarita, J.B.; Bornens, M. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 2005, 7, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Thery, M.; Jimenez-Dalmaroni, A.; Racine, V.; Bornens, M.; Julicher, F. Experimental and theoretical study of mitotic spindle orientation. Nature 2007, 447, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Tame, M.A.; Raaijmakers, J.A.; van den Broek, B.; Lindqvist, A.; Jalink, K.; Medema, R.H. Astral microtubules control redistribution of dynein at the cell cortex to facilitate spindle positioning. Cell Cycle 2014, 13, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Woodard, G.E.; Huang, N.N.; Cho, H.; Miki, T.; Tall, G.G.; Kehrl, J.H. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell Biol. 2010, 30, 3519–3530. [Google Scholar] [CrossRef] [PubMed]
- Kiyomitsu, T.; Cheeseman, I.M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012, 14, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Busso, C.; Gonczy, P. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 2014, 33, 1815–1830. [Google Scholar] [CrossRef] [PubMed]
- Burri, O.; Wolf, B.; Seitz, A.; Gonczy, P. TRACMIT: An effective pipeline for tracking and analyzing cells on micropatterns through mitosis. PLoS ONE 2017, 12, e0179752. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhu, H.B.; Wan, Q.W.; Liu, J.; Xiao, Z.N.; Siderovski, D.P.; Du, Q.S. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J. Cell Biol. 2010, 189, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Lechler, T.; Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 2005, 437, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Seldin, L.; Poulson, N.D.; Foote, H.P.; Lechler, T. NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell 2013, 24, 3651–3662. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Ngoc, T.; Afshar, K.; Gonczy, P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 2007, 9, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Couwenbergs, C.; Labbe, J.C.; Goulding, M.; Marty, T.; Bowerman, B.; Gotta, M. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 2007, 179, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, F.; Valon, L.; Li, Y.; Goiame, R.; Genovesio, A.; Morin, X. An RNAi Screen in a Novel Model of Oriented Divisions Identifies the Actin-Capping Protein Z beta as an Essential Regulator of Spindle Orientation. Curr. Biol. 2017, 27, 2452–2464. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Natsume, T.; Kanemaki, M.T.; Kiyomitsu, T. Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble. eLife 2018, 7, e36559. [Google Scholar] [CrossRef] [PubMed]
- Fielmich, L.E.; Schmidt, R.; Dickinson, D.J.; Goldstein, B.; Akhmanova, A.; van den Heuvel, S. Optogenetic dissection of mitotic spindle positioning in vivo. eLife 2018, 7, e38198. [Google Scholar] [CrossRef] [PubMed]
- Laan, L.; Pavin, N.; Husson, J.; Romet-Lemonne, G.; van Duijn, M.; Lopez, M.P.; Vale, R.D.; Julicher, F.; Reck-Peterson, S.L.; Dogterom, M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 2012, 148, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Taberner, N.; Weber, G.; You, C.; Dries, R.; Piehler, J.; Dogterom, M. Reconstituting functional microtubule-barrier interactions. Methods Cell Biol. 2014, 120, 69–90. [Google Scholar] [PubMed]
- Seldin, L.; Muroyama, A.; Lechler, T. NuMA-microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. eLife 2016, 5, e12504. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S. Mitotic Spindle: Illuminating Spindle Positioning with a Biological Lightsaber. Curr. Biol. 2018, 28, R1308–R1310. [Google Scholar] [CrossRef] [PubMed]
- Collins, E.S.; Balchand, S.K.; Faraci, J.L.; Wadsworth, P.; Lee, W.L. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell 2012, 23, 3380–3390. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Busso, C.; Gonczy, P. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 2013, 32, 2517–2529. [Google Scholar] [CrossRef] [PubMed]
- Kiyomitsu, T.; Cheeseman, I.M. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 2013, 154, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wan, Q.; Meixiong, G.; Du, Q. Cell cycle-regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol. Biol. Cell 2014, 25, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Collins, E.; Mann, B.J.; Wadsworth, P. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton (Hoboken) 2014, 71, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Bosveld, F.; Markova, O.; Guirao, B.; Martin, C.; Wang, Z.; Pierre, A.; Balakireva, M.; Gaugue, I.; Ainslie, A.; Christophorou, N.; et al. Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature 2016, 530, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Tsou, M.F.; Hayashi, A.; DeBella, L.R.; McGrath, G.; Rose, L.S. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development 2002, 129, 4469–4481. [Google Scholar] [PubMed]
- Wu, J.C.; Rose, L.S. PAR-3 and PAR-1 inhibit LET-99 localization to generate a cortical band important for spindle positioning in Caenorhabditis elegans embryos. Mol. Biol. Cell 2007, 18, 4470–4482. [Google Scholar] [CrossRef] [PubMed]
- Zwaal, R.R.; Ahringer, J.; van Luenen, H.G.; Rushforth, A.; Anderson, P.; Plasterk, R.H. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell 1996, 86, 619–629. [Google Scholar] [CrossRef]
- Thyagarajan, K.; Afshar, K.; Gonczy, P. Polarity mediates asymmetric trafficking of the Gbeta heterotrimeric G-protein subunit GPB-1 in C. elegans embryos. Development 2011, 138, 2773–2782. [Google Scholar] [CrossRef] [PubMed]
- Afshar, K.; Willard, F.S.; Colombo, K.; Johnston, C.A.; McCudden, C.R.; Siderovski, D.P.; Gonczy, P. RIC-8 is required for GPR-1/2-dependent Galpha function during asymmetric division of C. elegans embryos. Cell 2004, 119, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.W.N.; Monat, C.; Robitaille, M.; Lacomme, M.; Daulat, A.M.; Macleod, G.; McNeill, H.; Cayouette, M.; Angers, S. SAPCD2 Controls Spindle Orientation and Asymmetric Divisions by Negatively Regulating the Galphai-LGN-NuMA Ternary Complex. Dev. Cell 2016, 36, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Kress, E.; Schwager, F.; Holtackers, R.; Seiler, J.; Prodon, F.; Zanin, E.; Eiteneuer, A.; Toya, M.; Sugimoto, A.; Meyer, H.; et al. The UBXN-2/p37/p47 adaptors of CDC-48/p97 regulate mitosis by limiting the centrosomal recruitment of Aurora A. J. Cell Biol. 2013, 201, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Schwager, F.; Meraldi, P.; Gotta, M. p37/UBXN2B regulates spindle orientation by limiting cortical NuMA recruitment via PP1/Repo-Man. J. Cell Biol. 2018, 217, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Saadaoui, M.; Machicoane, M.; di Pietro, F.; Etoc, F.; Echard, A.; Morin, X. Dlg1 controls planar spindle orientation in the neuroepithelium through direct interaction with LGN. J. Cell Biol. 2014, 206, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Carminati, M.; Gallini, S.; Pirovano, L.; Alfieri, A.; Bisi, S.; Mapelli, M. Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation. Nat. Struct. Mol. Biol. 2016, 23, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Settele, F.; Kotak, S.; Sanchez-Pulido, L.; Ehret, L.; Ponting, C.P.; Gonczy, P.; Hoffmann, I. MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. J. Cell Biol. 2013, 200, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Kschonsak, Y.T.; Hoffmann, I. Activated ezrin controls MISP levels to ensure correct NuMA polarization and spindle orientation. J. Cell Sci. 2018, 131, jcs214544. [Google Scholar] [CrossRef] [PubMed]
- Gloerich, M.; Bianchini, J.M.; Siemers, K.A.; Cohen, D.J.; Nelson, W.J. Cell division orientation is coupled to cell-cell adhesion by the E-cadherin/LGN complex. Nat. Commun. 2017, 8, 13996. [Google Scholar] [CrossRef] [PubMed]
- Machicoane, M.; de Frutos, C.A.; Fink, J.; Rocancourt, M.; Lombardi, Y.; Garel, S.; Piel, M.; Echard, A. SLK-dependent activation of ERMs controls LGN-NuMA localization and spindle orientation. J. Cell Biol. 2014, 205, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Sugioka, K.; Bowerman, B. Combinatorial Contact Cues Specify Cell Division Orientation by Directing Cortical Myosin Flows. Dev. Cell 2018, 46, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Naganathan, S.R.; Furthauer, S.; Nishikawa, M.; Julicher, F.; Grill, S.W. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. eLife 2014, 3, e04165. [Google Scholar] [CrossRef] [PubMed]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Goldstein, B. Induction of gut in Caenorhabditis elegans embryos. Nature 1992, 357, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B. Establishment of gut fate in the E lineage of C. elegans: The roles of lineage-dependent mechanisms and cell interactions. Development 1993, 118, 1267–1277. [Google Scholar] [PubMed]
- Rocheleau, C.E.; Downs, W.D.; Lin, R.; Wittmann, C.; Bei, Y.; Cha, Y.H.; Ali, M.; Priess, J.R.; Mello, C.C. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 1997, 90, 707–716. [Google Scholar] [CrossRef]
- Hyman, A.A.; White, J.G. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J. Cell Biol. 1987, 105, 2123–2135. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B. Cell contacts orient some cell division axes in the Caenorhabditis elegans embryo. J. Cell Biol. 1995, 129, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, A.; Shelton, C.A.; Maloof, J.N.; Meneghini, M.; Bowerman, B. Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev. 1999, 13, 2028–2038. [Google Scholar] [CrossRef] [PubMed]
- Bei, Y.; Hogan, J.; Berkowitz, L.A.; Soto, M.; Rocheleau, C.E.; Pang, K.M.; Collins, J.; Mello, C.C. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev. Cell 2002, 3, 113–125. [Google Scholar] [CrossRef]
- Walston, T.; Tuskey, C.; Edgar, L.; Hawkins, N.; Ellis, G.; Bowerman, B.; Wood, W.; Hardin, J. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev. Cell 2004, 7, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Hyman, A.A. Centrosome movement in the early divisions of Caenorhabditis elegans: A cortical site determining centrosome position. J. Cell Biol. 1989, 109, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Liro, M.J.; Rose, L.S. Mitotic Spindle Positioning in the EMS Cell of Caenorhabditis elegans Requires LET-99 and LIN-5/NuMA. Genetics 2016, 204, 1177–1189. [Google Scholar] [CrossRef] [PubMed]
- Werts, A.D.; Roh-Johnson, M.; Goldstein, B. Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development 2011, 138, 4411–4422. [Google Scholar] [CrossRef] [PubMed]
- Heppert, J.K.; Pani, A.M.; Roberts, A.M.; Dickinson, D.J.; Goldstein, B. A CRISPR Tagging-Based Screen Reveals Localized Players in Wnt-Directed Asymmetric Cell Division. Genetics 2018, 208, 1147–1164. [Google Scholar] [CrossRef] [PubMed]
- Habib, S.J.; Chen, B.C.; Tsai, F.C.; Anastassiadis, K.; Meyer, T.; Betzig, E.; Nusse, R. A localized Wnt signal orients asymmetric stem cell division in vitro. Science 2013, 339, 1445–1448. [Google Scholar] [CrossRef] [PubMed]
- Besseling, J.; Bringmann, H. Engineered non-Mendelian inheritance of entire parental genomes in C. elegans. Nat. Biotechnol. 2016, 34, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Galli, M.; Munoz, J.; Portegijs, V.; Boxem, M.; Grill, S.W.; Heck, A.J.; van den Heuvel, S. aPKC phosphorylates NuMA-related LIN-5 to position the mitotic spindle during asymmetric division. Nat. Cell Biol. 2011, 13, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Du, Q.; Chen, X.; Zheng, Z.; Balsbaugh, J.L.; Maitra, S.; Shabanowitz, J.; Hunt, D.F.; Macara, I.G. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 2010, 20, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Tame, M.A.; Raaijmakers, J.A.; Afanasyev, P.; Medema, R.H. Chromosome misalignments induce spindle-positioning defects. EMBO Rep. 2016, 17, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Sana, S.; Keshri, R.; Rajeevan, A.; Kapoor, S.; Kotak, S. Plk1 regulates spindle orientation by phosphorylating NuMA in human cells. Life Sci. Alliance 2018, 1, e201800223. [Google Scholar] [CrossRef] [PubMed]
- Kettenbach, A.N.; Schweppe, D.K.; Faherty, B.K.; Pechenick, D.; Pletnev, A.A.; Gerber, S.A. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal 2011, 4, rs5. [Google Scholar] [CrossRef] [PubMed]
- Gallini, S.; Carminati, M.; De Mattia, F.; Pirovano, L.; Martini, E.; Oldani, A.; Asteriti, I.A.; Guarguaglini, G.; Mapelli, M. NuMA Phosphorylation by Aurora-A Orchestrates Spindle Orientation. Curr. Biol. 2016, 26, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Afshar, K.; Busso, C.; Gonczy, P. Aurora A kinase regulates proper spindle positioning in C. elegans and in human cells. J. Cell Sci. 2016, 129, 3015–3025. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, S.; Hamasaki, M.; Yamamoto, T.; Ebisuya, M.; Sato, M.; Nishida, E.; Toyoshima, F. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat. Commun. 2012, 3, 626. [Google Scholar] [CrossRef] [PubMed]
- Caussinus, E.; Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005, 37, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Quyn, A.J.; Appleton, P.L.; Carey, F.A.; Steele, R.J.; Barker, N.; Clevers, H.; Ridgway, R.A.; Sansom, O.J.; Nathke, I.S. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 2010, 6, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Hehnly, H.; Canton, D.; Bucko, P.; Langeberg, L.K.; Ogier, L.; Gelman, I.; Santana, L.F.; Wordeman, L.; Scott, J.D. A mitotic kinase scaffold depleted in testicular seminomas impacts spindle orientation in germ line stem cells. eLife 2015, 4, e09384. [Google Scholar] [CrossRef] [PubMed]
- Noatynska, A.; Gotta, M.; Meraldi, P. Mitotic spindle (DIS)orientation and DISease: Cause or consequence? J. Cell Biol. 2012, 199, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Fededa, J.P.; Esk, C.; Mierzwa, B.; Stanyte, R.; Yuan, S.; Zheng, H.; Ebnet, K.; Yan, W.; Knoblich, J.A.; Gerlich, D.W. MicroRNA-34/449 controls mitotic spindle orientation during mammalian cortex development. EMBO J. 2016, 35, 2386–2398. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotak, S. Mechanisms of Spindle Positioning: Lessons from Worms and Mammalian Cells. Biomolecules 2019, 9, 80. https://doi.org/10.3390/biom9020080
Kotak S. Mechanisms of Spindle Positioning: Lessons from Worms and Mammalian Cells. Biomolecules. 2019; 9(2):80. https://doi.org/10.3390/biom9020080
Chicago/Turabian StyleKotak, Sachin. 2019. "Mechanisms of Spindle Positioning: Lessons from Worms and Mammalian Cells" Biomolecules 9, no. 2: 80. https://doi.org/10.3390/biom9020080
APA StyleKotak, S. (2019). Mechanisms of Spindle Positioning: Lessons from Worms and Mammalian Cells. Biomolecules, 9(2), 80. https://doi.org/10.3390/biom9020080




