Lnc-EPB41-Protein Interactions Associated with Congenital Pouch Colon
Abstract
1. Introduction
2. Results and Discussion
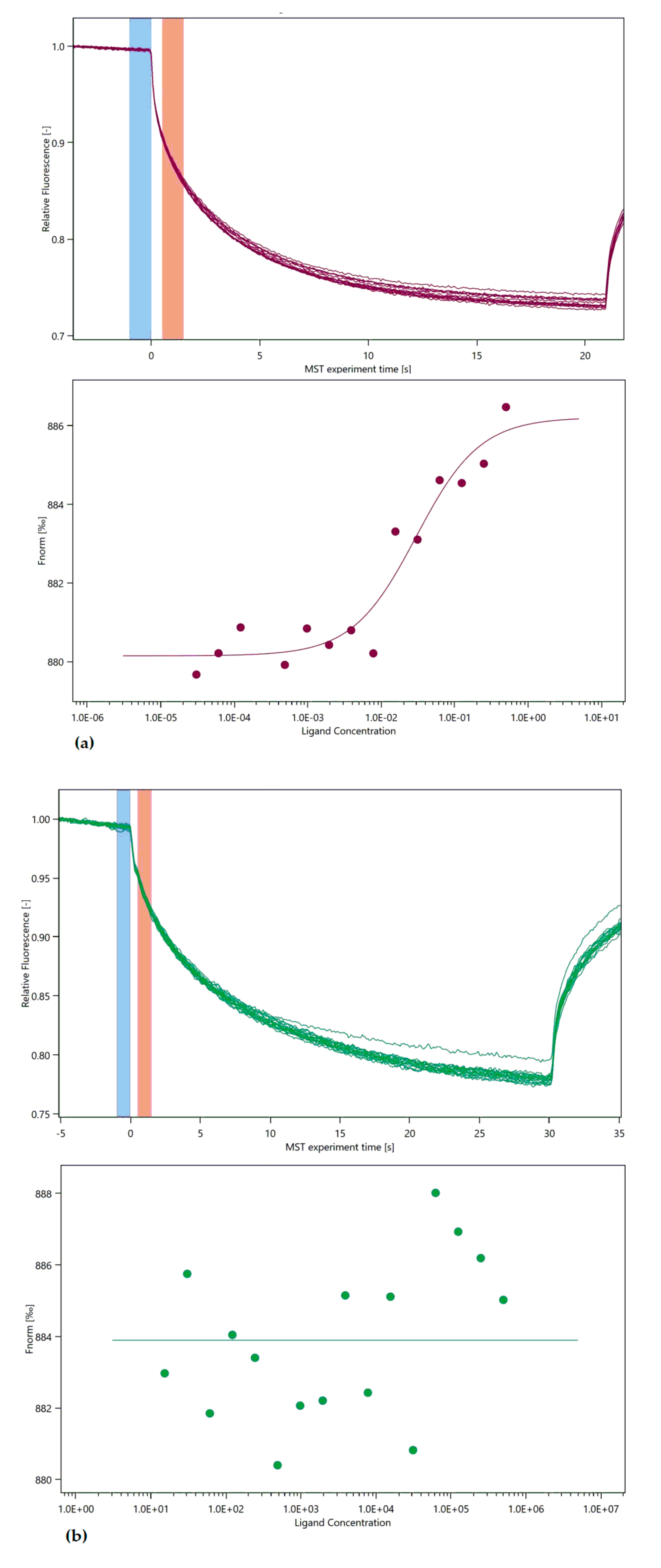
3. Microscale Thermophoresis
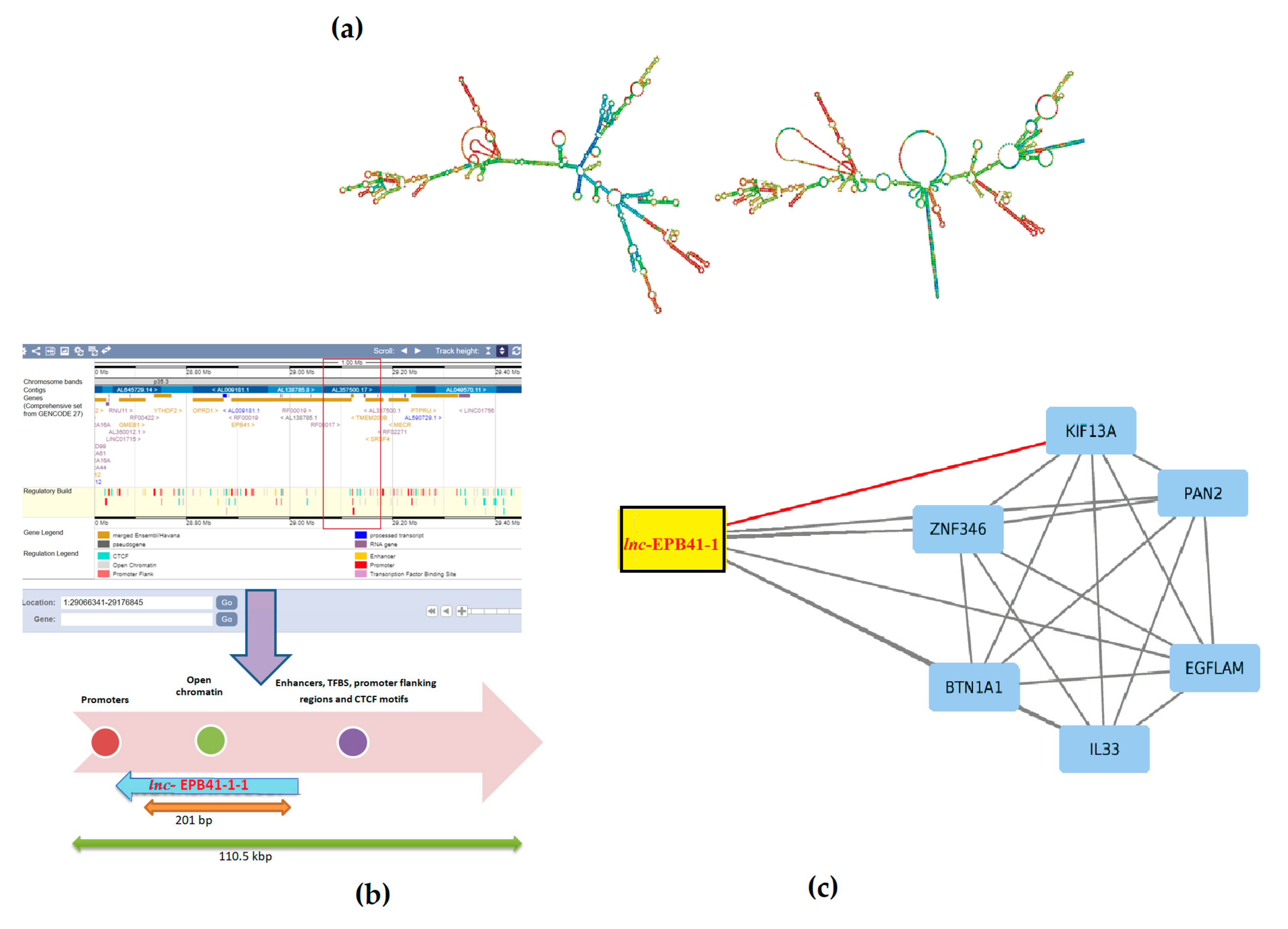
- The lnc-EPB41-1-1 harbors two potential open chromatin elements OCE, viz. ENSR00000003936 and ENSR00000003937 (E74-like factor 1, Ets family members respectively) linking it with a regulatory role [21]. In addition, the presence of transcription factor binding sites, promoters, CTCF motifs up/downstream provides evidence of its regulatory build for this region (Table S1) (Figure 1b). Furthermore, the primary role of CTCF motif is thought to be regulating the 3D structure of chromatin besides anchoring DNA to cellular structures, which influences the expression/repression of genes including lnc-EPB41-1-1. As lnc-EPB41-1-1 is one among a large number of conserved lncRNAs in mammalian/amniotic species [22], there is a growing significance that gene regulation could be associated with various phenotypes.
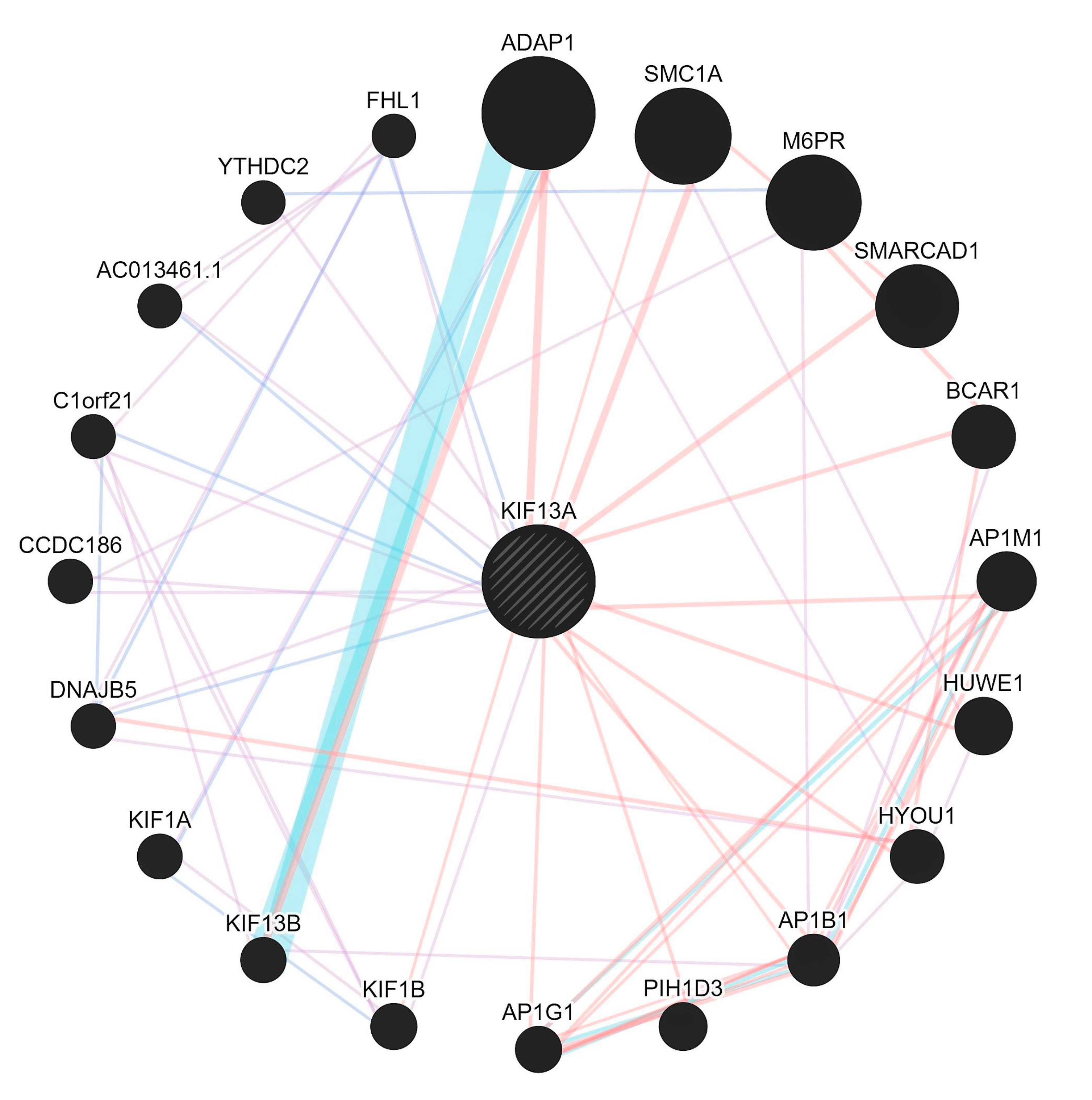
- Evidence shows that lnc-EPB41-1-1 are known to be largely expressed in prostate and non-functional pituitary adenomas (NFPA) supporting its regulatory role in urological/colonic tissues as seen from a RNA-Seq expression profile [23]. In addition, when we checked the gene ontology pathways, it was observed that the KIF13A is involved in the manifestation of colon related disorders particularly the anorectal malfunction [24].
- Due to the interactions in affected samples, there is a possibility that the pathways could be altered in CPC. We argue that with the mutations in essential genes tend to be causal for rare diseases [25] even as the mutations in non-coding genes could serve as drivers having higher prevalence rates. Furthermore, there appears to be selective pressure in those genes that share the pathways where they tend to be coexpressed, but not necessarily physically interacting/co-localizing (Figure 3).
4. Material and Methods
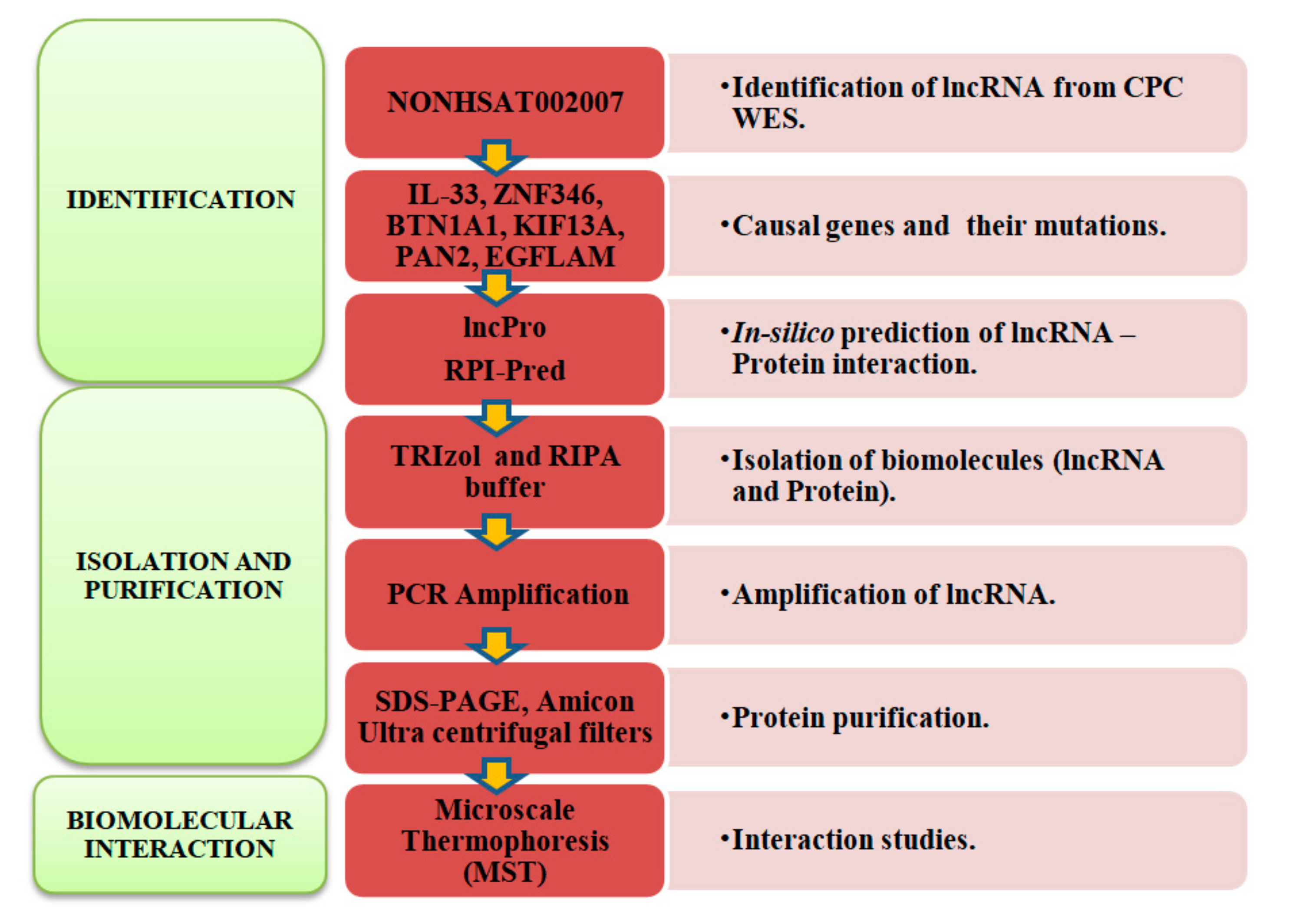
4.1. Identification of Long Non-Coding RNA
4.2. Extraction of Biomolecules
4.3. Label Free Thermal Shift Analysis
4.4. Micro Scale Thermophoresis Affinity Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mathur, P.; Gupta, R.; Simlot, A.; Goyal, R.B. Congenital Pouch Colon with Double Meckel’s Diverticulae. J. Neonatal Surg. 2013, 2, 48. [Google Scholar] [PubMed]
- Saxena, A.K.; Mathur, P. Congenital Pouch Colon. In New Born Surgery, 3rd ed.; Puri, P., Ed.; Hoddr Arnold: London, UK, 2011; Volume 62, pp. 582–589. [Google Scholar]
- Mathur, P.; Medicherla, K.M.; Chaudhary, S.; Patel, M.; Bagali, P.; Suravajhala, P. Whole exome sequencing reveals rare variants linked to congenital pouch colon. Sci. Rep. 2018, 8, 6646. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Hrdlickova, B.; Almeida, R.C.; Borek, Z.; Withoff, S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim. Biophys. Acta 2014, 1842, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Zappala, Z.; Montgomery, S.B. Non-coding loss-of-function variation in human genomes. Hum. Hered. 2016, 81, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, H.; Wu, Y.; Zheng, X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Suravajhala, P.; Kogelman, L.J.A.; Mazzoni, G.; Kadarmideen, H.N. Potential role of lncRNA cyp2c91–protein interactions on diseases of the immune system. Front. Genet. 2015, 6, 255. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Summersgill, B.; Yost, S.; Sultana, R.; Labreche, K.; Dudakia, D.; Renwick, A.; Seal, S.; Al-Saadi, R.; Broderick, P.; et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat. Commun. 2015, 6, 5973. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhou, L.; Ge, M.; Zhang, B.; Yang, X.; Xiong, X.; Fu, G.; Zhang, J.; Nie, X.; Li, H.; et al. Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum. Mol. Genet. 2016, 25, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- D’haene, E.; Jacobs, E.Z.; Volders, P.; Meyer, T.D.; Menten, B.; Vergult, S. Identification of long non-coding RNAs involved in neuronal development and intellectual disability. Sci. Rep. 2016, 6, 28396. [Google Scholar] [CrossRef] [PubMed]
- Barra, J.; Leucci, E. Probing Long Non-coding RNA-Protein Interactions. Front. Mol. Biosci. 2017, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.; Gorin, M.B. Challenges confronting precision medicine in the context of inherited retinal disorders. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 195–205. [Google Scholar] [CrossRef]
- Terni, E.; Giannini, N.; Brondi, M.; Montano, V.; Bonuccelli, U.; Mancuso, M. Genetics of ischaemic stroke in young adults. BBA Clin. 2015, 3, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Genin, E.; Feingold, J.; Clerget-Darpoux, F. Identifying modifier genes of monogenic disease: Strategies and difficulties. Hum. Genet. 2008, 124, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.D.; Nadeau, J.H. From peas to disease: Modifier genes, network resilience, and the genetics of health. Am. J. Hum. Genet. 2017, 101, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Gennari, L.; Gianfrancesco, F.; Di Stefano, M.; Rendina, D.; Merlotti, D.; Esposito, T.; Gallone, S.; Fusco, P.; Rainero, I.; Fenoglio, P.; et al. SQSTM1 gene analysis and gene-environment interaction in Paget’s disease of bone. J. Bone Miner. Res. 2010, 25, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- McHugh, A.C.; Russell, P.; Guttman, M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.Y.; Kellen, B.; Ferreira, F.; Alenquer, M.; Vale-Costa, S.; Raposo, G.; Delevoye, C.; Amorim, M.J. KIF13A mediates trafficking of influenza A virus ribonucleoproteins. J. Cell Sci. 2017, 130, 4038–4050. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Wang, X.; Zhao, H.L.; Chen, X.L.; Yuan, J.H.; Guo, J.Y.; Li, K.Q.; Li, G. Integrated analysis of transcription factor, microRNA and lncRNA in an animal model of obliterative bronchiolitis. Int. J. Clin. Exp. Pathol. 2015, 8, 7050–7058. [Google Scholar] [PubMed]
- Bu, D.; Luo, H.; Jiao, F.; Fang, S.; Tan, C.; Liu, Z.; Zhao, Y. Evolutionary annotation of conserved long non-coding RNAs in major mammalian species. Sci. China Life Sci. 2015, 58, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Noncode. Available online: http://www.noncode.org/show_rna.php?id=NONHSAT002007&version=2&utd=1# (accessed on 9 August 2018).
- Xu, X.; Pan, M.; Gasiewicz, A.E.; Li, R.; Kuo, S.M. Human and mouse microarrays-guided expression analysis of membrane protein trafficking-related genes in MDCK cells, a canine epithelial model for apical and basolateral differential protein targeting. Biochim. Open 2017, 4, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, C.; Jacomy, A.; Lu, J.L.; Jegga, A.G. The orphan disease networks. Am. J. Hum. Genet. 2011, 88, 755–766. [Google Scholar] [CrossRef] [PubMed]
- The UVA FASTA. Available online: https://fasta.bioch.virginia.edu/fasta_www2/fasta_list2.shtml# (accessed on 18 July 2018).
- Lu, Q.; Ren, S.; Lu, M.; Zhang, Y.; Zhu, D.; Zhang, X.; Li, T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genom. 2013, 14, 651. [Google Scholar] [CrossRef] [PubMed]
- Suresh, V.; Liu, L.; Adjeroh, D.; Zhou, X. RPI-Pred: Predicting ncRNA-protein interaction using sequence and structural information. Nucleic Acids Res. 2015, 43, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Generunner. Available online: www.generunner.net/ (accessed on 4 July 2018).
- Mohamadi, M.; Tschammer, N.; Breitsprecher, D. Quick Protein Binding Analysis by Label-Free Thermal Shift Analysis on the Tycho NT.6. 2017. Available online: http://www.mstechno.co.jp/html/applicationFile/ms5b1091a41026e/Tycho_Technote_Ty-002-01.pdf (accessed on 15 August 2018).
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, S.; Gupta, N.; Tiwari, P.; Menon, S.; Mathur, P.; Kothari, S.L.; Nallapeta, S.; Medicherla, K.M.; Suravajhala, P. Lnc-EPB41-Protein Interactions Associated with Congenital Pouch Colon. Biomolecules 2018, 8, 95. https://doi.org/10.3390/biom8030095
Gupta S, Gupta N, Tiwari P, Menon S, Mathur P, Kothari SL, Nallapeta S, Medicherla KM, Suravajhala P. Lnc-EPB41-Protein Interactions Associated with Congenital Pouch Colon. Biomolecules. 2018; 8(3):95. https://doi.org/10.3390/biom8030095
Chicago/Turabian StyleGupta, Sonal, Nidhi Gupta, Pradeep Tiwari, Saji Menon, Praveen Mathur, Shanker Lal Kothari, Sivaramaiah Nallapeta, Krishna Mohan Medicherla, and Prashanth Suravajhala. 2018. "Lnc-EPB41-Protein Interactions Associated with Congenital Pouch Colon" Biomolecules 8, no. 3: 95. https://doi.org/10.3390/biom8030095
APA StyleGupta, S., Gupta, N., Tiwari, P., Menon, S., Mathur, P., Kothari, S. L., Nallapeta, S., Medicherla, K. M., & Suravajhala, P. (2018). Lnc-EPB41-Protein Interactions Associated with Congenital Pouch Colon. Biomolecules, 8(3), 95. https://doi.org/10.3390/biom8030095






