Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants
Abstract
:1. Introduction
2. Structures of Plant LRR Proteins

3. Plant LRR@IR Proteins
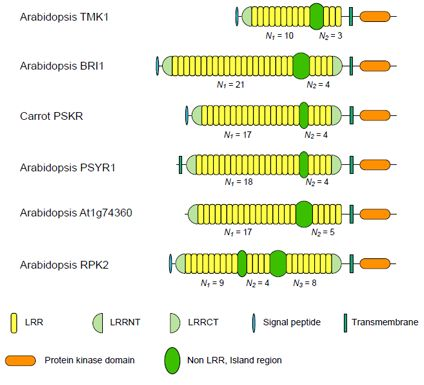
3.1. Six Families of LRR-RLKs
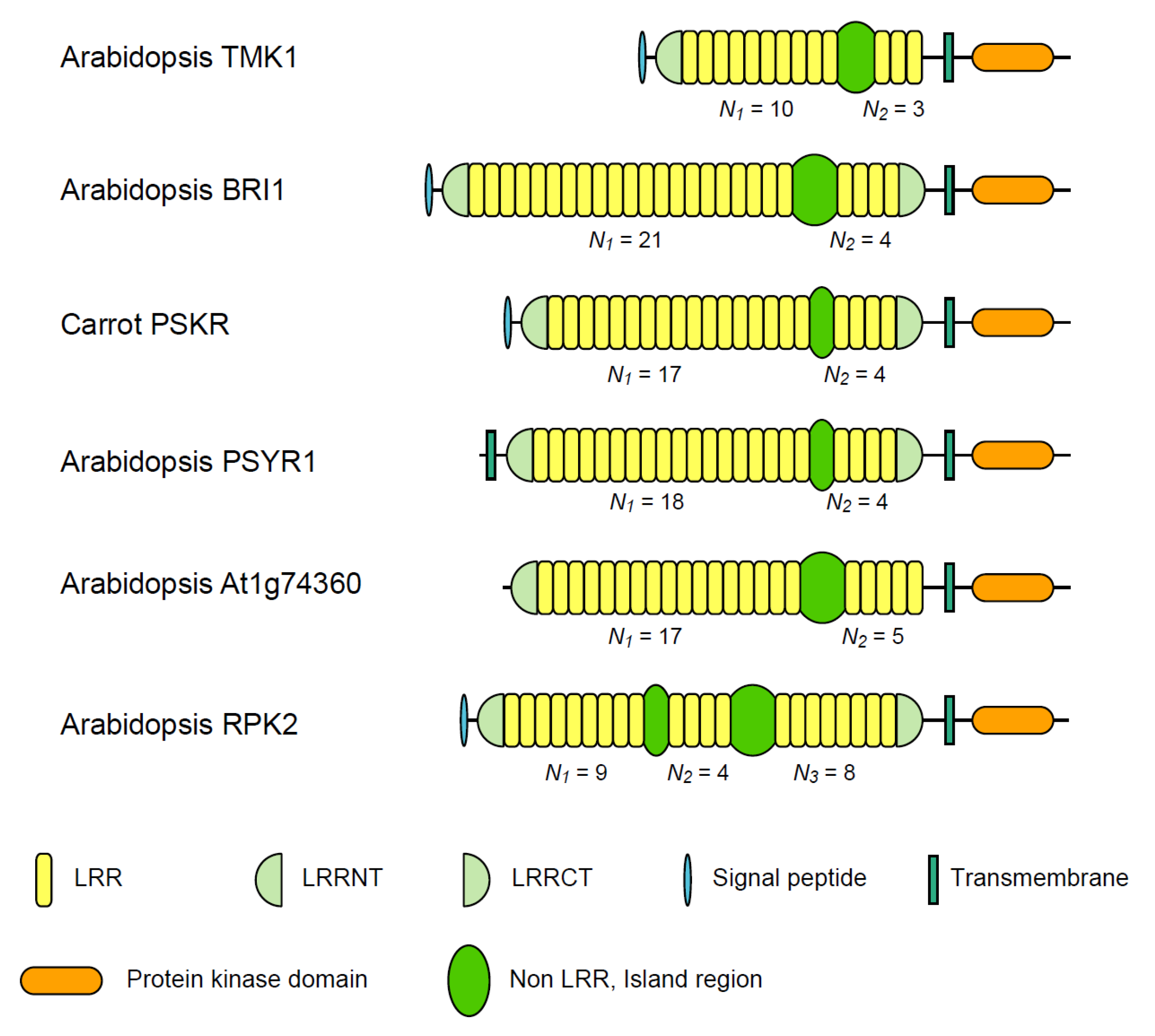
| Nineteen Families of Plant LRR Proteins | Species | Repeat number of LRRs | Lengths of non-LRR | |||
|---|---|---|---|---|---|---|
| N1a | N2b | N1/ N2c | Island | |||
| (A) | Six families of LRR-RLKs | |||||
| ArabidopsisTMK1/Soybean Rhg4 | 14 | 8~10 | 3 | 3.33 | 57~61 | |
| Arabidopsis BRI1 | 24 | 10~22 | 4 | 4.94 | 67~70 | |
| Carrot PSKR | 11 | 17~18 | 4 | 4.36 | 36~38 | |
| Arabidopsis PSYR1 | 9 | 17~18 | 4 | 4.47 | 37~38 | |
| Arabidopsis At1g74360 | 10 | 16~17 | 5 | 3.40 | 75~77 | |
| Arabidopsis RPK2 | 4 | 13d | 8d | 1.63 | 71~75 | |
| (B) | Eleven families of LRR-RLPs | |||||
| Tomato Cf-9/Cf-4 | 8 | 17~23 | 4 | 5.31 | 41~46 | |
| Tomato Cf-2/Cf-5 | 3 | 18~33 | 4 | 6.30 | 37~41 | |
| Tomato Ve | 12 | 28~30 | 4 | 7.27 | 41~49 | |
| Appl HcrVf | 1 | 22~28 | 4 | 6.52 | 39~46 | |
| Arabidopsis RPP27 | 2 | 12~26 | 4 | 5.78 | 65~71 | |
| Tomato EIXi | 1 | 27 | 4 | 6.75 | 47~49 | |
| Arabidopsis CLV2 | 11 | 18 | 4 | 4.50 | 41~44 | |
| Maize fascinated ear2 | 4 | 10~14 | 4 | 3.04 | 41~42 | |
| Arabidopsis AtRLP2 | 2 | 18 | 4 | 4.50 | 35~38 | |
| Rice Os10g0469700 | 6 | 6 | 4 | 1.50 | 39~40 | |
| Soybean disease resistance protein | 5 | 4~28 | 4 | 4.73 | 41~46 | |
| (C) | Two families of plant intracellular proteins | |||||
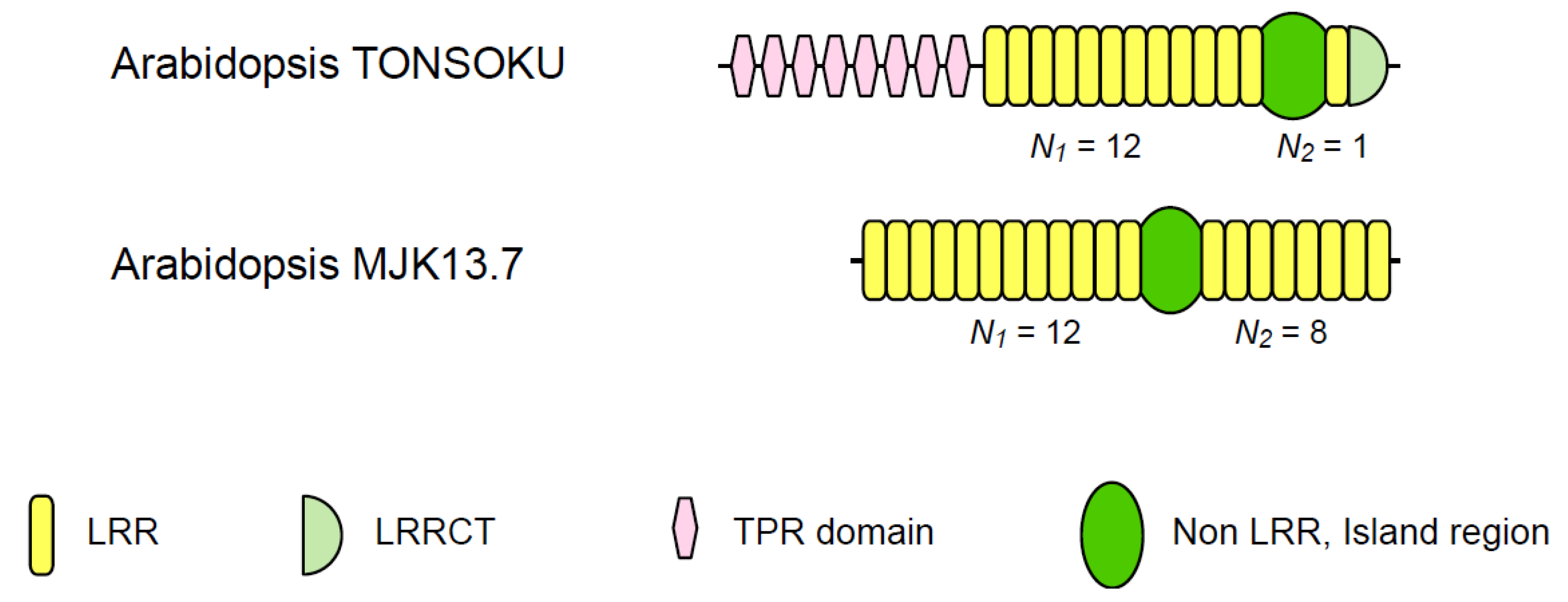
| Arabidopsis TONSOKU | 6 | 10~13 | 1 | 12.00 | 78~131 | |
| Arabidopsis MJK13.7 | 11 | 12 | 8 | 1.50 | 59~62 | |
3.2. Eleven Families of LRR-RLPs

3.3. Two Families of Plant Intracellular Proteins
3.4. An NBS-LRR Protein
4. Features, Structure, Function, and Evolution of the LRR Domains in Plant LRR@IR Proteins
4.1. Fundamental Features
4.2. Possible Structures
4.3. Possible Function(s)
4.4. Implications for Evolution
5. Evolution of Plant LRR@IR Proteins
6. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
References
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011, 40, D302–D305. [Google Scholar]
- Sigrist, C.J.; Cerutti, L.; de Castro, E.; Langendijk-Genevaux, P.S.; Bulliard, V.; Bairoch, A.; Hulo, N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010, 38, D161–D166. [Google Scholar]
- Burge, S.; Kelly, E.; Lonsdale, D.; Mutowo-Muellenet, P.; McAnulla, C.; Mitchell, A.; Sangrador-Vegas, A.; Yong, S.Y.; Mulder, N.; Hunter, S. Manual GO annotation of predictive protein signatures: The InterPro approach to GO curation. Database (Oxford) 2012, 2012. bar068. [Google Scholar]
- IPR001611 Leucine-rich repeat. Available online: http://www.ebi.ac.uk/interpro/IEntry?ac=IPR001611 (accessed on 5 May 2012).
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Matsushima, N.; Enkhbayar, P.; Kamiya, M.; Osaki, M.; Kretsinger, R. Leucine-Rich Repeats (LRRs): Structure, Function, Evolution and Interaction with Ligands. Drug. Design Rev. 2005, 2, 305–322. [Google Scholar] [CrossRef]
- Matsushima, N.; Tachi, N.; Kuroki, Y.; Enkhbayar, P.; Osaki, M.; Kamiya, M.; Kretsinger, R.H. Structural analysis of leucine-rich-repeat variants in proteins associated with human diseases. Cell Mol. Life Sci. 2005, 62, 2771–2791. [Google Scholar] [CrossRef]
- Bella, J.; Hindle, K.L.; McEwan, P.A.; Lovell, S.C. The leucine-rich repeat structure. Cell Mol. Life Sci. 2008, 65, 2307–2333. [Google Scholar] [CrossRef]
- Kajava, A.V. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998, 277, 519–527. [Google Scholar] [CrossRef]
- Ohyanagi, T.; Matsushima, N. Classification of tandem leucine-rich repeats within a great variety of proteins. FASEB J. 1997, 11, A949. [Google Scholar]
- Matsushima, N.; Miyashita, H.; Mikami, T.; Kuroki, Y. A nested leucine rich repeat (LRR) domain: The precursor of LRRs is a ten or eleven residue motif. BMC Microbiol. 2010, 10, 235. [Google Scholar] [CrossRef]
- Kajava, A.V.; Anisimova, M.; Peeters, N. Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: From plants to bacteria? PLoS One 2008, 3, e1694. [Google Scholar] [CrossRef]
- Di Matteo, A.; Federici, L.; Mattei, B.; Salvi, G.; Johnson, K.A.; Savino, C.; De Lorenzo, G.; Tsernoglou, D.; Cervone, F. The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc. Natl. Acad Sci. USA 2003, 100, 10124–10128. [Google Scholar]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–471. [Google Scholar]
- Chai, J.J.; She, J.; Han, Z.F.; Kim, T.W.; Wang, J.J.; Cheng, W.; Chang, J.B.; Shi, S.A.; Wang, J.W.; Yang, M.J.; et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–496. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Gish, L.A.; Clark, S.E. The RLK/Pelle family of kinases. Plant J. 2011, 66, 117–127. [Google Scholar] [CrossRef]
- Dievart, A.; Clark, S.E. LRR-containing receptors regulating plant development and defense. Development 2004, 131, 251–261. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar]
- Di Matteo, A.; Bonivento, D.; Tsernoglou, D.; Federici, L.; Cervone, F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: A structural view. Phytochemistry 2006, 67, 528–533. [Google Scholar] [CrossRef]
- Di, C.; Zhang, M.; Xu, S.; Cheng, T.; An, L. Role of poly-galacturonase inhibiting protein in plant defense. Crit. Rev. Microbiol. 2006, 32, 91–100. [Google Scholar] [CrossRef]
- Wang, G.; Fiers, M. Receptor-like proteins: Searching for functions. Plant Signal. Behav. 2010, 5, 540–542. [Google Scholar]
- Jones, D.; Jones, J. The role of leucine-rich repeat proteins in plant defenses. Adv. Bot. Res. 1997, 24, 89–167. [Google Scholar] [CrossRef]
- Jaillais, Y.; Belkhadir, Y.; Balsemao-Pires, E.; Dangl, J.L.; Chory, J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 2011, 108, 8503–8507. [Google Scholar]
- Tor, M.; Lotze, M.T.; Holton, N. Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 2009, 60, 3645–3654. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Meyers, B.C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998, 8, 1113–1130. [Google Scholar]
- Couch, B.C.; Spangler, R.; Ramos, C.; May, G. Pervasive purifying selection characterizes the evolution of I2 homologs. Mol. Plant Microbe Interact. 2006, 19, 288–303. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Dai, L.; Wang, G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genomics 2007, 34, 765–776. [Google Scholar] [CrossRef]
- Friedman, A.R.; Baker, B.J. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 2007, 17, 493–499. [Google Scholar] [CrossRef]
- McDowell, J.M.; Simon, S.A. Molecular diversity at the plant-pathogen interface. Dev. Comp. Immunol. 2008, 32, 736–744. [Google Scholar] [CrossRef]
- Wulff, B.B.; Chakrabarti, A.; Jones, D.A. Recognitional specificity and evolution in the tomato-Cladosporium fulvum pathosystem. Mol. Plant Microbe Interact 2009, 22, 1191–1202. [Google Scholar] [CrossRef]
- Hulbert, S.H.; Webb, C.A.; Smith, S.M.; Sun, Q. Resistance gene complexes: Evolution and utilization. Annu. Rev. Phytopathol. 2001, 39, 285–312. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.M.; Teh, T.; Wang, C.-I.A.; Ayliffe, M.A.; Kobe, B. Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar]
- Parniske, M.; Hammond-Kosack, K.E.; Golstein, C.; Thomas, C.M.; Jones, D.A.; Harrison, K.; Wulff, B.B.; Jones, J.D. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 1997, 91, 821–832. [Google Scholar] [CrossRef]
- Wei, F.; Wing, R.A.; Wise, R.P. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 2002, 14, 1903–1917. [Google Scholar] [CrossRef]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef]
- A, Baumgarten; Cannon, S.; Spangler, R.; May, G. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 2003, 165, 309–319. [Google Scholar]
- Zhou, B.; Dolan, M.; Sakai, H.; Wang, G.L. The genomic dynamics and evolutionary mechanism of the Pi2/9 locus in rice. Mol. Plant Microbe Interact 2007, 20, 63–71. [Google Scholar] [CrossRef]
- Mondragon-Palomino, M.; Gaut, B.S. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 2005, 22, 2444–2456. [Google Scholar] [CrossRef]
- Dixon, M.S.; Hatzixanthis, K.; Jones, D.A.; Harrison, K.; Jones, J.D. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 1998, 10, 1915–1925. [Google Scholar]
- Bryan, G.T.; Wu, K.S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2033–2046. [Google Scholar]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. Embo J. 2000, 19, 4004–4014. [Google Scholar]
- Fritz-Laylin, L.K.; Krishnamurthy, N.; Tor, M.; Sjolander, K.V.; Jones, J.D. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005, 138, 611–623. [Google Scholar] [CrossRef]
- Matsushima, N.; Mikami, T.; Tanaka, T.; Miyashita, H.; Yamada, K.; Kuroki, Y. Analyses of non-leucine-rich repeat (non-LRR) regions intervening between LRRs in proteins. Biochim. Biophys. Acta 1790, 1217–1237. [Google Scholar]
- Torii, K.U. Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathway. Int. Rev. Cytol. 2004, 234, 1–46. [Google Scholar] [CrossRef]
- van der Hoorn, R.A.; Wulff, B.B.; Rivas, S.; Durrant, M.C.; van der Ploeg, A.; de Wit, P.J.; Jones, J.D. Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 2005, 17, 1000–1015. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.M.; Woods-Tor, A.; Zipfel, C.; et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Nomura, T.; Bishop, G.J.; Kaneta, T.; Reid, J.B.; Chory, J.; Yokota, T. The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J. 2003, 36, 291–300. [Google Scholar] [CrossRef]
- Scheer, J.M.; Ryan, C.A., Jr. The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 2002, 99, 9585–9590. [Google Scholar]
- Montoya, T.; Nomura, T.; Farrar, K.; Kaneta, T.; Yokota, T.; Bishop, G.J. Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 2002, 14, 3163–3176. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Morita, A.; Sakagami, Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 2002, 296, 1470–1472. [Google Scholar] [CrossRef]
- Jones, D.A.; Thomas, C.M.; Hammond-Kosack, K.E.; Balint-Kurti, P.J.; Jones, J.D. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 1994, 266, 789–793. [Google Scholar]
- Kruijt, M.; Kip, D.J.; Joosten, M.H.; Brandwagt, B.F.; de Wit, P.J. The Cf-4 and Cf-9 resistance genes against Cladosporium fulvum are conserved in wild tomato species. Mol. Plant Microbe Interact 2005, 18, 1011–1021. [Google Scholar] [CrossRef]
- Jeong, S.; Trotochaud, A.E.; Clark, S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 1999, 11, 1925–1934. [Google Scholar]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef]
- de Kock, J.D.M.; Brandwagt, F.B.; Bonnema, G.; de Wit, P.J.G.M.; Lindhout, P. The tomato Orion locus comprises a unique class of Hcr9 genes. Mol. Breed. 2005, 15, 409–422. [Google Scholar] [CrossRef]
- Chow, B.; McCourt, P. Plant hormone receptors: Perception is everything. Genes Dev. 2006, 20, 1998–2008. [Google Scholar] [CrossRef]
- Kinoshita, T.; Cano-Delgado, A.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005, 433, 167–171. [Google Scholar]
- Shinohara, H.; Ogawa, M.; Sakagami, Y.; Matsubayashi, Y. Identification of ligand binding site of phytosulfokine receptor by on-column photoaffinity labeling. J. Biol. Chem. 2007, 282, 124–131. [Google Scholar]
- Wang, G.; Long, Y.; Thomma, B.; de Wit, P.; Angenent, G.; Fiers, M. Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol. 2010, 152, 320–331. [Google Scholar] [CrossRef]
- Matsushima, N.; Tanaka, T.; Enkhbayar, P.; Mikami, T.; Taga, M.; Yamada, K.; Kuroki, Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics 2007, 8, 124. [Google Scholar] [CrossRef]
- Mikami, T.; Miyashita, H.; Takatsuka, S.; Kuroki, Y.; Matsushima, N. Molecular evolution of vertebrate Toll-like receptors: Evolutionary rate difference between their leucine-rich repeats and their TIR domains. Gene 2012, 503, 235–243. [Google Scholar] [CrossRef]
- Gay, N.J.; Gangloff, M.; Weber, A.N. Toll-like receptors as molecular switches. Nat. Rev. Immunol. 2006, 6, 693–698. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition in the innate immune response. Biochem. J. 2009, 420, 1–16. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef]
- Botos, I.; Segal, D.M.; Davies, D.R. The Structural Biology of Toll-like Receptors. Structure 2011, 19, 447–459. [Google Scholar]
- Matsushima, N.; Miyashita, H.; Mikami, T.; Yamada, K. A New Method for the Identification of Leucine-Rich Repeats by Incorpolating Protein Secondar Structure Prediction. In Bioinformatics: Genome Bioinformatics and Computational Biology; NOVA Sience Pulishers: Hauppauge, NY, USA, 2011. [Google Scholar]
- Enkhbayar, P.; Kamiya, M.; Osaki, M.; Matsumoto, T.; Matsushima, N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins 2004, 54, 394–403. [Google Scholar]
- Tan, X.; Calderon-Villalobos, L.I.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Eechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar]
- Hayashi, K.; Tan, X.; Zheng, N.; Hatate, T.; Kimura, Y.; Kepinski, S.; Nozaki, H. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 5632–5637. [Google Scholar]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar]
- Lehti-Shiu, M.D.; Zou, C.; Hanada, K.; Shiu, S.H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009, 150, 12–26. [Google Scholar] [CrossRef]
- Hwang, S.G.; Kim, D.S.; Jang, C.S. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica 2011, 139, 1023–1032. [Google Scholar] [CrossRef]
- Nadeau, J.A.; Sack, F.D. Control of stomatal distribution on the Arabidopsis leaf surface. Science 2002, 296, 1697–1700. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, re22. [Google Scholar]
- van der Knaap, E.; Song, W.Y.; Ruan, D.L.; Sauter, M.; Ronald, P.C.; Kende, H. Expression of a gibberellin-induced leucine-rich repeat receptor-like protein kinase in deepwater rice and its interaction with kinase-associated protein phosphatase. Plant Physiol. 1999, 120, 559–570. [Google Scholar] [CrossRef]
- Cho, H.S.; Pai, H.S. Cloning and characterization of ntTMK1 gene encoding a TMK1-homologous receptor-like kinase in tobacco. Mol. Cells 2000, 10, 317–324. [Google Scholar]
- Meksem, K.; Ruben, E.; Hyten, D.L.; Schmidt, M.E.; Lightfoot, D.A. High-throughput genotyping for a polymorphism linked to soybean cyst nematode resistance gene Rhg4 by using Taqman (TM) probes. Mol. Breed. 2001, 7, 63–71. [Google Scholar] [CrossRef]
- Friedrichsen, D.; Chory, J. Steroid signaling in plants: From the cell surface to the nucleus. Bioessays 2001, 23, 1028–1036. [Google Scholar] [CrossRef]
- Bishop, G.J. Brassinosteroid Mutants of Crops. J. Plant Growth Regul. 2003, 22, 325–335. [Google Scholar] [CrossRef]
- Napier, R. Plant hormone binding sites. 2004, 93, 227–233. [Google Scholar]
- Belkhadir, Y.; Wang, X.; Chory, J. Brassinosteroid signaling pathway. 2006, 2006. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Chory, J. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science 2006, 314, 1410–1411. [Google Scholar] [CrossRef]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 2009, 47, 1–8. [Google Scholar] [CrossRef]
- Tang, W.; Deng, Z.; Wang, Z.Y. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr. Opin. Plant Biol. 2010, 13, 27–33. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Kihara, H.; Niwa, M.; Sakagami, Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006, 142, 45–53. [Google Scholar] [CrossRef]
- Irving, H.R.; Kwezi, L.; Ruzvidzo, O.; Wheeler, J.I.; Govender, K.; Iacuone, S.; Thompson, P.E.; Gehring, C. The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J. Biol. Chem. 2011, 286, 22580–22588. [Google Scholar]
- Amano, Y.; Tsubouchi, H.; Shinohara, H.; Ogawa, M.; Matsubayashi, Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18333–18338. [Google Scholar]
- Nodine, M.D.; Yadegari, R.; Tax, F.E. RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev. Cell 2007, 12, 943–956. [Google Scholar] [CrossRef]
- Nodine, M.D.; Tax, F.E. Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev. Biol. 2008, 314, 161–170. [Google Scholar] [CrossRef]
- Mizuno, S.; Osakabe, Y.; Maruyama, K.; Ito, T.; Osakabe, K.; Sato, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2007, 50, 751–766. [Google Scholar] [CrossRef]
- Kinoshita, A.; Betsuyaku, S.; Osakabe, Y.; Mizuno, S.; Nagawa, S.; Stahl, Y.; Simon, R.; Yamaguchi-Shinozaki, K.; Fukuda, H.; Sawa, S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010, 137, 3911–3920. [Google Scholar]
- Betsuyaku, S.; Takahashi, F.; Kinoshita, A.; Miwa, H.; Shinozaki, K.; Fukuda, H.; Sawa, S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2010, 52, 14–29. [Google Scholar]
- Sawa, S.; Tabata, R. RPK2 functions in diverged CLE signaling. Plant Signal. Behav. 2011, 6, 86–88. [Google Scholar] [CrossRef]
- Kruijt, M.; de Kock, M.J.D.; de Wit, P.J.G.M. Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 2005, 6, 85–97. [Google Scholar]
- Dixon, M.S.; Jones, D.A.; Keddie, J.S.; Thomas, C.M.; Harrison, K.; Jones, J.D. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 1996, 84, 451–459. [Google Scholar]
- Thomas, C.M.; Jones, D.A.; Parniske, M.; Harrison, K.; Balint-Kurti, P.J.; Hatzixanthis, K.; Jones, J.D. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 1997, 9, 2209–2224. [Google Scholar]
- Kawchuk, L.M.; Hachey, J.; Lynch, D.R.; Kulcsar, F.; van Rooijen, G.; Waterer, D.R.; Robertson, A.; Kokko, E.; Byers, R.; Howard, R.J.; et al. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 6511–6515. [Google Scholar]
- Vinatzer, B.A.; Patocchi, A.; Gianfranceschi, L.; Tartarini, S.; Zhang, H.B.; Gessler, C.; Sansavini, S. Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol. Plant Microbe Interact 2001, 14, 508–515. [Google Scholar] [CrossRef]
- Tor, M.; Brown, D.; Cooper, A.; Woods-Tor, A.; Sjolander, K.; Jones, J.D.; Holub, E.B. Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9. Plant Physiol. 2004, 135, 1100–1112. [Google Scholar] [CrossRef]
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef]
- Sharfman, M.; Bar, M.; Ehrlich, M.; Schuster, S.; Melech-Bonfil, S.; Ezer, R.; Sessa, G.; Avni, A. Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. Plant J. 2011, 68, 413–423. [Google Scholar]
- Fiers, M.; Golemiec, E.; Xu, J.; van der Geest, L.; Heidstra, R.; Stiekema, W.; Liu, C.M. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 2005, 17, 2542–2553. [Google Scholar] [CrossRef]
- Fiers, M.; Golemiec, E.; van der Schors, R.; van der Geest, L.; Li, K.W.; Stiekema, W.J.; Liu, C.M. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 2006, 141, 1284–1292. [Google Scholar]
- Song, X.; Guo, P.; Li, C.; Liu, C.M. The cysteine pairs in CLV2 are not necessary for sensing the CLV3 peptide in shoot and root meristems. J. Integr. Plant Biol. 2010, 52, 774–781. [Google Scholar] [CrossRef]
- Krusell, L.; Sato, N.; Fukuhara, I.; Koch, B.E.; Grossmann, C.; Okamoto, S.; Oka-Kira, E.; Otsubo, Y.; Aubert, G.; Nakagawa, T.; et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011, 65, 861–871. [Google Scholar] [CrossRef]
- Reid, D.E.; Ferguson, B.J.; Hayashi, S.; Lin, Y.H.; Gresshoff, P.M. Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 2011, 108, 789–795. [Google Scholar]
- Takemoto, D.; Hayashi, M.; Doke, N.; Mishimura, M.; Kawakita, K. Isolation of the gene for EILP, an elicitor-inducible LRR receptor-like protein, from tobacco by differential display. Plant Cell Physiol. 2000, 41, 458–464. [Google Scholar] [CrossRef]
- Wulff, B.B.; Kruijt, M.; Collins, P.L.; Thomas, C.M.; Ludwig, A.A.; De Wit, P.J.; Jones, J.D. Gene shuffling-generated and natural variants of the tomato resistance gene Cf-9 exhibit different auto-necrosis-inducing activities in Nicotiana species. Plant J. 2004, 40, 942–956. [Google Scholar] [CrossRef]
- Chai, Y.; Zhao, L.; Liao, Z.; Sun, X.; Zuo, K.; Zhang, L.; Wang, S.; Tang, K. Molecular cloning of a potential Verticillium dahliae resistance gene SlVe1 with multi-site polyadenylation from Solanum licopersicoides. DNA Seq. 2003, 14, 375–384. [Google Scholar] [CrossRef]
- Fei, J.; Chai, Y.; Wang, J.; Lin, J.; Sun, X.; Sun, C.; Zuo, K.; Tang, K. CDNA cloning and characterization of the Ve homologue gene StVe from Solanum torvum Swartz. DNA Seq. 2004, 15, 88–95. [Google Scholar] [CrossRef]
- Patocchi, A.; Vinatzer, B.A.; Gianfranceschi, L.; Tartarini, S.; Zhang, H.B.; Sansavini, S.; Gessler, C. Construction of a 550 kb BAC contig spanning the genomic region containing the apple scab resistance gene Vf. Mol. Gen. Genet 1999, 262, 884–891. [Google Scholar] [CrossRef]
- Joshi, S.G.; Schaart, J.G.; Groenwold, R.; Jacobsen, E.; Schouten, H.J.; Krens, F.A. Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol. Biol. 2011, 75, 579–591. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef]
- Guyomarc’h, S.; Vernoux, T.; Traas, J.; Zhou, D.X.; Delarue, M. MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J. Exp. Bot. 2004, 55, 673–684. [Google Scholar]
- Suzuki, T.; Inagaki, S.; Nakajima, S.; Akashi, T.; Ohto, M.A.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. 2004, 38, 673–684. [Google Scholar] [CrossRef]
- Guyomarc’h, S.; Benhamed, M.; Lemonnier, G.; Renou, J.P.; Zhou, D.X.; Delarue, M. MGOUN3: Evidence for chromatin-mediated regulation of FLC expression. J. Exp. Bot. 2006, 57, 2111–2119. [Google Scholar]
- Takeda, S.; Tadele, Z.; Hofmann, I.; Probst, A.V.; Angelis, K.J.; Kaya, H.; Araki, T.; Mengiste, T.; Mittelsten Scheid, O.; Shibahara, K.; et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004, 18, 782–793. [Google Scholar] [CrossRef]
- Kobe, B.; Deisenhofer, J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 1993, 366, 751–756. [Google Scholar]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.G.; Lee, H.; Lee, J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef]
- Dumas, J.J.; Kumar, R.; Seehra, J.; Somers, W.S.; Mosyak, L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science 2003, 301, 222–226. [Google Scholar] [CrossRef]
- Uff, S.; Clemetson, J.M.; Harrison, T.; Clemetson, K.J.; Emsley, J. Crystal structure of the platelet glycoprotein Ib(alpha) N-terminal domain reveals an unmasking mechanism for receptor activation. J. Biol. Chem. 2002, 277, 35657–35663. [Google Scholar]
- McEwan, P.A.; Andrews, R.K.; Emsley, J. Glycoprotein Ibalpha inhibitor complex structure reveals a combined steric and allosteric mechanism of von Willebrand factor antagonism. Blood 2009, 114, 4883–4885. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.H.; Ramet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef]
- Wei, T.; Gong, J.; Jamitzky, F.; Heckl, W.M.; Stark, R.W.; Rossle, S.C. Homology modeling of human Toll-like receptors TLR7, 8, and 9 ligand-binding domain. Protein Sci. 2009, 18, 1684–1691. [Google Scholar] [CrossRef]
- Afzal, A.J.; Lightfoot, D.A. Soybean disease resistance protein RHG1-LRR domain expressed, purified and refolded from Escherichia coli inclusion bodies: Preparation for a functional analysis. Protein Expr. Purif. 2007, 53, 346–355. [Google Scholar] [CrossRef]
- Seear, P.J.; Dixon, M.S. Variable leucine-rich repeats of tomato disease resistance genes Cf-2 and Cf-5 determine specificity. Mol. Plant Pathol. 2003, 4, 199–202. [Google Scholar] [CrossRef]
- van der Hoorn, R.A.; Roth, R.; de Wit, P.J. Identification of distinct specificity determinants in resistance protein Cf-4 allows construction of a Cf-9 mutant that confers recognition of avirulence protein Avr4. Plant Cell 2001, 13, 273–285. [Google Scholar]
- Wulff, B.B.; Thomas, C.M.; Smoker, M.; Grant, M.; Jones, J.D. Domain swapping and gene shuffling identify sequences required for induction of an Avr-dependent hypersensitive response by the tomato Cf-4 and Cf-9 proteins. Plant Cell 2001, 13, 255–272. [Google Scholar]
- Chakrabarti, A.; Panter, S.N.; Harrison, K.; Jones, J.D.; Jones, D.A. Regions of the Cf-9B disease resistance protein able to cause spontaneous necrosis in Nicotiana benthamiana lie within the region controlling pathogen recognition in tomato. Mol. Plant Microbe Interact 2009, 22, 1214–1226. [Google Scholar] [CrossRef]
- Wulff, B.B.; Heese, A.; Tomlinson-Buhot, L.; Jones, D.A.; de la Pena, M.; Jones, J.D. The major specificity-determining amino acids of the tomato Cf-9 disease resistance protein are at hypervariable solvent-exposed positions in the central leucine-rich repeats. Mol. Plant Microbe Interact 2009, 22, 1203–1213. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Meisenhelder, J.; Hunter, T.; Yoshida, S.; Asami, T.; Chory, J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 2005, 8, 855–865. [Google Scholar] [CrossRef]
- Matsushima, N.; Ohyanagi, T.; Tanaka, T.; Kretsinger, R.H. Super-motifs and evolution of tandem leucine-rich repeats within the small proteoglycans—biglycan, decorin, lumican, fibromodulin, PRELP, keratocan, osteoadherin, epiphycan, and osteoglycin. Proteins 2000, 38, 210–225. [Google Scholar] [CrossRef]
- Matsushima, N.; Kamiya, M.; Suzuki, N.; Tanaka, T. Super-motifs of leucine-rich repeats (LRRs) proteins. Genome Inform. 2000, 11, 343–345. [Google Scholar]
- Haigis, M.C.; Haag, E.S.; Raines, R.T. Evolution of ribonuclease inhibitor by exon duplication. Mol. Biol. Evol. 2002, 19, 959–963. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Matsushima, N.; Miyashita, H. Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules 2012, 2, 288-311. https://doi.org/10.3390/biom2020288
Matsushima N, Miyashita H. Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules. 2012; 2(2):288-311. https://doi.org/10.3390/biom2020288
Chicago/Turabian StyleMatsushima, Norio, and Hiroki Miyashita. 2012. "Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants" Biomolecules 2, no. 2: 288-311. https://doi.org/10.3390/biom2020288
APA StyleMatsushima, N., & Miyashita, H. (2012). Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules, 2(2), 288-311. https://doi.org/10.3390/biom2020288