Investigation of the Effect of 2,3-Dihydrobenzoic Acid Acid (2,3-DHBA) on the Lipid Profiles of MCF-7 and MDA-MB-231 Human Breast Cancer Cells via an Untargeted Lipidomic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Reagents
2.2. Growth Condition of the Cells
2.3. In Vitro Cytotoxic Activity Assay of 2,3-DHBA
2.4. Lipidomics Analysis
2.4.1. Lipid Isolation with the Bligh–Dyer Method
2.4.2. Lipid Phosphorus Assay
2.4.3. Determination of Lipid Types via Electrospray Ionization Mass Spectrometry (ESI-MS)
2.4.4. Multivariate Statistical Analysis of Lipids
2.5. Statistical Analysis
3. Results
3.1. In Vitro Cytotoxic Activity of 2,3-DHBA Against MCF-7 and MDA-MB-231 Cells
3.2. Lipidomic Analysis
3.2.1. Determination of Inorganic Phosphate
3.2.2. ESI-MS Analysis
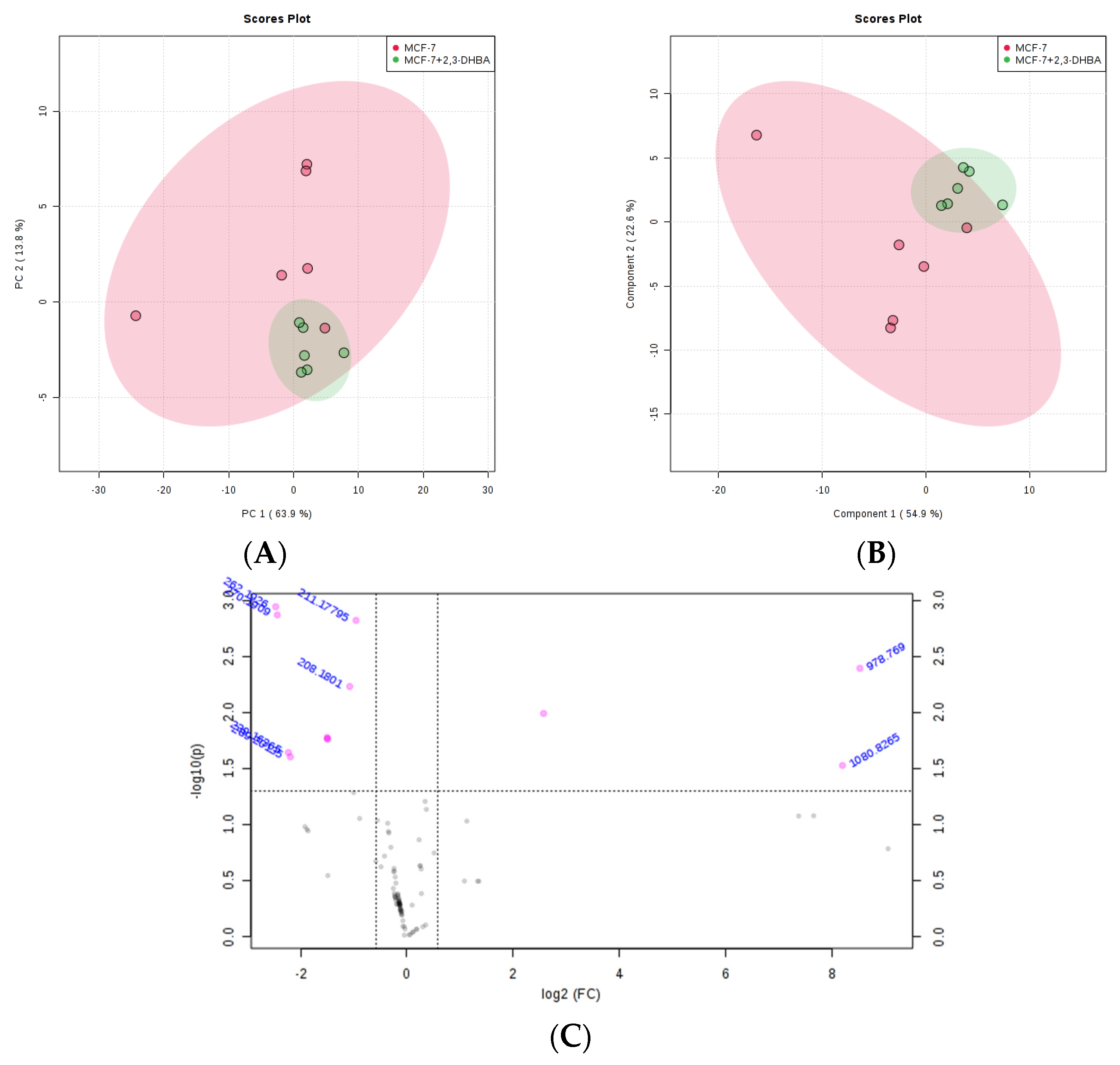
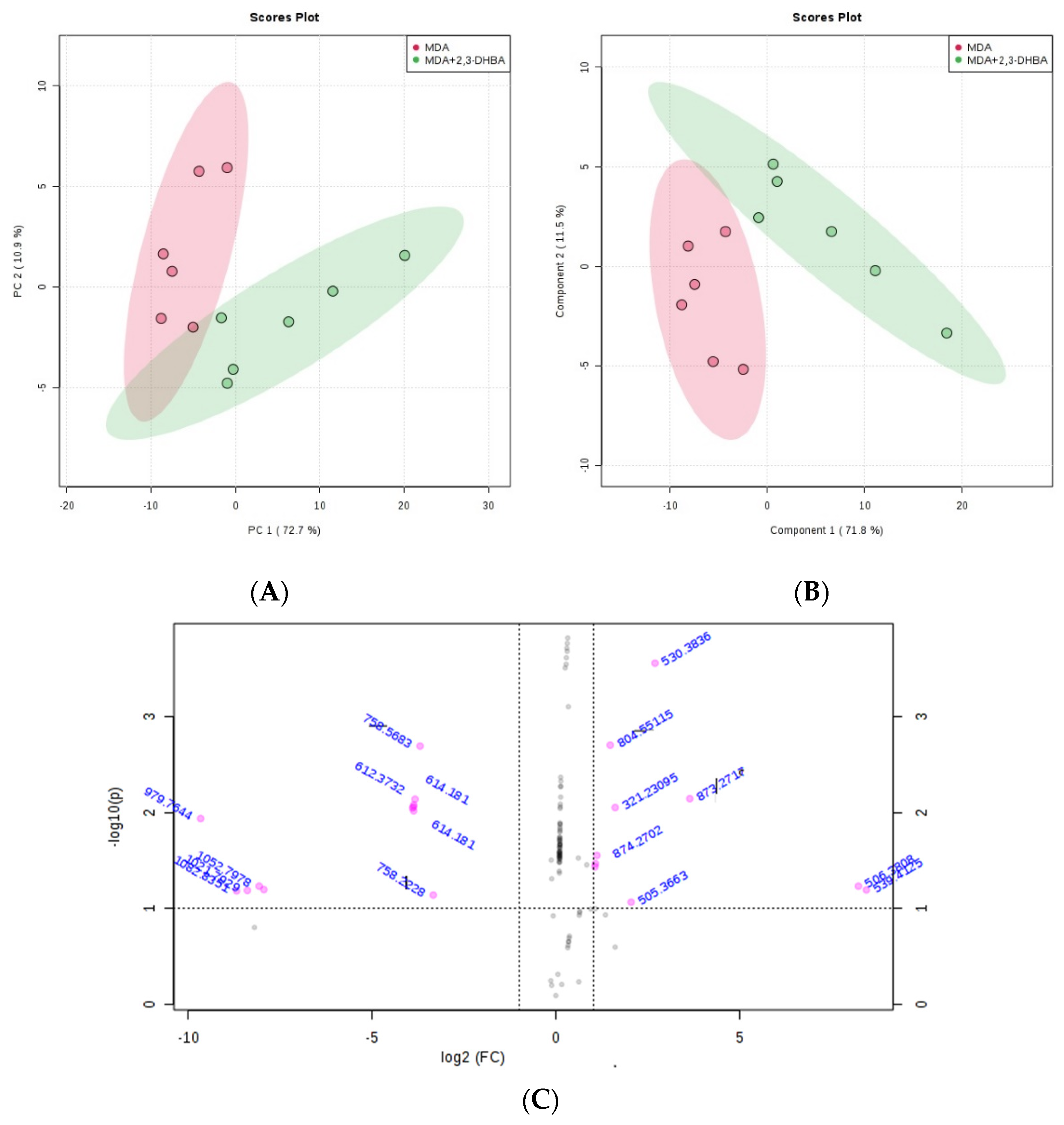
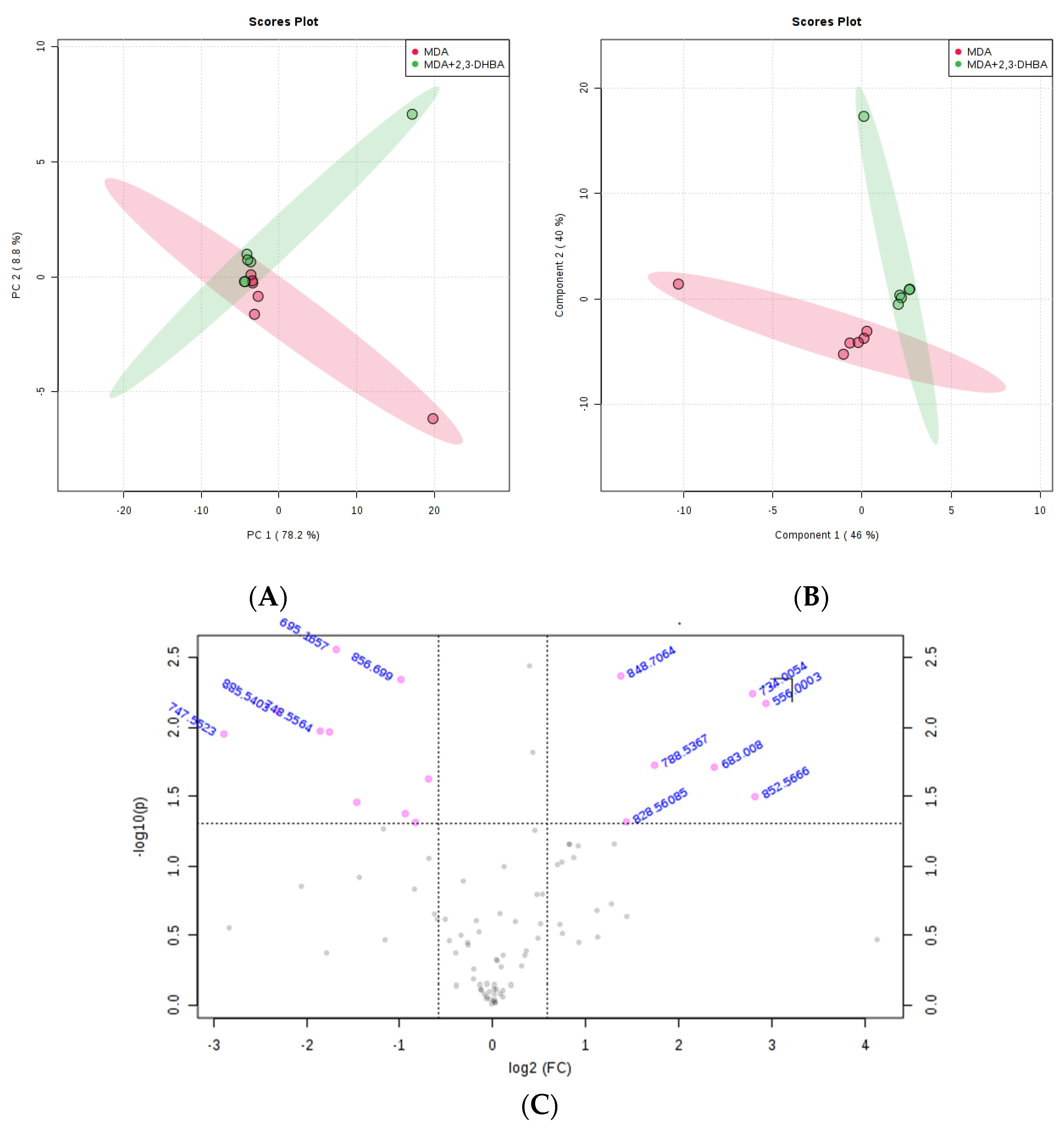
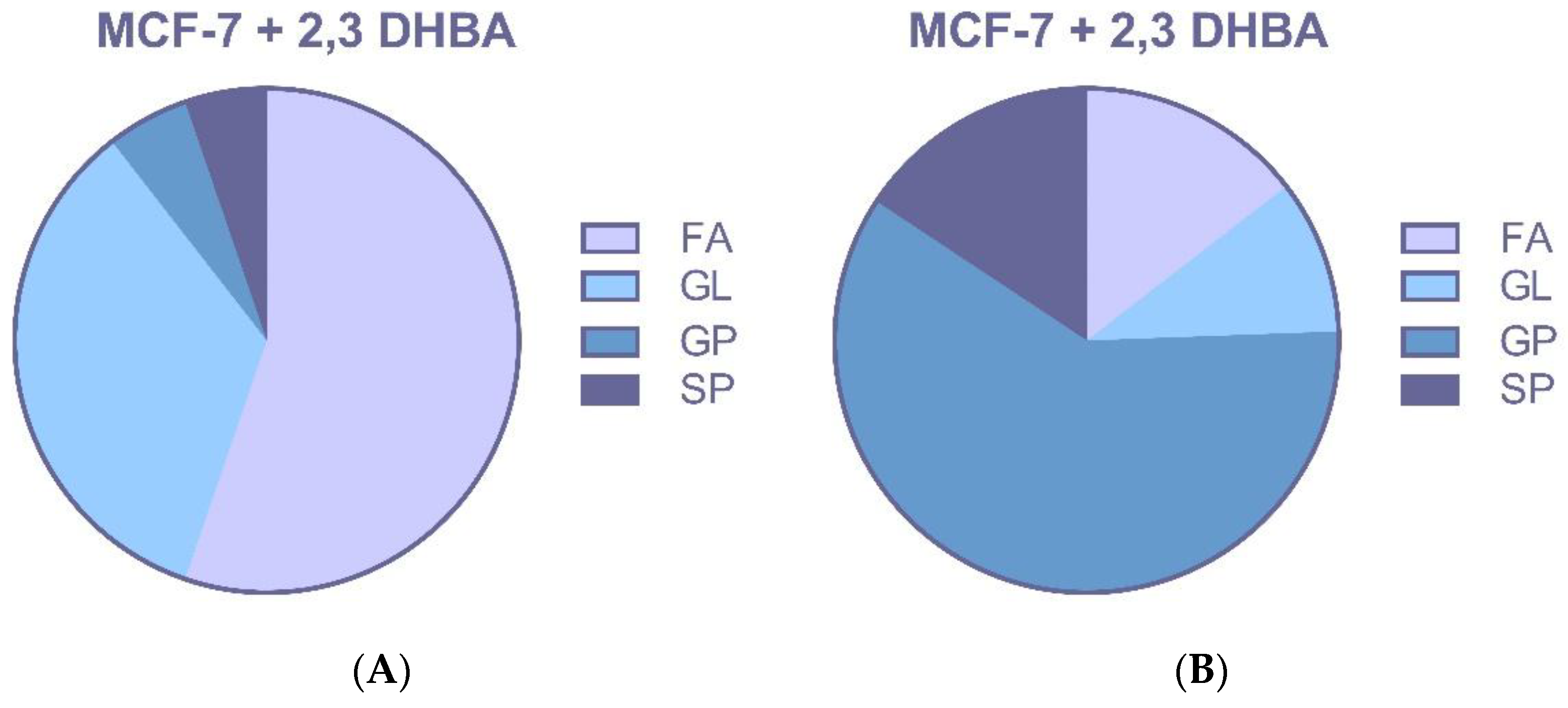
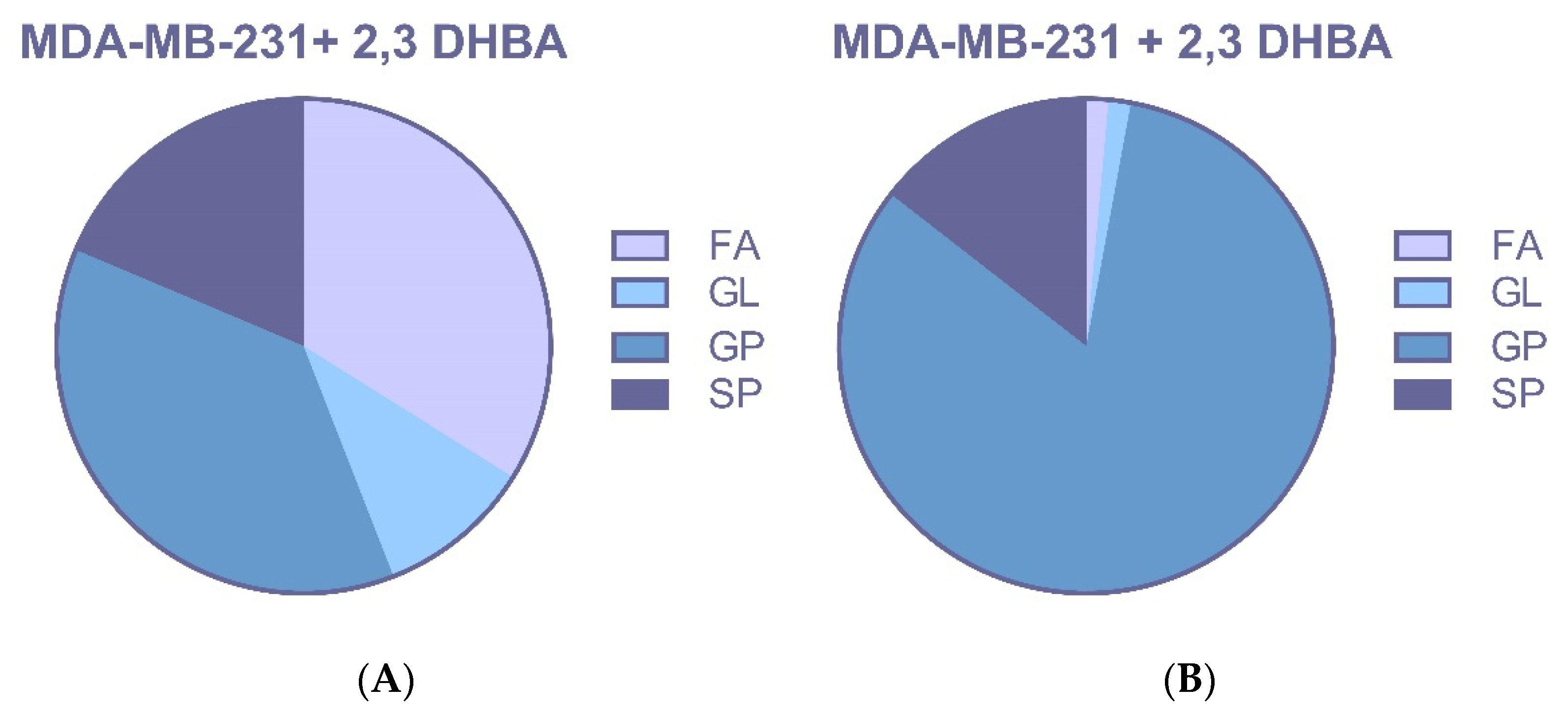
3.2.3. Multivariate Statistical Analysis for Lipids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juurlink, B.H.J.; Azouz, H.J.; Aldalati, A.M.Z.; AlTinawi, B.M.H.; Ganguly, P. Hydroxybenzoic acid isomers and the cardiovascular system. Nutr. J. 2014, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, J.G.; Mahdi, A.J.; Mahdi, A.J.; Bowen, I.D. The historical analysis of aspirin discovery, its relation to the willow tree and antiproliferative and anticancer potential. Cell Prolif. 2006, 39, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Robbins, R.J. Phenolic acids in foods: An overview of analytical methodology. J. Agric. Food Chem. 2003, 51, 2866–2887. [Google Scholar] [CrossRef] [PubMed]
- Joyeux, M.; Lobstein, A.; Anton, R.; Mortier, F. Comparative antilipoperoxidant, antinecrotic and scavanging properties of terpenes and biflavones from Ginkgo and some flavonoids. Planta Medica 1995, 61, 126–129. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free. Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Dinelli, G.; Carretero, A.S.; Di Silvestro, R.; Marotti, I.; Fu, S.; Benedettelli, S.; Ghiselli, L.; Gutiérrez, A.F. Determination of phenolic compounds in modern and old varieties of durum wheat using liquid chromatography coupled with time-of-flight mass spectrometry. J. Chromatogr. A 2009, 1216, 7229–7240. [Google Scholar] [CrossRef]
- Bacon, J.R.; Rhodes, M.J.C. Binding affinity of hydrolyzable tannins to parotid saliva and to proline-rich proteins derived from it. J. Agric. Food Chem. 2000, 48, 838–843. [Google Scholar] [CrossRef]
- Sabally, K. Lipase-Catalyzed Synthesis of Selected Phenolic Lipids in Organic Solvent Media. Ph.D. Thesis, McGill University, Montreal, QC, Canada, 2011. [Google Scholar]
- Stamatis, H.; Sereti, V.; Kolisis, F.N. Enzymatic synthesis of hydrophilic and hydrophobic derivatives of natural phenolic acids in organic media. J. Mol. Catal.—B Enzym. 2001, 11, 323–328. [Google Scholar] [CrossRef]
- Türk, H. Bazı Sofralık Üzüm Çeşitlerinde Farklı Dönemlerde Alınan Yapraklardaki Fenolik ve Mineral Madde Değişimlerinin Belirlenmesi. Ph.D. Thesis, Süleyman Demirel University, Isparta, Turkey, 2009. [Google Scholar]
- Grootveld, M.; Halliwell, B. 2,3-Dihydroxybenzoic acid is a product of human aspirin metabolism. Biochem. Pharmacol. 1988, 37, 271–280. [Google Scholar] [CrossRef]
- Liu, D.; Su, Z.; Wang, C.; Gu, M. Separation of five isomers of dihydroxybenzoic acid by high-speed counter-current chromatography with dual-rotation elution method. J. Chromatogr. Sci. 2009, 47, 345–348. [Google Scholar] [CrossRef][Green Version]
- Torres, A.M.; Mau-Lastovicka, T.; Rezaaiyan, R. Total phenolics and high-performance liquid chromatography of phenolic acids of avocado. J. Agric. Food Chem. 1987, 35, 921–925. [Google Scholar] [CrossRef]
- Zhang, K.; Zuo, Y. GC-MS Determination of Flavonoids and Phenolic and Benzoic Acids in Human Plasma after Consumption of Cranberry Juice. J. Agric. Food Chem. 2004, 52, 222–227. [Google Scholar] [CrossRef]
- Zuo, Y.; Wang, C.; Zhan, J. Separation, characterization, and quantitation of benzoic and phenolic antioxidants in American cranberry fruit by GC-MS. J. Agric. Food Chem. 2002, 50, 3789–3794. [Google Scholar] [CrossRef] [PubMed]
- Blatt, J.; Taylor, S.R.; Kontoghiorghes, G.J. Comparison of Activity of Deferoxamine with That of Oral Iron Chelators against Human Neuroblastoma Cell Lines. Cancer Res. 1989, 49, 2925–2927. [Google Scholar] [PubMed]
- Graziano, J.H.; Miller, D.R.; Grady, R.W.; Cerami, A. Inhibition of Membrane Peroxidation in Thalassaemic Erythrocytes by 2,3-Dihydroxybenzoic Acid. Br. J. Haematol. 1976, 32, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Neilands, J.B. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Ejsing, C.S.; Sampaio, J.L.; Surendranath, V.; Duchoslav, E.; Ekroos, K.; Klemm, R.W.; Simons, K.; Shevchenko, A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 2136–2141. [Google Scholar] [CrossRef]
- Navas-Iglesias, N.; Carrasco-Pancorbo, A.; Cuadros-Rodríguez, L. From lipids analysis towards lipidomics, a new challenge for the analytical chemistry of the 21st century. Part II: Analytical lipidomics. TrAC Trends Anal. Chem. 2009, 28, 393–403. [Google Scholar] [CrossRef]
- Vaz, F.M.; Pras-Raves, M.; Bootsma, A.H.; van Kampen, A.H. Principles and practice of lipidomics. J. Inherit. Metab. Dis. 2015, 38, 41–52. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Cheng, X.L.; Lin, R.C. Lipidomics applications for discovering biomarkers of diseases in clinical chemistry. Int. Rev. Cell Mol. Biol. 2014, 313, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Shui, G. Lipidomics as a principal tool for advancing biomedical research. J. Genet. Genom. 2013, 40, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Min, H.K.; Lim, S.; Chung, B.C.; Moon, M.H. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal. Bioanal. Chem. 2011, 399, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Taïb, B.; Aboussalah, A.M.; Moniruzzaman, M.; Chen, S.; Haughey, N.J.; Kim, S.F.; Ahima, R.S. Lipid accumulation and oxidation in glioblastoma multiforme. Sci. Rep. 2019, 9, 19593. [Google Scholar] [CrossRef]
- Zhou, X.; Mao, J.; He, Z.; Henegar, J. Lipidomics in identifying lipid biomarkers of prostate cancer. FASEB J. 2010, 24, 354–356. [Google Scholar] [CrossRef]
- Zhou, X.; Mao, J.; Ai, J.; Deng, Y.; Roth, M.R.; Pound, C.; Henegar, J.; Welti, R.; Bigler, S.A.; Addison, C.L. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS ONE 2012, 7, e48889. [Google Scholar] [CrossRef]
- Kvasnička, A.; Najdekr, L.; Dobešová, D.; Piskláková, B.; Ivanovová, E.; Friedecký, D. Clinical lipidomics in the era of the big data. Clin. Chem. Lab. Med. 2023, 61, 587–598. [Google Scholar] [CrossRef]
- Yang, L.; Li, M.; Shan, Y.; Shen, S.; Bai, Y.; Liu, H. Recent advances in lipidomics for disease research. J. Sep. Sci. 2016, 39, 38–50. [Google Scholar] [CrossRef]
- Kim, J.; Harper, A.; McCormack, V.; Sung, H.; Houssami, N.; Morgan, E.; Mutebi, M.; Garvey, G.; Soerjomataram, I.; Fidler-Benaoudia, M.M. Global patterns and trends in breast cancer incidence and mortality across 185 countries. Nat. Med. 2025, 31, 1154–1162. [Google Scholar] [CrossRef]
- Valuate Reports. Alternative Cancer Treatment—Global Market Share and Ranking, Overall Sales and Demand Forecast 2024–2030. 2024. Available online: https://reports.valuates.com/market-reports/QYRE-Auto-25P12376/global-alternative-cancer-treatment (accessed on 19 June 2025).
- Boon, H.; Brown, J.B.; Gavin, A.; Kennard, A.A.; Stewart, M. Breast cancer survivors’ perceptions of complementary/alternative medicine (CAM): Making the decision to use or not to use. Qual. Health Res. 1999, 9, 639–653. [Google Scholar] [CrossRef]
- Rana, A.; Samtiya, M.; Dhewa, T.; Mishra, V.; Aluko, R.E. Health benefits of polyphenols: A concise review. J. Food Biochem. 2022, 46, e14264. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.L.; Davis, S. An organic phosphorus assay which avoids the use of hazardous perchloric acid. Clin. Chim. Acta 1982, 121, 111–116. [Google Scholar] [CrossRef]
- Peterson, B.; Stovall, K.; Monian, P.; Franklin, J.L.; Cummings, B.S. Alterations in phospholipid and fatty acid lipid profiles in primary neocortical cells during oxidant-induced cell injury. Chem.-Biol. Interact. 2008, 174, 163–176. [Google Scholar] [CrossRef]
- Taguchi, R.; Hayakawa, J.; Takeuchi, Y.; Ishida, M. Two-dimensional analysis of phospholipids by capillary liquid chromatography/electrospray ionization mass spectrometry. J. Mass Spectrom. 2000, 35, 953–966. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35 (Suppl. S2), W606–W612. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Kinsey, G.R.; Blum, J.L.; Covington, M.D.; Cummings, B.S.; McHowat, J.; Schnellmann, R.G. Decreased iPLA2γ expression induces lipid peroxidation and cell death and sensitizes cells to oxidant-induced apoptosis. J. Lipid Res. 2008, 49, 1477–1487. [Google Scholar] [CrossRef]
- Zhang, L.; Peterson, B.L.; Cummings, B.S. The effect of inhibition of Ca2+-independent phospholipase A2 on chemotherapeutic-induced death and phospholipid profiles in renal cells. Biochem. Pharmacol. 2005, 70, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Dehelean, C.A.; Marcovici, I.; Soica, C.; Mioc, M.; Coricovac, D.; Iurciuc, S.; Cretu, O.M.; Pinzaru, I. Plant-derived anticancer compounds as new perspectives in drug discovery and alternative therapy. Molecules 2021, 26, 1109. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H. Health benefits of fruit and vegetables are from additive and synergistic combinations of phytochemicals. Am. J. Clin. Nutr. 2003, 78 (Suppl. S3), 517S–520S. [Google Scholar] [CrossRef] [PubMed]
- Dachineni, R.; Kumar, D.R.; Callegari, E.; Kesharwani, S.S.; Sankaranarayanan, R.; Seefeldt, T.; Tummala, H.; Bhat, G.J. Salicylic acid metabolites and derivatives inhibit CDK activity: Novel insights into aspirin’s chemopreventive effects against colorectal cancer. Int. J. Oncol. 2017, 51, 1661–1673. [Google Scholar] [CrossRef]
- Sankaranarayanan, R.; Valiveti, C.K.; Dachineni, R.; Kumar, D.R.; Lick, T.; Bhat, G.J. Aspirin metabolites 2, 3-DHBA and 2, 5-DHBA inhibit cancer cell growth: Implications in colorectal cancer prevention. Mol. Med. Rep. 2020, 21, 20–34. [Google Scholar] [CrossRef]
- Rezaei-Seresht, H.; Cheshomi, H.; Falanji, F.; Movahedi-Motlagh, F.; Hashemian, M.; Mireskandari, E. Cytotoxic activity of caffeic acid and gallic acid against MCF-7 human breast cancer cells: An in silico and in vitro study. Avicenna J. Phytomed. 2019, 9, 574–586. [Google Scholar] [CrossRef]
- Kalinowska, M.; Gołębiewska, E.; Świderski, G.; Męczyńska-Wielgosz, S.; Lewandowska, H.; Pietryczuk, A.; Cudowski, A.; Astel, A.; Świsłocka, R.; Samsonowicz, M.; et al. Plant-derived and dietary hydroxybenzoic acids—A comprehensive study of structural, anti-/pro-oxidant, lipophilic, antimicrobial, and cytotoxic activity in MDA-MB-231 and MCF-7 cell lines. Nutrients 2021, 13, 3107. [Google Scholar] [CrossRef]
- Huang, X.; Liu, B.; Shen, S. Lipid Metabolism in Breast Cancer: From Basic Research to Clinical Application. Cancers 2025, 17, 650. [Google Scholar] [CrossRef]
- Cai, X.-X.; Zhang, Z.-Z.; Yang, X.-X.; Shen, W.-R.; Yuan, L.-W.; Ding, X.; Yu, Y.; Cai, W.-Y. Unveiling the impact of lipid metabolism on triple-negative breast cancer growth and treatment options. Front. Oncol. 2025, 15, 1579423. [Google Scholar] [CrossRef]
- Lupu, R.; Menendez, J.A. Pharmacological inhibitors of fatty acid synthase (FASN)-catalyzed endogenous fatty acid biogenesis: A new family of anti-cancer agents? Curr. Pharm. Biotechnol. 2006, 7, 483–494. [Google Scholar] [CrossRef]
- Janardhan, S.; Srivani, P.; Sastry, G.N. Choline kinase: An important target for cancer. Curr. Med. Chem. 2006, 13, 1169–1186. [Google Scholar] [CrossRef]
- Danilo, C.; Frank, P.G. Cholesterol and breast cancer development. Curr. Opin. Pharmacol. 2012, 12, 677–682. [Google Scholar] [CrossRef]
- Bandu, R.; Mok, H.J.; Kim, K.P. Phospholipids as cancer biomarkers: Mass spectrometry-based analysis. Mass Spectrom. Rev. 2018, 37, 107–138. [Google Scholar] [CrossRef]
- Postle, A.D. Lipidomics. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, L.; Liu, N.; He, C.; Li, Z. Decreased serum levels of free fatty acids are associated with breast cancer. Clin. Chim. Acta 2014, 437, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Dória, M.L.; Cotrim, C.Z.; Simões, C.; Macedo, B.; Domingues, P.; Domingues, M.R.; Helguero, L.A. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J. Cell. Physiol. 2013, 228, 457–468. [Google Scholar] [CrossRef] [PubMed]
- More, T.H.; Bagadi, M.; RoyChoudhury, S.; Dutta, M.; Uppal, A.; Mane, A.; Santra, M.K.; Chaudhury, K.; Rapole, S. Comprehensive quantitative lipidomic approach to investigate serum phospholipid alterations in breast cancer. Metabolomics 2017, 13, 3. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, G.; Pan, L.; Yan, C.; Zhang, L.; Weng, Y.; Wang, W.; Chen, X.; Yang, G. Potential plasma lipid biomarkers in early-stage breast cancer. Biotechnol. Lett. 2017, 39, 1657–1666. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, X.; Sun, Y.; Jin, T.; Tang, R.; Zhou, X.; Xu, M.; Gan, Y.; Wang, R.; Luo, H.; et al. ACSL4-mediated membrane phospholipid remodeling induces integrin β1 activation to facilitate triple-negative breast cancer metastasis. Cancer Res. 2024, 84, 1856–1871. [Google Scholar] [CrossRef]
- Stoica, C.; Ferreira, A.K.; Hannan, K.; Bakovic, M. Bilayer forming phospholipids as targets for cancer therapy. Int. J. Mol. Sci. 2022, 23, 5266. [Google Scholar] [CrossRef]
- Cheng, M.; Bhujwalla, Z.M.; Glunde, K. Targeting phospholipid metabolism in cancer. Front. Oncol. 2016, 6, 266. [Google Scholar] [CrossRef]
- Fazio, A.; Obeng, E.O.; Rusciano, I.; Marvi, M.V.; Zoli, M.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; McCubrey, J.A.; Cocco, L.; et al. Subcellular localization relevance and cancer-associated mechanisms of diacylglycerol kinases. Int. J. Mol. Sci. 2020, 21, 5297. [Google Scholar] [CrossRef]
- Mérida, I.; Torres-Ayuso, P.; Ávila-Flores, A.; Arranz-Nicolás, J.; Andrada, E.; Tello-Lafoz, M.; Liébana, R.; Arcos, R. Diacylglycerol kinases in cancer. Adv. Biol. Regul. 2017, 63, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Sakane, F.; Hoshino, F.; Ebina, M.; Sakai, H.; Takahashi, D. The roles of diacylglycerol kinase α in cancer cell proliferation and apoptosis. Cancers 2021, 13, 5190. [Google Scholar] [CrossRef] [PubMed]
- Ramu, D.; Kim, E. Exosomal lipids in cancer progression and metastasis. Cancer Med. 2025, 14, e70687. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.V.; Riley, D.; Cosper, K.E.; Finlay-Schultz, J.; Brechbuhl, H.M.; Libby, A.E.; Hill, K.B.; Varshney, R.R.; Kabos, P.; Rudolph, M.C.; et al. Lipid metabolic reprogramming drives triglyceride storage and variable sensitivity to FASN inhibition in endocrine-resistant breast cancer cells. Breast Cancer Res. 2025, 27, 32. [Google Scholar] [CrossRef]
- Li, R.-Z.; Wang, X.-R.; Wang, J.; Xie, C.; Wang, X.-X.; Pan, H.-D.; Meng, W.-Y.; Liang, T.-L.; Li, J.-X.; Yan, P.-Y.; et al. The key role of sphingolipid metabolism in cancer: New therapeutic targets, diagnostic and prognostic values, and anti-tumor immunotherapy resistance. Front. Oncol. 2022, 12, 941643. [Google Scholar] [CrossRef]
- Janneh, A.H.; Ogretmen, B. Targeting sphingolipid metabolism as a therapeutic strategy in cancer treatment. Cancers 2022, 14, 2183. [Google Scholar] [CrossRef]
- Pal, P.; Atilla-Gokcumen, G.E.; Frasor, J. Emerging roles of ceramides in breast cancer biology and therapy. Int. J. Mol. Sci. 2022, 23, 11178. [Google Scholar] [CrossRef]
- Ohya, Y.; Ogiso, Y.; Matsuda, M.; Sakae, H.; Nishida, K.; Miki, Y.; Fox, T.E.; Kester, M.; Sakamoto, W.; Nabe, T.; et al. Pronecroptotic therapy using ceramide nanoliposomes is effective for triple-negative breast cancer cells. Cells 2024, 13, 405. [Google Scholar] [CrossRef]
- Schiffmann, S.; Sandner, J.; Birod, K.; Wobst, I.; Angioni, C.; Ruckhäberle, E.; Kaufmann, M.; Ackermann, H.; Lötsch, J.; Schmidt, H.; et al. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis 2009, 30, 745–752. [Google Scholar] [CrossRef]
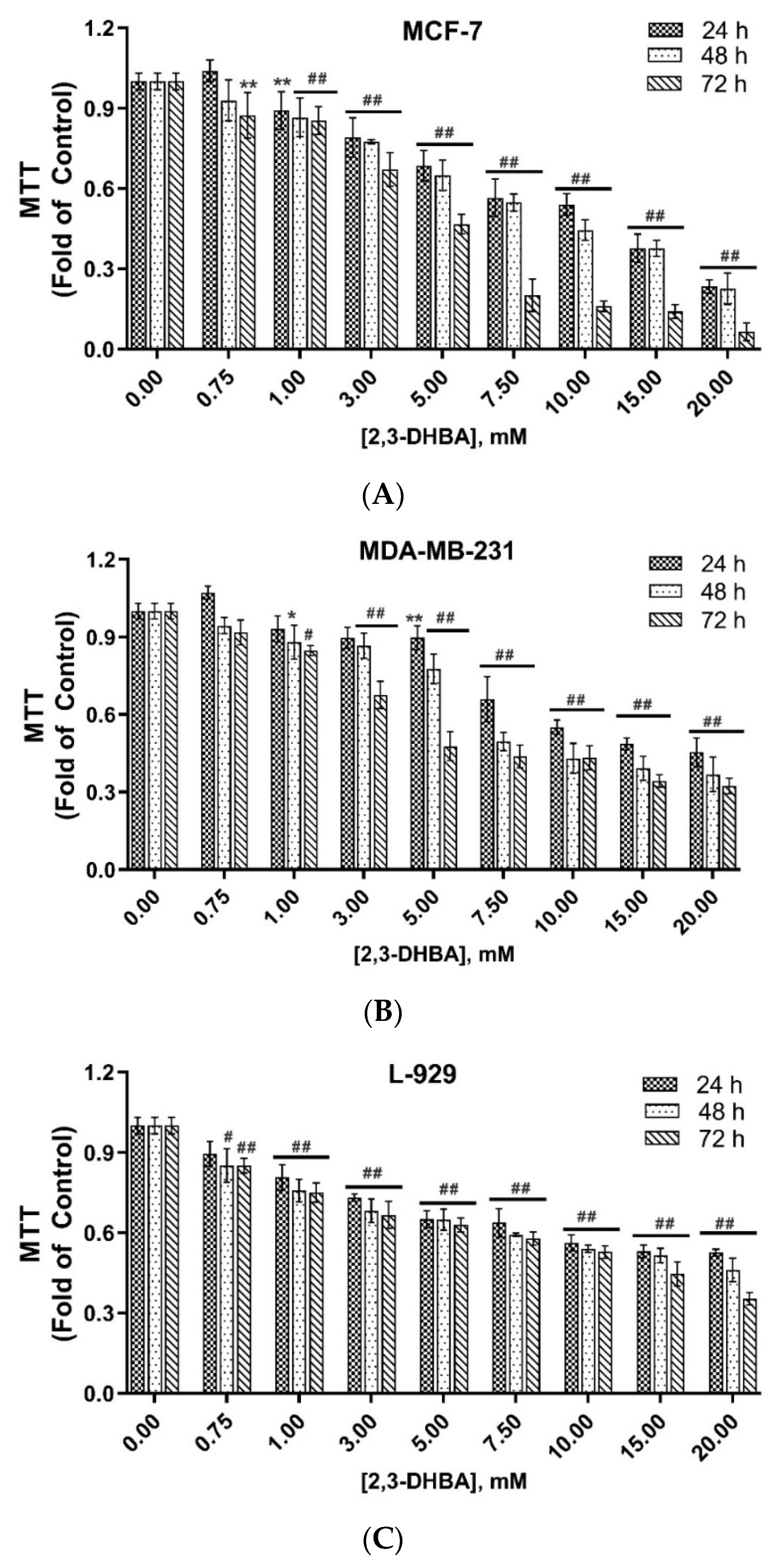

| (IC50, mM) a | ||||||
|---|---|---|---|---|---|---|
| 24 h | SI-24 h | 48 h | SI-48 h | 72 h | SI-72 h | |
| MCF-7 | 13.35 ± 0.14 | >1.49 | 8.61 ± 0.21 | 1.90 | 4.20 ± 0.12 | 3.45 |
| MDA-MB-231 | 10.52 ± 0.20 | >1.90 | 5.84 ± 0.10 | 2.80 | 4.09 ± 0.10 | 3.54 |
| L-929 b | >20 | 16.35 ± 0.13 | 14.51 ± 0.10 | |||
| m/z | Lipid Class Code a | Lipid Class Name | Formula b |
|---|---|---|---|
| 208.18 | LMFA08010014 | Faty acid | C8H18NS2 |
| 211.178 | LMFA00000040 | Faty acid | C5H12O2S |
| LMFA01030244 | Faty acid | C13H23O2 | |
| LMFA01030963 | Faty acid | C6H5O4Cl2 | |
| LMFA01150037 | Faty acid | C12H19O3 | |
| LMFA01170107 | Faty acid | C6H11O8 | |
| LMFA07010604 | Faty acid | C13H23O2 | |
| 215.151 | LMFA01031065 | Faty acid | C14H15O2 |
| LMFA01050168 | Faty acid | C12H23O3 | |
| 239.164 | LMFA01060185 | Faty acid | C14H23O3 |
| LMFA01150051 | Faty acid | C12H15O5 | |
| LMFA12000368 | Faty acid | C11H11O2S2 | |
| 262.193 | LMFA07070041 | Faty acid | C12H24NO5 |
| LMFA07070073 | Faty acid | C12H24NO5 | |
| LMFA07070081 | Faty acid | C11H20NO6 | |
| LMFA08030039 | Faty acid | C14H16NO4 | |
| 270.191 | LMFA01120009 | Faty acid | C14H24NO4 |
| 273.189 | LMFA01030510 | Faty acid | C18H25O2 |
| 274.181 | LMFA07070065 | Faty acid | C12H20NO6 |
| 289.201 | LMFA01031034 | Faty acid | C18H25O3 |
| LMFA12000356 | Faty acid | C15H13O2S2 | |
| 786.599 | LMGP01010764 | Glycerophospholipid | C44H85NO8P |
| LMGP01020225 | Glycerophospholipid | C45H89NO7P | |
| LMGP01080001 | Glycerophospholipid | C46H77NO7P | |
| LMGP02010709 | Glycerophospholipid | C45H73NO8P | |
| LMGP03010136 | Glycerophospholipid | C42H77NO10P | |
| LMGP03030066 | Glycerophospholipid | C43H81NO9P | |
| 832.241 | LMGP01010847 | Glycerophospholipid | C48H83NO8P |
| LMGP03010454 | Glycerophospholipid | C46H75NO10P | |
| LMGP20010046 | Glycerophospholipid | C44H83NO11P | |
| 875.266 | LMGP01010663 | Glycerophospholipid | C50H101NO8P |
| LMGP03010525 | Glycerophospholipid | C48H93NO10P | |
| LMSP02040010 | Sphingolipid | C57H112NO4 | |
| 977.775 | LMGL03011720 | Glycerolipid | C64H113O6 |
| LMGL03012263 | Glycerolipid | C65H101O6 | |
| LMGP06010691 | Glycerophospholipid | C53H102O13P13P | |
| 978.769 | LMGP13010007 | Glycerophospholipid | C46H82N3O15P2 |
| 1080.8265 | LMSP0502AA03 | Sphingolipid | C56H106NO18 |
| m/z | Lipid Classes Code a | Lipid Class Name | Formula b |
|---|---|---|---|
| 205.0902 | LMFA01050442 | Fatty acid | C7H9O7 |
| LMFA01070034 | Fatty acid | C11H9O4 | |
| LMFA01130001 | Fatty acid | C8H13O2S2 | |
| LMFA12000343 | Fatty acid | C11H9O2S | |
| 217.1225 | LMGL03012615 | Glycerolipid | C9H13O6 |
| 229.1412 | LMFA12000352 | Fatty acid | C13H9S2 |
| 255.2317 | LMFA01010001 | Fatty acid | C16H31O2 |
| LMFA01060047 | Fatty acid | C15H27O3 | |
| LMFA01150060 | Fatty acid | C12H15O6 | |
| LMFA05000009 | Fatty acid | C17H35O | |
| LMFA06000252 | Fatty acid | C16H31O2 | |
| 355.0776 | LMFA01030186 | Fatty acid | C24H35O2 |
| LMFA01040040 | Fatty acid | C19H31O6 | |
| LMFA01050077 | Fatty acid | C22H43O3 | |
| LMFA01170036 | Fatty acid | C21H39O4 | |
| LMFA03010069 | Fatty acid | C20H35O5 | |
| LMFA05000082 | Fatty acid | C23H47O2 | |
| LMFA06000269 | Fatty acid | C26H27O | |
| LMFA08020219 | Fatty acid | C19H35N2O4 | |
| LMFA08020264 | Fatty acid | C20H39N2O3 | |
| 539.3697 | LMFA03120034 | Fatty acid | C27H36O9Cl |
| LMFA08020206 | Fatty acid | C30H39N2O7 | |
| 551.3998 | LMFA01030844 | Fatty acid | C38H63O2 |
| LMGL02010360 | Glycerolipid | C34H63O5 | |
| 556.0005 | LMFA01030830 | Fatty acid | C38H67O2 |
| 605.4828 | LMGL02030029 | Glycerolipid | C41H65O3 |
| LMGP10010048 | Glycerophospholipid | C32H62O8P | |
| LMGP10020004 | Glycerophospholipid | C33H66O7P | |
| LMGP20070024 | Glycerophospholipid | C30H54O10P | |
| LMGP20070032 | Glycerophospholipid | C29H50O11P | |
| 606.3652 | LMGP20020016 | Glycerophospholipid | C28H49NO11P |
| LMGP20020023 | Glycerophospholipid | C29H53NO10P | |
| LMGP20020028 | Glycerophospholipid | C30H57NO9P | |
| LMGP20040019 | Glycerophospholipid | C28H49NO11P | |
| 606.4846 | LMGP01050057 | Glycerophospholipid | C32H65NO7P |
| LMGP02010058 | Glycerophospholipid | C31H61NO8P | |
| LMSP00000016 | Sphingolipid | C40H80NO2 | |
| LMSP02010017 | Sphingolipid | C39H76NO3 | |
| LMSP02010080 | Sphingolipid | C38H72NO4 | |
| LMGL02010007 | Glycerolipid | C37H69O5 | |
| 607.4824 | LMGL02010329 | Glycerolipid | C39H59O5 |
| LMGP20060022 | Glycerophospholipid | C29H52O11P | |
| 608.9148 | LMSP02010072 | Sphingolipid | C38H74NO4 |
| LMGP03060024 | Glycerophospholipid | C30H59NO9P | |
| LMGP20020017 | Glycerophospholipid | C28H51NO11P | |
| 615.5169 | LMFA08020175 | Fatty acid | C25H47N6O8 |
| LMGL02010063 | Glycerolipid | C39H67O5 | |
| LMGP10010051 | Glycerophospholipid | C33H60O8P | |
| LMGP10030006 | Glycerophospholipid | C34H64O7P | |
| LMSP04000001 | Sphingolipid | C34H68N2O5P | |
| 616.5143 | LMGL02070005 | Glycerolipid | C40H71O4 |
| LMGP20020040 | Glycerophospholipid | C30H51NO10P | |
| LMSP00000022 | Sphingolipid | C34H66NO6S | |
| LMSP02050002 | Sphingolipid | C34H67NO6P | |
| 617.5099 | LMGP06050026 | Glycerophospholipid | C29H46O12P |
| LMGP10020007 | Glycerophospholipid | C34H66O7P | |
| LMGP20070002 | Glycerophospholipid | C31H54O10P | |
| LMSP03020066 | Sphingolipid | C32H62N2O7P | |
| 619.3821 | LMGL02070007 | Glycerolipid | C41H63O4 |
| LMGP06050006 | Glycerophospholipid | C29H48O12P | |
| LMGP10010890 | Glycerophospholipid | C33H64O8P | |
| LMGP10020006 | Glycerophospholipid | C34H68O7P | |
| LMGP20070001 | Glycerophospholipid | C31H56O10P | |
| 634.8158 | LMGP01010001 | Glycerophospholipid | C33H65NO8P |
| LMGP02020022 | Glycerophospholipid | C34H69NO7P | |
| LMGP20010012 | Glycerophospholipid | C31H57NO10P | |
| LMGP20020043 | Glycerophospholipid | C30H53NO11P | |
| LMGP20040010 | Glycerophospholipid | C29H49NO12P | |
| LMSP00000015 | Sphingolipid | C42H84NO2 | |
| LMSP02010021 | Sphingolipid | C41H80NO3 | |
| LMSP02010093 | Sphingolipid | C40H76NO4 | |
| 638.8238 | LMSP02020032 | Sphingolipid | C40H80NO4 |
| 644.8418 | LMGP02010365 | Glycerophospholipid | C34H63NO8P |
| LMGP02030010 | Glycerophospholipid | C35H67NO7P | |
| 655.4137 | LMFA13030004 | Fatty acid | C36H63O10 |
| LMGP06050024 | Glycerophospholipid | C31H60O12P | |
| LMGP10010083 | Glycerophospholipid | C36H64O8P | |
| LMGP10020079 | Glycerophospholipid | C37H68O7P | |
| LMSP03020033 | Sphingolipid | C36H68N2O6P | |
| 656.8771 | LMGP02010367 | Glycerophospholipid | C35H63NO8P |
| LMSP00000025 | Sphingolipid | C37H70NO6S | |
| 666.0187 | LMGP20040007 | Glycerophospholipid | C30H53NO13P |
| LMGP20040014 | Glycerophospholipid | C31H57NO12P | |
| LMSP0501AA54 | Sphingolipid | C38H68NO8 | |
| 667.021 | LMGP10010065 | Glycerophospholipid | C37H64O8P |
| LMGP20050014 | Glycerophospholipid | C31H56O13P | |
| 683.0088 | LMGP10010151 | Glycerophospholipid | C38H68O8P |
| LMGP20050015 | Glycerophospholipid | C31H56O14P | |
| 706.8112 | LMGP02010445 | Glycerophospholipid | C39H65NO8P |
| LMGP03010889 | Glycerophospholipid | C36H69NO10P | |
| LMGP03020005 | Glycerophospholipid | C37H73NO9P | |
| LMSP02020028 | Sphingolipid | C46H92NO3 | |
| 734.0059 | LMGP02010533 | Glycerophospholipid | C41H69NO8P |
| LMGP03010029 | Glycerophospholipid | C38H73NO10P | |
| 747.5135 | LMGL03012646 | Glycerolipid | C47H87O6 |
| LMGP04010002 | Glycerophospholipid | C40H76O10P | |
| LMGP04020011 | Glycerophospholipid | C41H80O9P | |
| LMGP10010039 | Glycerophospholipid | C43H72O8P | |
| LMGP15040002 | Glycerophospholipid | C32H60O17P | |
| 748.5144 | LMGP20040036 | Glycerophospholipid | C36H63NO13P |
| LMGP01010447 | Glycerophospholipid | C42H71NO8P | |
| 750.5376 | LMSP03030150 | Sphingolipid | C38H73NO11P |
| 752.8087 | LMGP01010506 | Glycerophospholipid | C42H75NO8P |
| LMGP01020026 | Glycerophospholipid | C43H79NO7P | |
| LMGP01040092 | Glycerophospholipid | C44H83NO6P | |
| LMGP02020037 | Glycerophospholipid | C43H79NO7P | |
| LMGP03010131 | Glycerophospholipid | C40H67NO10P | |
| 768.851 | LMSP05010052 | Sphingolipid | C44H82NO9 |
| LMSP0501AA31 | Sphingolipid | C45H86NO8 | |
| LMGP01011423 | Glycerophospholipid | C43H79NO8P | |
| LMGP01020053 | Glycerophospholipid | C44H83NO7P | |
| LMGP03010091 | Glycerophospholipid | C41H71NO10P | |
| LMGP03020031 | Glycerophospholipid | C42H75NO9P | |
| 770.8552 | LMSP05010070 | Sphingolipid | C44H84NO9 |
| LMGP01011360 | Glycerophospholipid | C43H81NO8P | |
| LMGP01020188 | Glycerophospholipid | C44H85NO7P | |
| LMGP01040088 | Glycerophospholipid | C46H77NO6P | |
| LMGP02080002 | Glycerophospholipid | C45H73NO7P | |
| LMGP03010153 | Glycerophospholipid | C41H73NO10P | |
| LMGP03020029 | Glycerophospholipid | C42H77NO9P | |
| LMGL00000134 | Glycerolipid | C46H76NO8 | |
| 773.5216 | LMGP04110002 | Glycerophospholipid | C44H70O9P |
| LMGP06010024 | Glycerophospholipid | C39H66O13P | |
| LMGP15040007 | Glycerophospholipid | C34H62O17P | |
| LMSP05010076 | Sphingolipid | C44H86NO9 | |
| LMGP10010835 | Glycerophospholipid | C45H74O8P | |
| LMGP04100003 | Glycerophospholipid | C42H78O10P | |
| LMGL05010001 | Glycerolipid | C45H73O10 | |
| 774.7834 | LMGP01010401 | Glycerophospholipid | C43H85NO8P |
| LMGP01020023 | Glycerophospholipid | C44H89NO7P | |
| LMSP02040003 | Sphingolipid | C50H96NO4 | |
| 775.4956 | LMGP04110003 | Glycerophospholipid | C44H72O9P |
| LMGP06010022 | Glycerophospholipid | C39H68O13P | |
| LMGP15040006 | Glycerophospholipid | C34H64O17P | |
| 775.5 | LMGP04010037 | Glycerophospholipid | C42H80O10P |
| LMGP04020033 | Glycerophospholipid | C43H84O9P | |
| LMGP10010585 | Glycerophospholipid | C45H76O8P | |
| 776.5165 | LMGP01010512 | Glycerophospholipid | C44H75NO8P |
| LMGP02020104 | Glycerophospholipid | C45H79NO7P | |
| LMGP03010088 | Glycerophospholipid | C41H79NO10P | |
| LMGP03020070 | Glycerophospholipid | C42H83NO9P | |
| 786.8336 | LMGP01010750 | Glycerophospholipid | C44H85NO8P |
| LMGP01020208 | Glycerophospholipid | C45H89NO7P | |
| LMGP01080003 | Glycerophospholipid | C46H77NO7P | |
| LMGP02010767 | Glycerophospholipid | C45H73NO8P | |
| LMSP05010044 | Sphingolipid | C44H84NO10 | |
| 792.8503 | LMGP01011460 | Glycerophospholipid | C45H79NO8P |
| LMGP01020066 | Glycerophospholipid | C46H83NO7P | |
| LMGP03010160 | Glycerophospholipid | C43H71NO10P | |
| LMGP03020088 | Glycerophospholipid | C44H75NO9P | |
| 806.5769 | LMGP01010645 | Glycerophospholipid | C46H81NO8P |
| LMGP02030091 | Glycerophospholipid | C47H85NO7P | |
| LMGP03010043 | Glycerophospholipid | C44H73NO10P | |
| LMGP20020004 | Glycerophospholipid | C45H77NO9P | |
| LMSP03030154 | Sphingolipid | C42H81NO11P | |
| LMSP06020001 | Sphingolipid | C42H80NO11S | |
| 807.981 | LMGL03012734 | Glycerolipid | C52H87O6 |
| LMGP04010306 | Glycerophospholipid | C45H76O10P | |
| LMGP04020083 | Glycerophospholipid | C46H80O9P | |
| LMGP06010027 | Glycerophospholipid | C41H76O13P | |
| LMGP06020009 | Glycerophospholipid | C42H80O12P | |
| LMGP10010723 | Glycerophospholipid | C47H84O8P | |
| LMGP20050035 | Glycerophospholipid | C39H68O15P | |
| LMSP03030007 | Sphingolipid | C42H83NO11P | |
| 885.541 | LMGP06010010 | Glycerophospholipid | C47H82O13P |
| 886.5455 | LMGP03010786 | Glycerophospholipid | C50H81NO10P |
| 964.7116 | LMSP03030023 | Sphingolipid | C52H103NO12P |
| 1003.977 | LMGL03012162 | Glycerolipid | C66H115O6 |
| LMGL03012500 | Glycerolipid | C67H103O6 | |
| 1033.682 | LMGL03012468 | Glycerolipid | C68H121O6 |
| LMGL03012580 | Glycerolipid | C69H109O6 | |
| 1047.906 | LMGL03012466 | Glycerolipid | C69H123O6 |
| m/z | Lipid Classes Code a | Lipid Class Name | Formula b |
|---|---|---|---|
| 235.1762 | LMFA01090008 | Fatty acid | C12H24O2Cl |
| LMFA01090080 | Fatty acid | C9H16O2 | |
| LMFA01140064 | Fatty acid | C15H23O2 | |
| LMFA06000156 | Fatty acid | C14H19O3 | |
| LMFA12000354 | Fatty acid | C12H11OS2 | |
| 245.1809 | LMFA00000009 | Fatty acid | C12H25N2O3 |
| LMFA01030703 | Fatty acid | C16H21O2 | |
| LMFA01050445 | Fatty acid | C10H17N2O5 | |
| LMFA01150045 | Fatty acid | C14H13O4 | |
| LMFA01170010 | Fatty acid | C13H25O4 | |
| LMFA05000019 | Fatty acid | C17H25O | |
| 285.2044 | LMFA01050459 | Fatty acid | C18H21O3 |
| LMFA01150068 | Fatty acid | C14H21O6 | |
| LMFA12000363 | Fatty acid | C12H13O4S2 | |
| 321.231 | LMFA01030678 | Fatty acid | C22H25O2 |
| LMFA01090112 | Fatty acid | C16H18O2 | |
| 511.3819 | LMGP20070018 | Glycerophospholipid | C23H44O10P |
| 512.3858 | LMGP03010021 | Glycerophospholipid | C22H43NO10P |
| LMGP03050030 | Glycerophospholipid | C23H47NO9P | |
| 612.3732 | LMGP03060022 | Glycerophospholipid | C30H63NO9P |
| LMGP20040016 | Glycerophospholipid | C27H51NO12P | |
| LMSP02020031 | Sphingolipid | C38H78NO4 | |
| 614.181 | LMSP02030033 | Sphingolipid | C37H76NO5 |
| 619.443 | LMFA00000026 | Fatty acid | C24H44O5SCl5 |
| LMFA00000049 | Fatty acid | C32H59O11 | |
| LMFA08020171 | Fatty acid | C27H51N6O10 | |
| LMFA13010023 | Fatty acid | C34H67O9 | |
| LMGP06050026 | Glycerophospholipid | C29H48O12P | |
| LMGP10010072 | Glycerophospholipid | C33H64O8P | |
| LMGP10020007 | Glycerophospholipid | C34H68O7P | |
| LMGP20070002 | Glycerophospholipid | C31H56O10P | |
| LMSP03020066 | Sphingolipid | C32H64N2O7P | |
| 758.5683 | LMGP01010585 | Glycerophospholipid | C42H81NO8P |
| LMGP01020201 | Glycerophospholipid | C43H85NO7P | |
| LMGP02040014 | Glycerophospholipid | C45H77NO6P | |
| LMGP03030038 | Glycerophospholipid | C41H77NO9P | |
| LMSP02050010 | Sphingolipid | C44H89NO6P | |
| LMSP05010059 | Sphingolipid | C43H84NO9 | |
| LMSP0501AA20 | Sphingolipid | C44H88NO8 | |
| 804.5512 | LMGP00000048 | Glycerophospholipid | C43H83NO10P |
| LMGP01010696 | Glycerophospholipid | C46H79NO8P | |
| LMGP01010975 | Glycerophospholipid | C45H91NO8P | |
| LMGP01020060 | Glycerophospholipid | C46H95NO7P | |
| LMGP02030092 | Glycerophospholipid | C47H83NO7P | |
| LMGP03020015 | Glycerophospholipid | C44H87NO9P | |
| LMSP02040004 | Sphingolipid | C52H102NO4 | |
| LMSP05010083 | Sphingolipid | C45H90NO10 | |
| 873.2717 | LMGL03016884 | Glycerolipid | C55H69O9 |
| 978.7616 | LMGP13010007 | Glycerophospholipid | C46H82N3O15P2 |
| LMGP13010007 | Glycerophospholipid | C46H82N3O15P2 | |
| 979.7644 | LMGL03011721 | Glycerolipid | C64H115O6 |
| LMGL03012217 | Glycerolipid | C65H103O6 | |
| LMGL03015976 | Glycerolipid | C64H115O6 | |
| LMGL03016504 | Glycerolipid | C65H103O6 | |
| LMGP06010915 | Glycerophospholipid | C53H104O13P | |
| 1019.787 | LMGL03012213 | Glycerolipid | C67H119O6 |
| LMSP0601AA02 | Sphingolipid | C53H99N2O16 | |
| 1020.79 | LMSP05010036 | Sphingolipid | C58H102NO13 |
| 1080.829 | LMSP0502AA03 | Sphingolipid | C56H106NO18 |
| m/z | Lipid Classes Code a | Lipid Class Name | Formula b |
|---|---|---|---|
| 556.0003 | LMFA01030830 | Fatty acid | C38H67O2 |
| 647.2407 | LMGP06050013 | Glycerophospholipid | C31H52O12P |
| LMGP10010012 | Glycerophospholipid | C35H68O8P | |
| LMGP20060025 | Glycerophospholipid | C31H52O12P | |
| LMSP02010009 | Sphingolipid | C42H80NO3 | |
| LMSP03020059 | Sphingolipid | C34H68N2O7P | |
| 683.008 | LMGP10010151 | Glycerophospholipid | C38H68O8P |
| LMGP20050015 | Glycerophospholipid | C31H56O14P | |
| 695.1657 | LMGP10010025 | Glycerophospholipid | C39H68O8P |
| LMGP20050022 | Glycerophospholipid | C32H56O14P | |
| LMSP02020018 | Sphingolipid | C44H88NO4 | |
| 734.0054 | LMGP02010533 | Glycerophospholipid | C41H69NO8P |
| LMGP03010029 | Glycerophospholipid | C38H73NO10P | |
| 747.5523 | LMGL03012646 | Glycerolipid | C47H87O6 |
| LMGP04010002 | Glycerophospholipid | C40H76O10P | |
| LMGP04020011 | Glycerophospholipid | C41H80O9P | |
| LMGP10010039 | Glycerophospholipid | C43H72O8P | |
| LMGP15040002 | Glycerophospholipid | C32H60O17P | |
| 748.5564 | LMGP20040036 | Glycerophospholipid | C36H63NO13P |
| LMGP01010447 | Glycerophospholipid | C42H71NO8P | |
| 750.532 | LMSP03030150 | Sphingolipid | C38H73NO11P |
| 788.5367 | LMGP01010006 | Glycerophospholipid | C44H87NO8P |
| LMGP01011461 | Glycerophospholipid | C45H75NO8P | |
| LMGP01020080 | Glycerophospholipid | C45H91NO7P | |
| LMGP01030015 | Glycerophospholipid | C46H79NO7P | |
| LMGP03010025 | Glycerophospholipid | C42H79NO10P | |
| LMGP03020033 | Glycerophospholipid | C43H83NO9P | |
| LMSP05010079 | Sphingolipid | C44H86NO10 | |
| 806.2032 | LMGP01010645 | Glycerophospholipid | C46H81NO8P |
| LMGP02030091 | Glycerophospholipid | C47H85NO7P | |
| LMGP03010043 | Glycerophospholipid | C44H73NO10P | |
| LMGP20020004 | Glycerophospholipid | C45H77NO9P | |
| LMSP03030154 | Sphingolipid | C42H81NO11P | |
| LMSP06020001 | Sphingolipid | C42H80NO11S | |
| 828.5609 | LMGP01010947 | Glycerophospholipid | C48H79NO8P |
| LMGP01011518 | Glycerophospholipid | C47H91NO8P | |
| LMGP01020213 | Glycerophospholipid | C48H95NO7P | |
| LMGP02010287 | Glycerophospholipid | C47H91NO8P | |
| LMGP03010246 | Glycerophospholipid | C45H83NO10P | |
| LMGP03010396 | Glycerophospholipid | C46H71NO10P | |
| LMGP03020039 | Glycerophospholipid | C46H87NO9P | |
| LMGP20010043 | Glycerophospholipid | C44H79NO11P | |
| 832.5941 | LMGP01010821 | Glycerophospholipid | C48H83NO8P |
| LMGP03010045 | Glycerophospholipid | C46H75NO10P | |
| LMGP03010244 | Glycerophospholipid | C45H87NO10P | |
| LMGP03010395 | Glycerophospholipid | C46H75NO10P | |
| LMGP03020059 | Glycerophospholipid | C46H91NO9P | |
| LMSP0501AB12 | Sphingolipid | C44H82NO13 | |
| 848.7064 | LMGP01011785 | Glycerophospholipid | C49H87NO8P |
| LMGP01030103 | Glycerophospholipid | C50H91NO7P | |
| LMGP03010479 | Glycerophospholipid | C47H79NO10P | |
| LMGP03020093 | Glycerophospholipid | C48H83NO9P | |
| 852.5666 | LMGP01011058 | Glycerophospholipid | C50H79NO8P |
| LMGP01011784 | Glycerophospholipid | C49H91NO8P | |
| LMGP01020252 | Glycerophospholipid | C50H95NO7P | |
| LMGP03010478 | Glycerophospholipid | C47H83NO10P | |
| LMGP03010840 | Glycerophospholipid | C48H71NO10P | |
| LMGP03020068 | Glycerophospholipid | C48H87NO9P | |
| LMGP04010556 | Glycerophospholipid | C48H84O10P | |
| LMSP03030014 | Sphingolipid | C44H87NO12P | |
| 856.699 | LMGP01011752 | Glycerophospholipid | C49H95NO8P |
| LMGP01011865 | Glycerophospholipid | C50H83NO8P | |
| LMGP01020241 | Glycerophospholipid | C50H99NO7P | |
| LMGP03010477 | Glycerophospholipid | C47H87NO10P | |
| LMGP03010619 | Glycerophospholipid | C48H75NO10P | |
| LMGP03020067 | Glycerophospholipid | C48H91NO9P | |
| LMSP05010046 | Sphingolipid | C49H94NO10 | |
| 885.5403 | LMGP06010010 | Glycerophospholipid | C47H82O13P |
| 886.5437 | LMGP03010786 | Glycerophospholipid | C50H81NO10P |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daş, B.; Şahin, S. Investigation of the Effect of 2,3-Dihydrobenzoic Acid Acid (2,3-DHBA) on the Lipid Profiles of MCF-7 and MDA-MB-231 Human Breast Cancer Cells via an Untargeted Lipidomic Approach. Biomolecules 2025, 15, 1341. https://doi.org/10.3390/biom15091341
Daş B, Şahin S. Investigation of the Effect of 2,3-Dihydrobenzoic Acid Acid (2,3-DHBA) on the Lipid Profiles of MCF-7 and MDA-MB-231 Human Breast Cancer Cells via an Untargeted Lipidomic Approach. Biomolecules. 2025; 15(9):1341. https://doi.org/10.3390/biom15091341
Chicago/Turabian StyleDaş, Büşra, and Serap Şahin. 2025. "Investigation of the Effect of 2,3-Dihydrobenzoic Acid Acid (2,3-DHBA) on the Lipid Profiles of MCF-7 and MDA-MB-231 Human Breast Cancer Cells via an Untargeted Lipidomic Approach" Biomolecules 15, no. 9: 1341. https://doi.org/10.3390/biom15091341
APA StyleDaş, B., & Şahin, S. (2025). Investigation of the Effect of 2,3-Dihydrobenzoic Acid Acid (2,3-DHBA) on the Lipid Profiles of MCF-7 and MDA-MB-231 Human Breast Cancer Cells via an Untargeted Lipidomic Approach. Biomolecules, 15(9), 1341. https://doi.org/10.3390/biom15091341







