Exploring the Inhibitory Potential of Six Porphyrin Compounds Against α-Amylase and α-Glucosidase Linked to Diabetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Inhibitory Activity Assay
2.2.1. α-Amylase Inhibitory Activity Assay
2.2.2. α-Glucosidase Inhibitory Activity Assay
2.2.3. Calculation of IC50
2.3. Mode of α-AMY and α-GLU Inhibition
2.4. In Vitro Stability Studies
2.5. Fluorescence Quenching Method
2.6. Synchronous Fluorescence Spectroscopy
2.7. Three-Dimensional Fluorescence Spectroscopy
2.8. Fourier Transform Infrared Spectroscopy
2.9. Molecular Docking
2.10. Molecular Dynamics Simulation
2.11. Cytotoxicity Assay
2.12. Pharmacokinetic Analysis
2.13. Statistical Analysis
3. Results
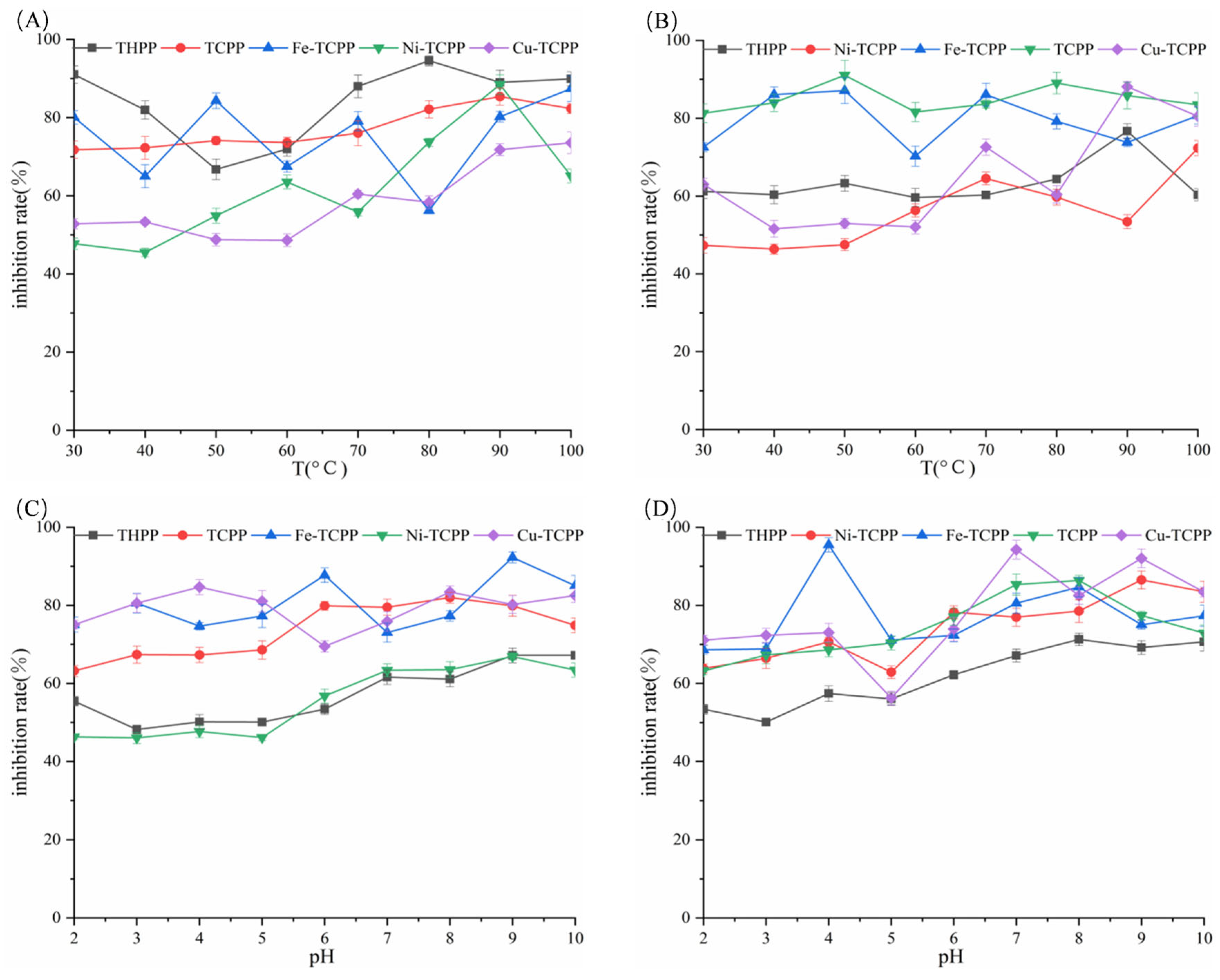
3.1. Inhibition of α-AMY and α-GLU by Six Porphyrin Compounds
3.2. Inhibition Modes of Six Porphyrin Compounds Against Two Enzymes
3.3. Study of Stability In Vitro
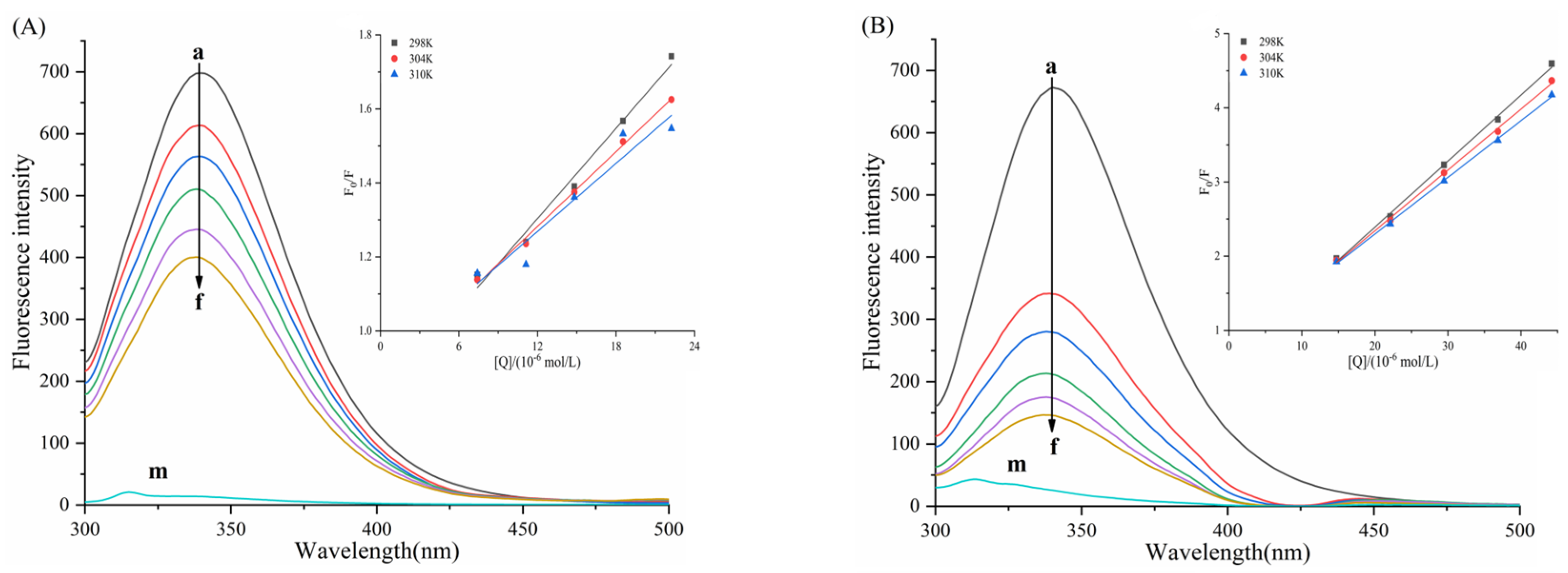
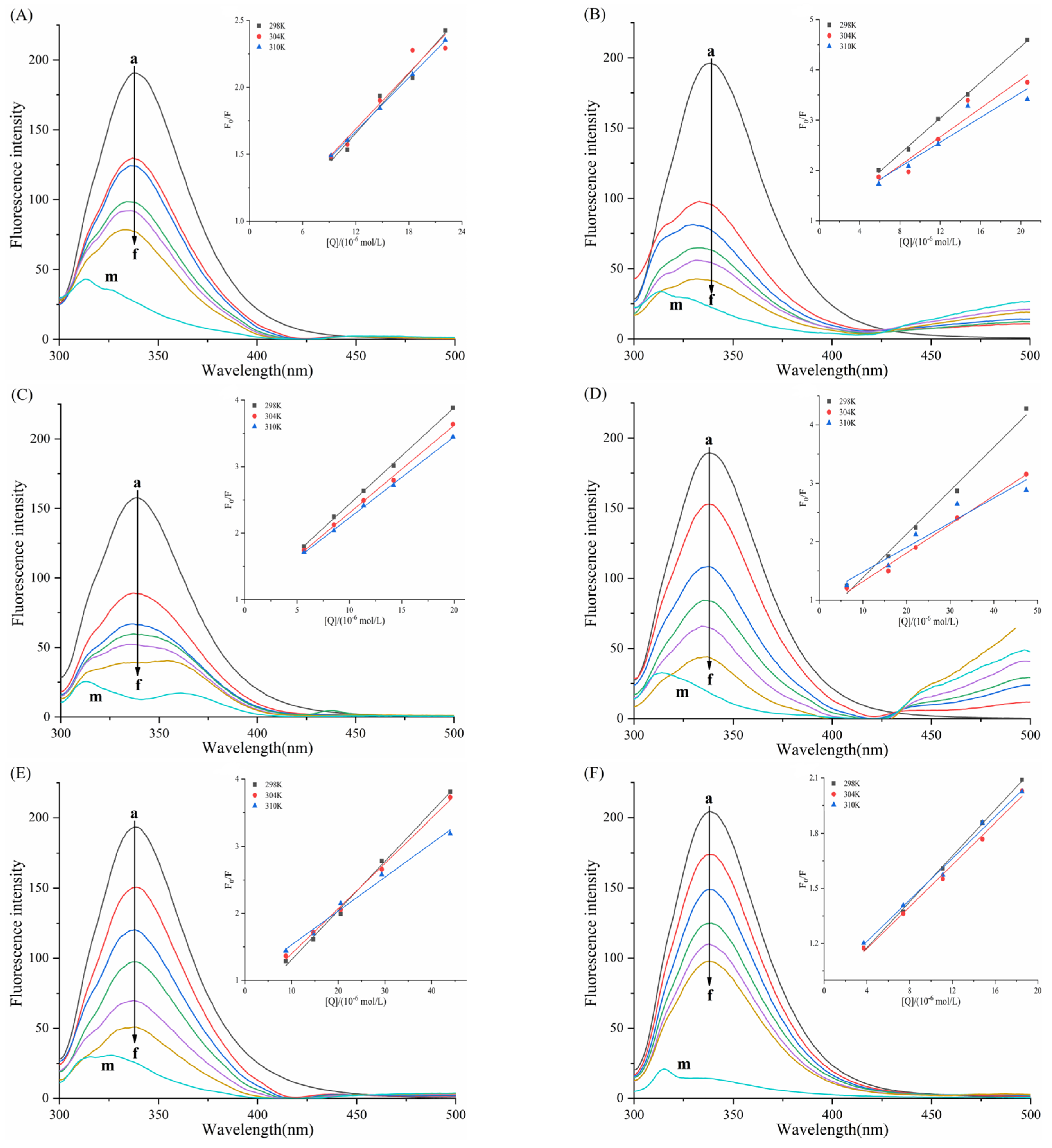
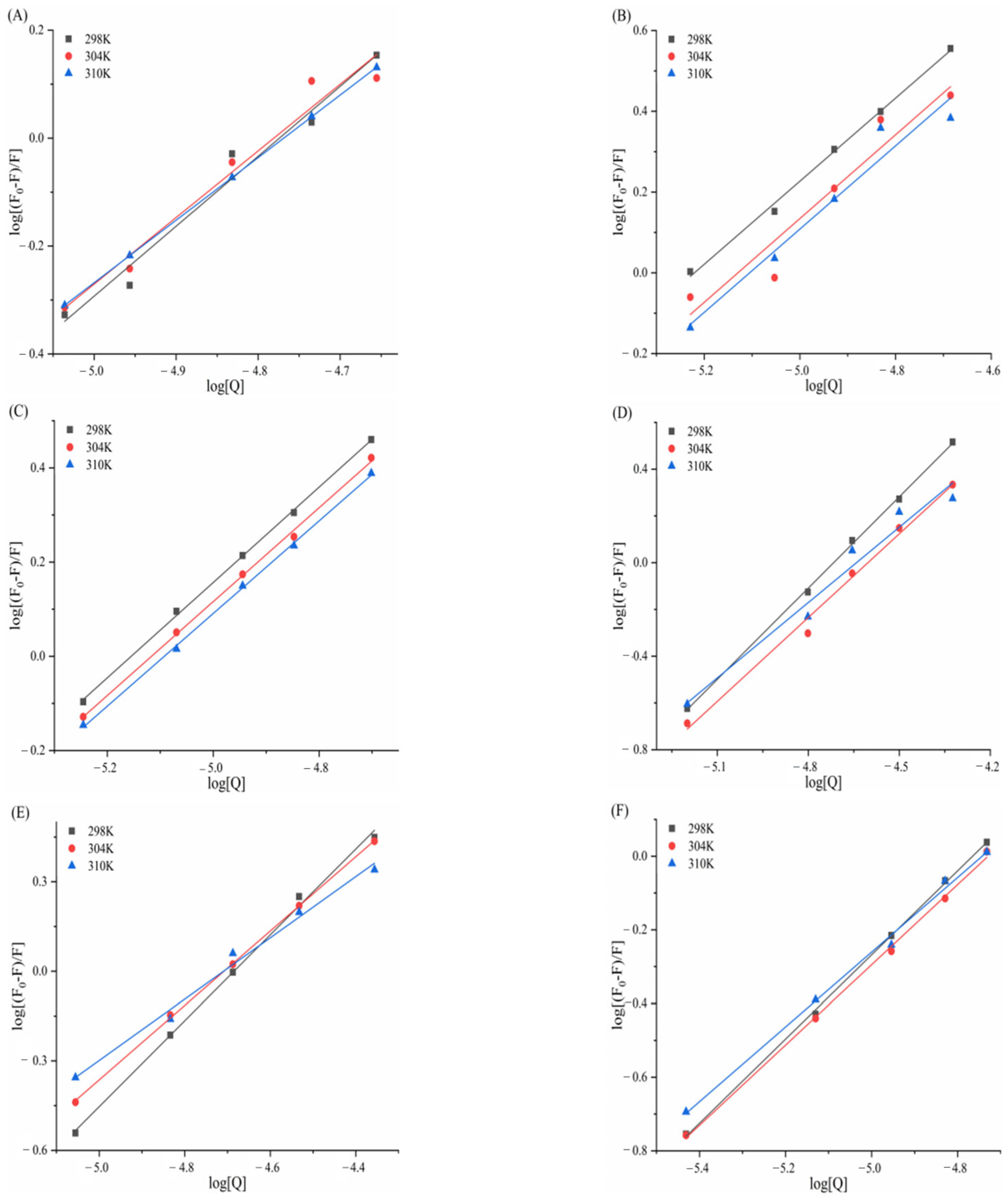
3.4. Fluorescence Quenching and Binding of Enzymes by Six Porphyrin Compounds
3.5. Determination of Thermodynamic Parameters and Main Forces
3.6. Effect of Porphyrins on the Synchronized Fluorescence Spectra
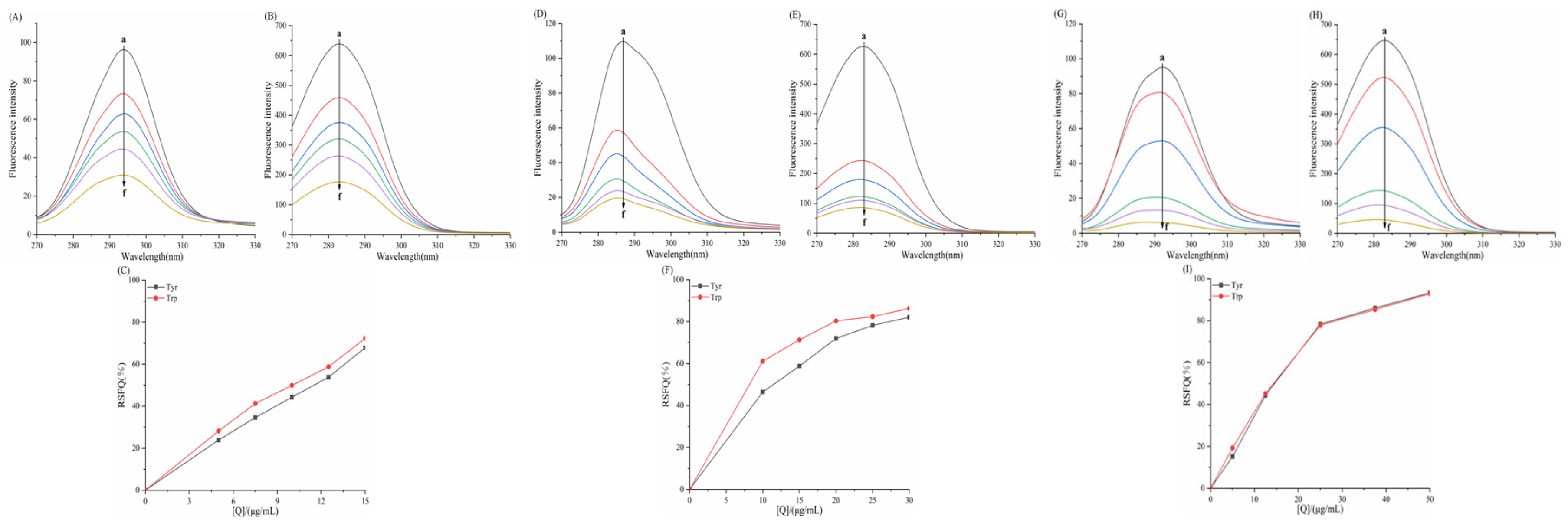
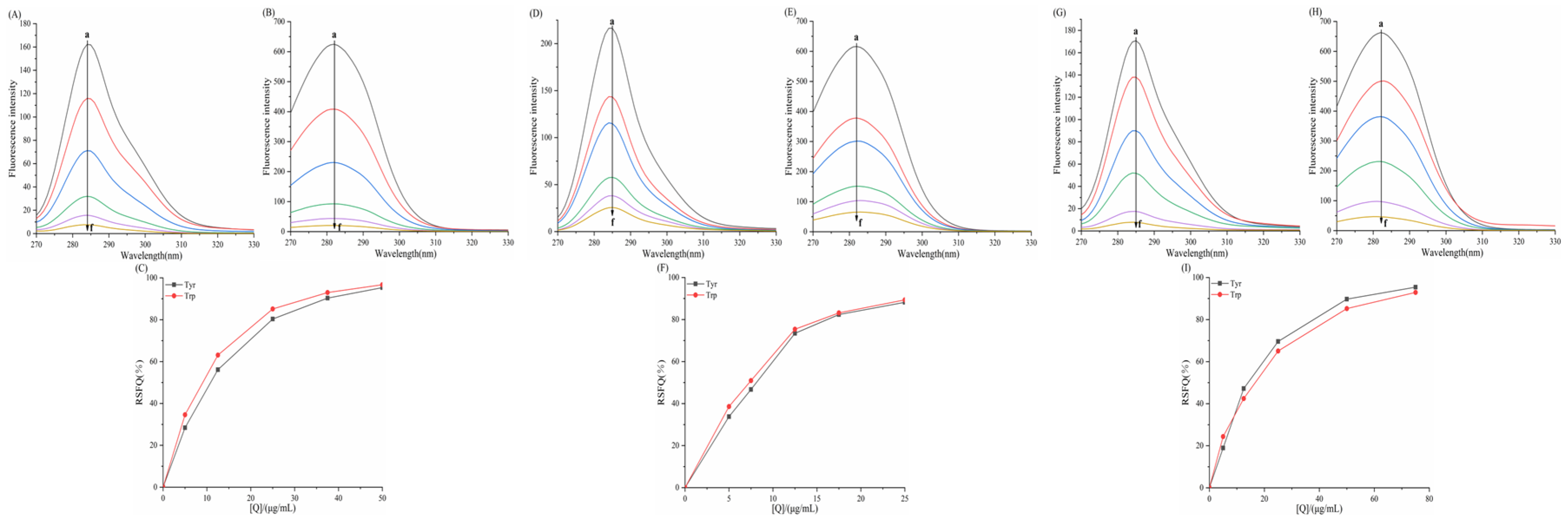
3.6.1. Effect of Porphyrins on the Synchronized Fluorescence Spectra of α-AMY
3.6.2. Effect of Porphyrins on the Synchronized Fluorescence Spectra of α-GLU
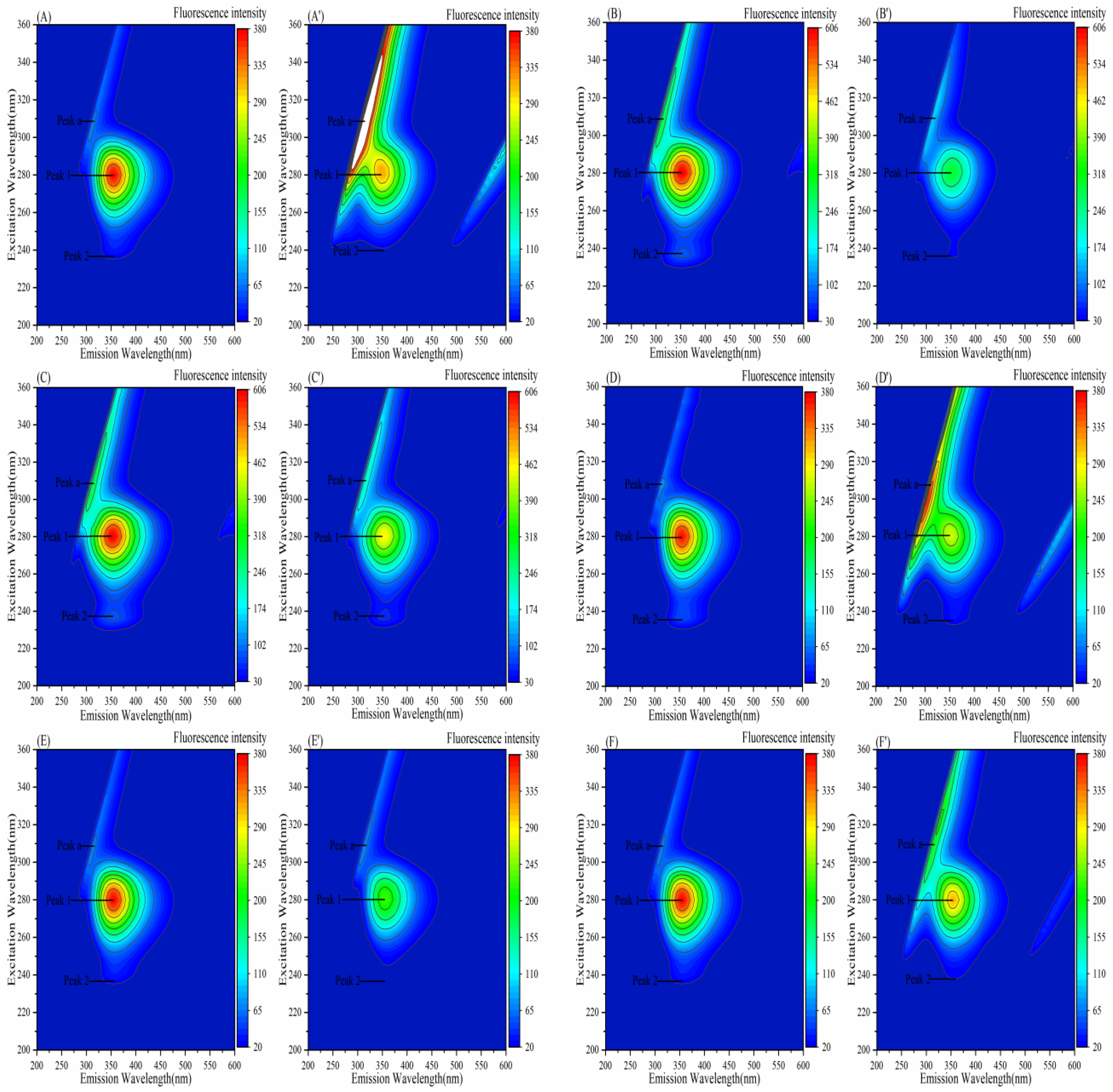
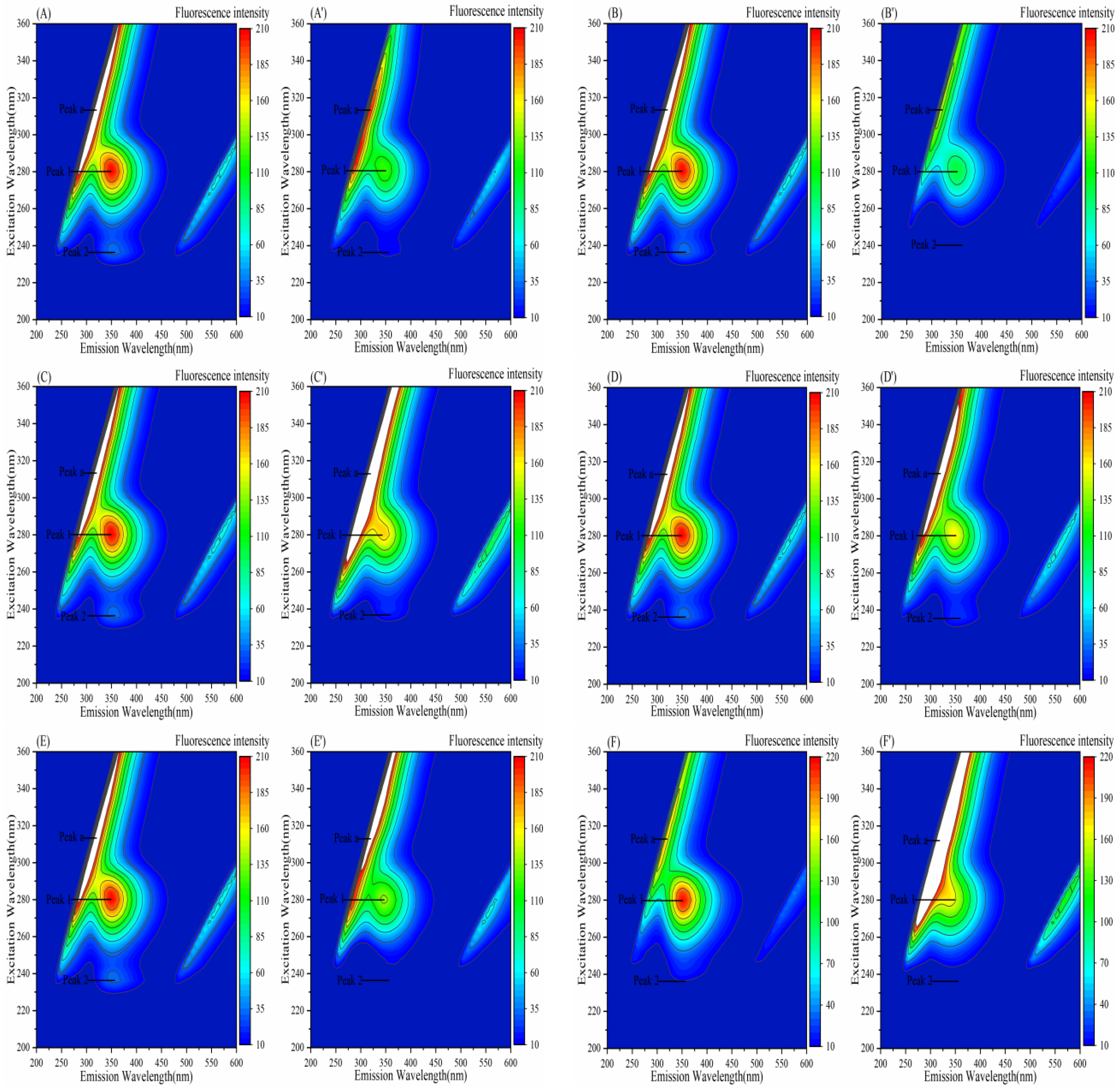
3.7. Effect of Porphyrins on the Three-Dimensional Fluorescence Spectra of Enzymes
3.7.1. Effect of Porphyrins on the Three-Dimensional Fluorescence Spectra of α-Amylase
3.7.2. Effect of Porphyrins on the Three-Dimensional Fluorescence Spectra of α-Glucosidase
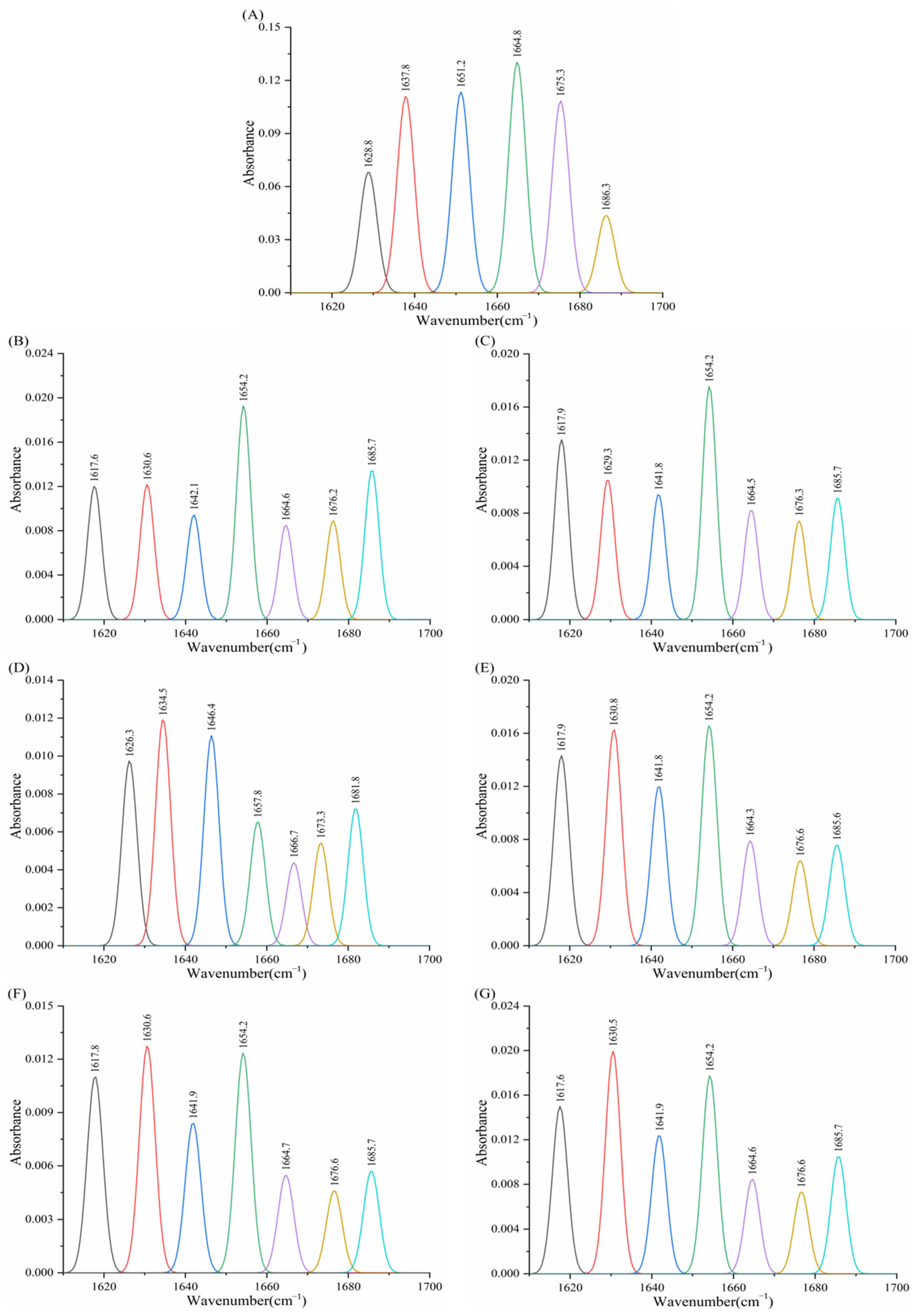
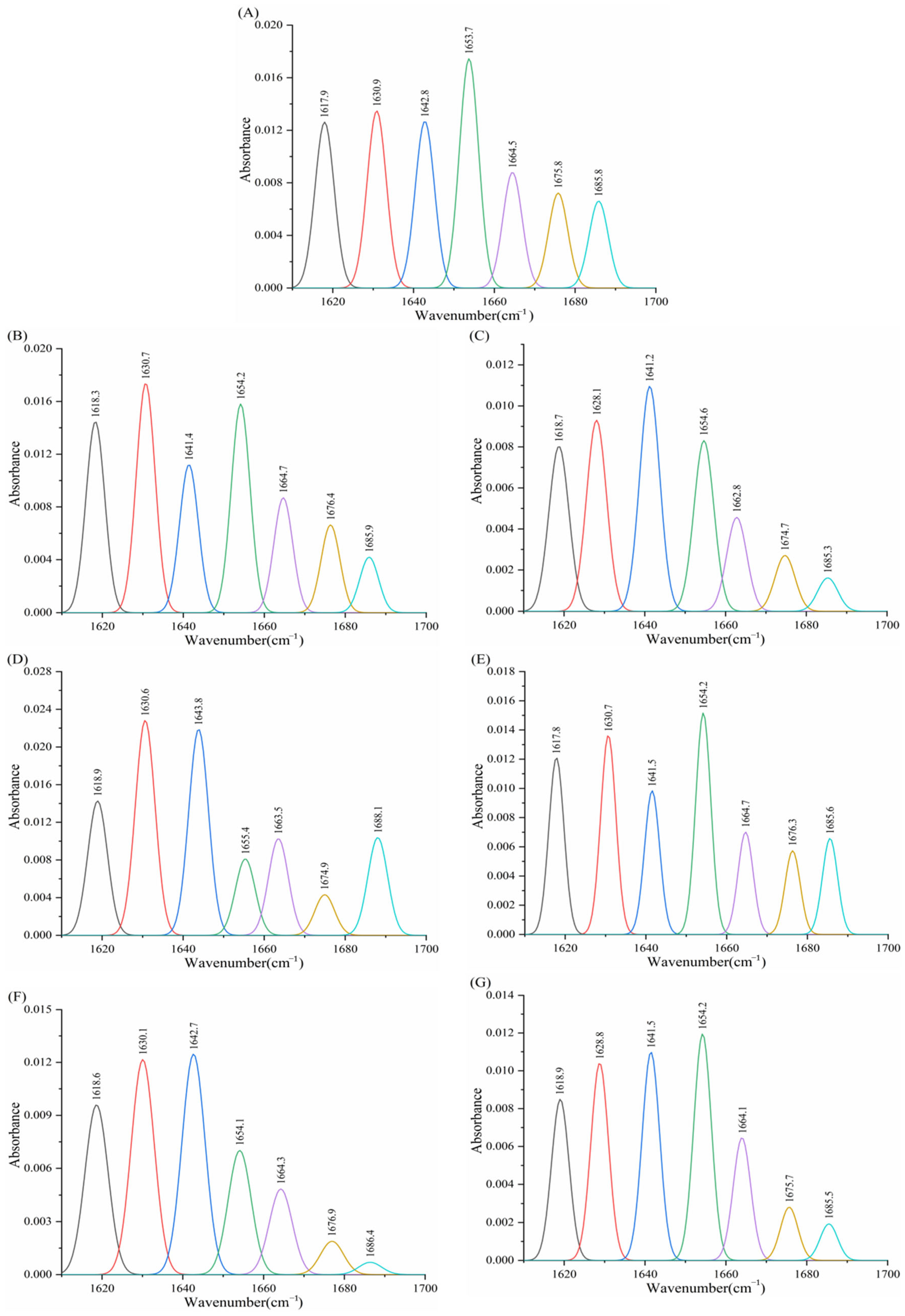
3.8. Effect of Porphyrins on the Infrared Spectrum of Enzymes
3.8.1. Effect of Porphyrins on the Infrared Spectrum of α-Amylase
3.8.2. Effect of Porphyrins on the Infrared Spectroscopy of α-Glucosidase
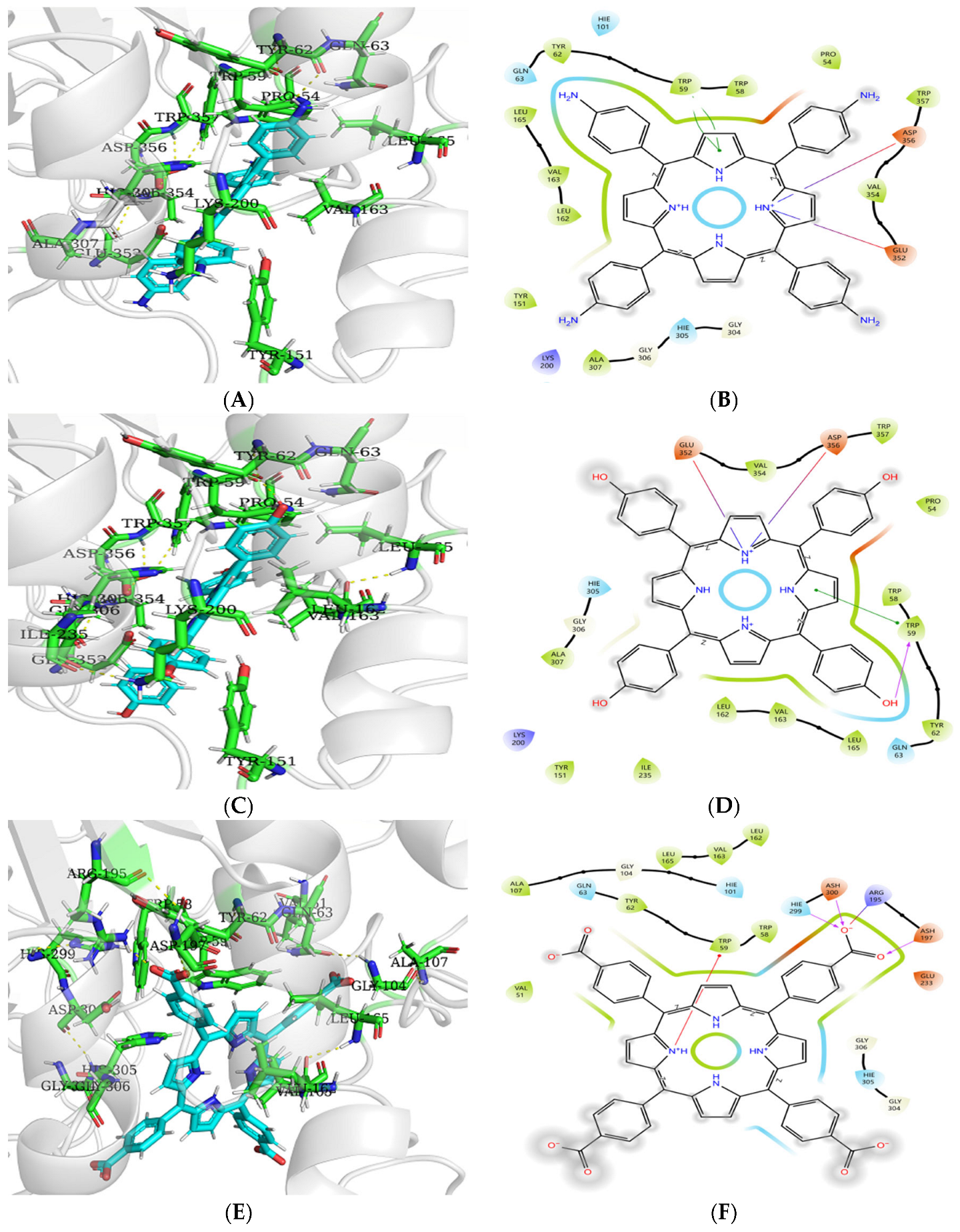
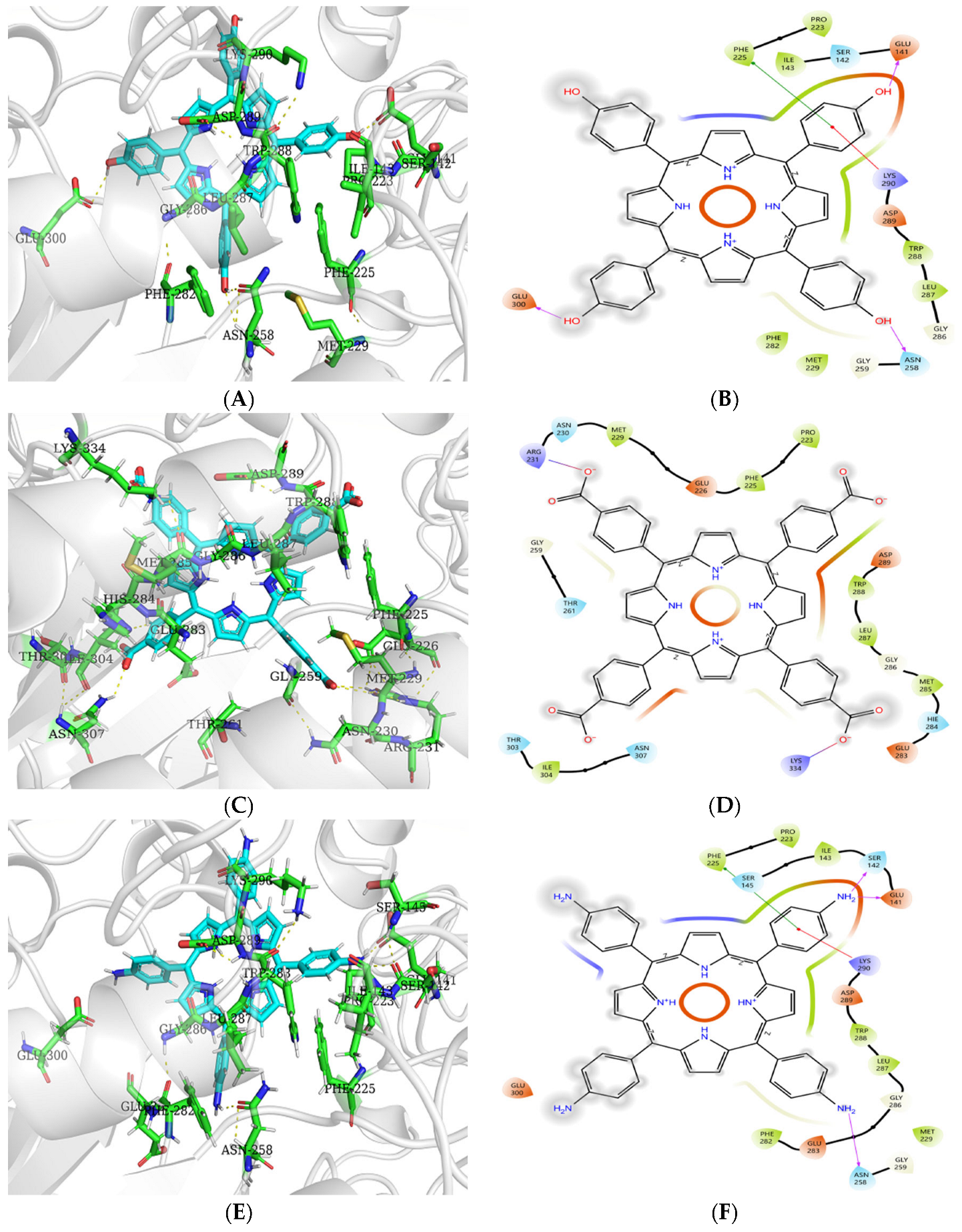
3.9. Molecular Docking Analysis of Porphyrins to Enzymes
3.9.1. Molecular Docking Analysis of Porphyrins with α-Amylase
3.9.2. Molecular Docking Analysis of Porphyrins with α-Glucosidase
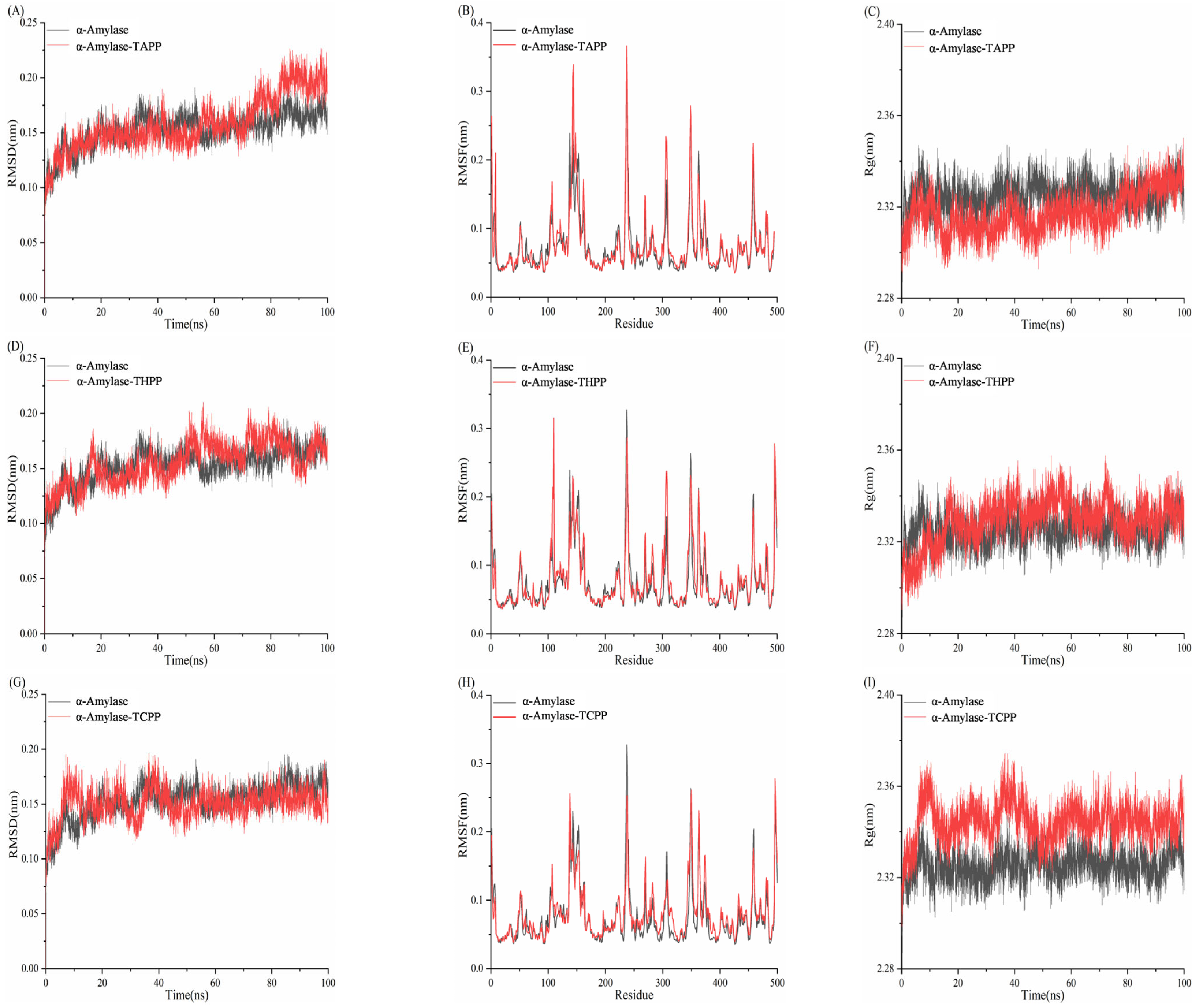
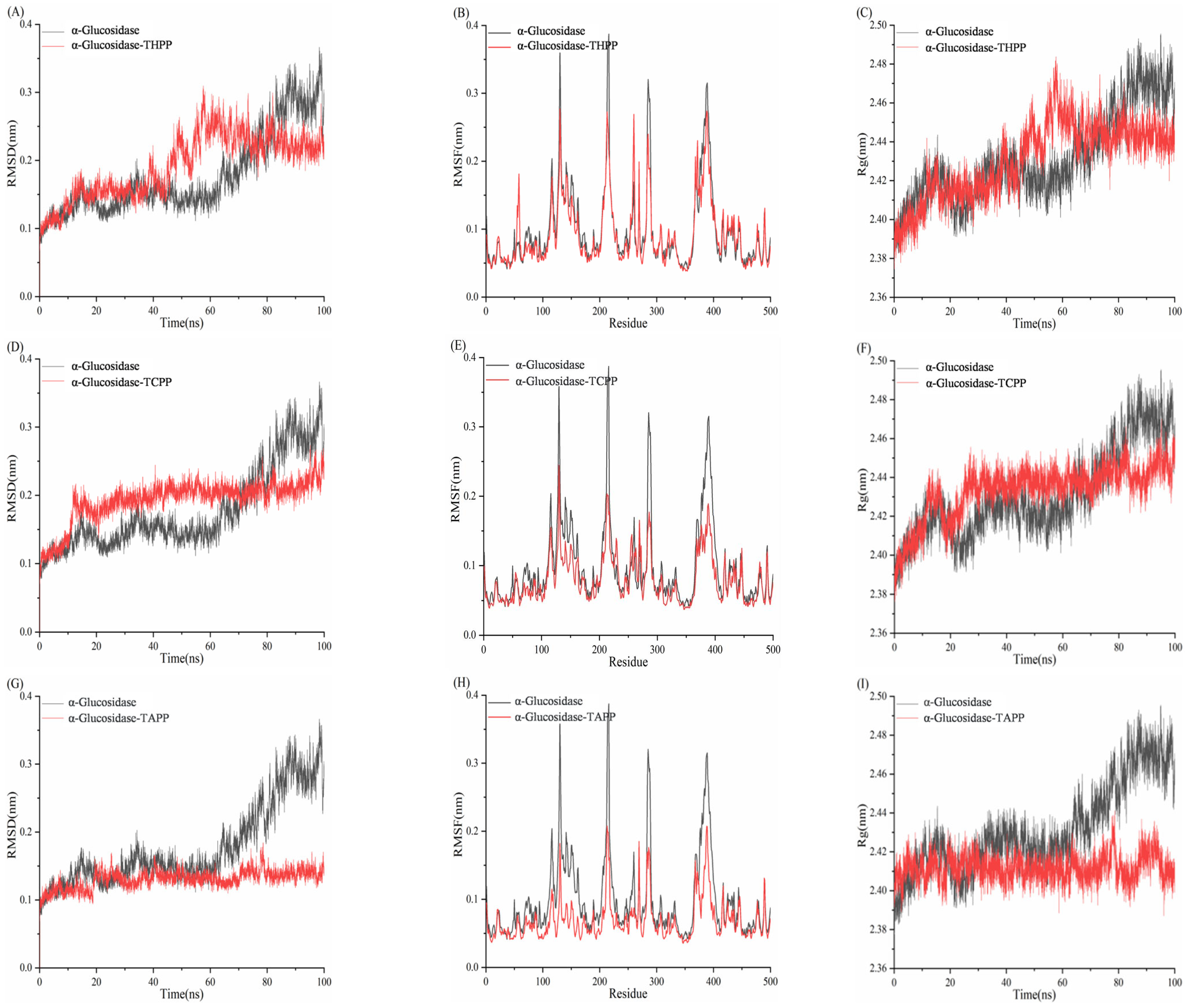
3.10. Molecular Dynamics Simulation Analysis of Porphyrins and Enzymes
3.10.1. Molecular Dynamics Simulation Analysis of Porphyrins and α-Amylase
3.10.2. Molecular Dynamics Simulation Analysis of Porphyrins and α-Glucosidase
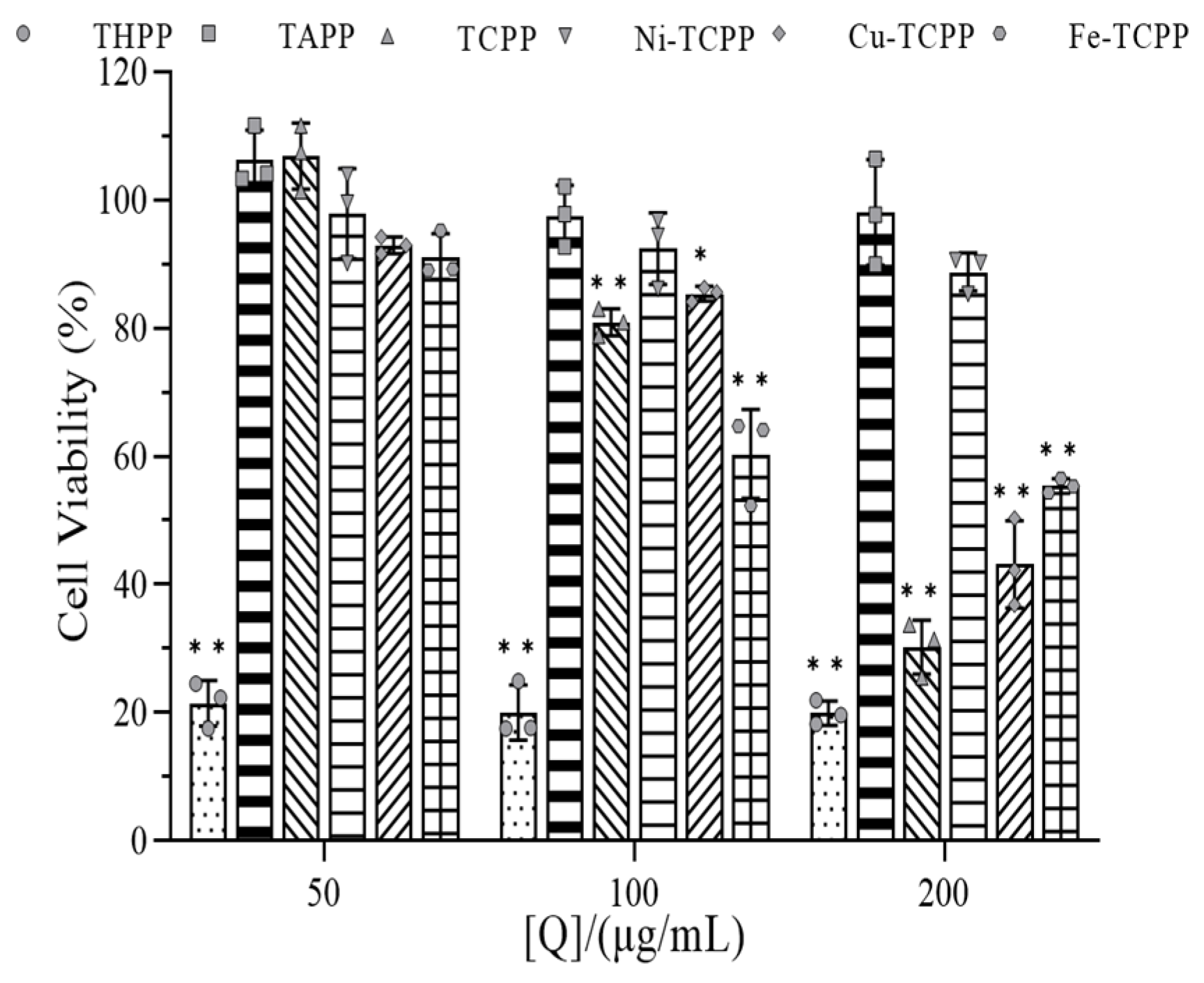
3.11. Cytotoxicity Analysis of Porphyrins
3.12. Pharmacokinetic Analysis Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| α-AMY | α-amylas |
| α-GLU | α-glucosidase |
References
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef]
- Cole, J.B.; Florez, J.C. Genetics of diabetes mellitus and diabetes complications. Nat. Rev. Nephrol. 2020, 16, 377–390. [Google Scholar] [CrossRef]
- Taha, M.; Hayat, S.; Rahim, F.; Uddin, N.; Wadood, A.; Nawaz, M.; Gollapalli, M.; Rehman, A.U.; Khan, K.M.; Farooq, R.K. Exploring thiazole-based Schiff base analogs as potent α-glucosidase and α-amylase inhibitor: Their synthesis and in-silico study. J. Mol. Struct. 2023, 1287, 135672. [Google Scholar] [CrossRef]
- Xiong, Y.; Ng, K.; Zhang, P.; Warner, R.D.; Shen, S.; Tang, H.-Y.; Liang, Z.; Fang, Z. In vitro α-glucosidase and α-amylase inhibitory activities of free and bound phenolic extracts from the bran and kernel fractions of five sorghum grain genotypes. Foods 2020, 9, 1301. [Google Scholar] [CrossRef] [PubMed]
- Seino, Y.; Nanjo, K.; Tajima, N.; Kadowaki, T.; Kashiwagi, A.; Araki, E.; Ito, C.; Inagaki, N.; Iwamoto, Y.; Kasuga, M. Report of the Committee on the classification and diagnostic criteria of diabetes mellitus. J. Diabetes Investig. 2010, 1, 212–228. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Zhong, X.; Huang, Y.; Peng, Z.; Wang, G. Synthesis, biological evaluation and molecular docking studies of chromone derivatives as potential α-glucosidase inhibitors. J. Mol. Struct. 2023, 1274, 134575. [Google Scholar] [CrossRef]
- Kousaxidis, A.; Petrou, A.; Lavrentaki, V.; Fesatidou, M.; Nicolaou, I.; Geronikaki, A. Aldose reductase and protein tyrosine phosphatase 1B inhibitors as a promising therapeutic approach for diabetes mellitus. Eur. J. Med. Chem. 2020, 207, 112742. [Google Scholar] [CrossRef]
- Chen, X.; Gao, M.; Jian, R.; Hong, W.D.; Tang, X.; Li, Y.; Zhao, D.; Zhang, K.; Chen, W.; Zheng, X. Design, synthesis and α-glucosidase inhibition study of novel embelin derivatives. J. Enzym. Inhib. Med. Chem. 2020, 35, 565–573. [Google Scholar] [CrossRef]
- Saleem, F.; Khan, K.M.; Chigurupati, S.; Andriani, Y.; Solangi, M.; Hameed, S.; Hafez, A.A.M.A.; Begum, F.; Lodhi, M.A.; Taha, M. Dicyanoanilines as potential and dual inhibitors of α-amylase and α-glucosidase enzymes: Synthesis, characterization, in vitro, in silico, and kinetics studies. Arab. J. Chem. 2022, 15, 103651. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, L.; Dong, Y.; Fang, Z.; Nisar, T.; Zhao, T.; Wang, Z.-C.; Guo, Y. Chemical compositions and α-glucosidase inhibitory effects of anthocyanidins from blueberry, blackcurrant and blue honeysuckle fruits. Food Chem. 2019, 299, 125102. [Google Scholar] [CrossRef]
- Nan, X.; Jia, W.; Zhang, Y.; Wang, H.; Lin, Z.; Chen, S. An on-line detection system for screening small molecule inhibitors of α-Amylase and α-Glucosidase in Prunus mume. J. Chromatogr. A 2022, 1663, 462754. [Google Scholar] [CrossRef]
- Ambery, P.; Parker, V.E.; Stumvoll, M.; Posch, M.G.; Heise, T.; Plum-Moerschel, L.; Tsai, L.-F.; Robertson, D.; Jain, M.; Petrone, M. MEDI0382, a GLP-1 and glucagon receptor dual agonist, in obese or overweight patients with type 2 diabetes: A randomised, controlled, double-blind, ascending dose and phase 2a study. Lancet 2018, 391, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Brunner, K.T.; Henneberg, C.J.; Wilechansky, R.M.; Long, M.T. Nonalcoholic fatty liver disease and obesity treatment. Curr. Obes. Rep. 2019, 8, 220–228. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J.-l.; Shen, L.-h.; Feng, L.-j.; Zhou, Q. Inhibition mechanism of diacylated anthocyanins from purple sweet potato (Ipomoea batatas L.) against α-amylase and α-glucosidase. Food Chem. 2021, 359, 129934. [Google Scholar] [CrossRef]
- Kang, X.; Gao, W.; Wang, B.; Yu, B.; Zhang, H.; Cui, B.; Abd El-Aty, A. Effects of proteins on the structure, physicochemical properties, and in vitro digestibility of wheat starch-lauric acid complexes under various cooking methods. Int. J. Biol. Macromol. 2021, 182, 1112–1119. [Google Scholar] [CrossRef]
- D’Costa, A.; Bordenave, N. Inhibition of starch digestion by flavonoids: Role of flavonoid-amylase binding kinetics. Food Chem. 2021, 341, 128256. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Shang, J.; Bai, J.; Wu, K.; Liu, J.; Yang, Z.; Ou, H.; Shao, L. Metabolites extracted from microorganisms as potential inhibitors of glycosidases (α-glucosidase and α-amylase): A review. Front. Microbiol. 2022, 13, 1050869. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, J.; Tian, J.; Chen, S.; Ye, X.; Hu, Y.; Chen, J. Inhibitory mechanism of novel allosteric inhibitor, Chinese bayberry (Myrica rubra Sieb. et Zucc.) leaves proanthocyanidins against α-glucosidase. J. Funct. Foods 2019, 56, 286–294. [Google Scholar] [CrossRef]
- Wang, L.; Ai, C.; Jin, C.; Mou, J.; Deng, Y. Xanthones as potential α-glucosidase non-competition inhibitors: Synthesis, inhibitory activities, and in silico studies. Chem. Biol. Drug Des. 2023, 102, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Huang, D.; Chen, S.; Xia, Y.; Zhu, S. The inhibitory mechanism of chlorogenic acid and its acylated derivatives on α-amylase and α-glucosidase. Food Chem. 2022, 372, 131334. [Google Scholar] [CrossRef]
- Yang, J.; Wang, X.; Zhang, C.; Ma, L.; Wei, T.; Zhao, Y.; Peng, X. Comparative study of inhibition mechanisms of structurally different flavonoid compounds on α-glucosidase and synergistic effect with acarbose. Food Chem. 2021, 347, 129056. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, L.; Fang, Z.; Nisar, T.; Zou, L.; Li, D.; Guo, Y. Lycium ruthenicum Murray anthocyanins effectively inhibit α-glucosidase activity and alleviate insulin resistance. Food Biosci. 2021, 41, 100949. [Google Scholar] [CrossRef]
- Radzuan, S.N.M.; Phongphane, L.; Bakar, M.H.A.; Omar, M.T.C.; Shahril, N.S.N.; Supratman, U.; Harneti, D.; Wahab, H.A.; Azmi, M.N. Synthesis, biological activities, and evaluation molecular docking-dynamics studies of new phenylisoxazole quinoxalin-2-amine hybrids as potential α-amylase and α-glucosidase inhibitors. RSC Adv. 2024, 14, 7684–7698. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, B.; Sun, W.; Xing, Y. Intervention of Prediabetes by Flavonoids from Oroxylum indicum. In Bioactive Food as Dietary Interventions for Diabetes; Elsevier: Amsterdam, The Netherlands, 2019; pp. 559–575. [Google Scholar]
- Zhang, J.; Zhang, L.; Lai, C.; Liang, Y.; Gao, L.; Kaliaperumal, K.; Jiang, Y. Nutraceutical potential of navel orange peel in diabetes management: The chemical profile, antioxidant, α-glucosidase inhibitory and antiglycation effects of its flavonoids. Food Biosci. 2022, 49, 101943. [Google Scholar] [CrossRef]
- Cho, M.; Han, J.H.; You, S. Inhibitory effects of fucan sulfates on enzymatic hydrolysis of starch. LWT-Food Sci. Technol. 2011, 44, 1164–1171. [Google Scholar] [CrossRef]
- Avcı, D.; Altürk, S.; Sönmez, F.; Tamer, Ö.; Başoğlu, A.; Atalay, Y.; Kurt, B.Z.; Dege, N. Novel Cu (II), Co (II) and Zn (II) metal complexes with mixed-ligand: Synthesis, crystal structure, α-glucosidase inhibition, DFT calculations, and molecular docking. J. Mol. Struct. 2019, 1197, 645–655. [Google Scholar] [CrossRef]
- Pandeya, K.B.; Tripathi, I.P.; Mishra, M.K.; Dwivedi, N.; Pardhi, Y.; Kamal, A.; Gupta, P.; Dwivedi, N.; Mishra, C. A critical review on traditional herbal drugs: An emerging alternative drug for diabetes. Int. J. Org. Chem. 2013, 03, 1–22. [Google Scholar] [CrossRef]
- Hilário-Souza, E.; Cuillel, M.; Mintz, E.; Charbonnier, P.; Vieyra, A.; Cassio, D.; Lowe, J. Modulation of hepatic copper-ATPase activity by insulin and glucagon involves protein kinase A (PKA) signaling pathway. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 2086–2097. [Google Scholar] [CrossRef]
- Toyokuni, S. Role of iron in carcinogenesis: Cancer as a ferrotoxic disease. Cancer Sci. 2009, 100, 9–16. [Google Scholar] [CrossRef]
- Sankar, R.; Sharmila, T. Co, Cu, Ni, and Zn complexes of N-[(3-phenoxy phenyl) methylidene]-l-valine as α-glycosidase and α-amylase inhibitors: Synthesis, molecular docking & antimicrobial evaluation. Bioorganic Chem. 2025, 154, 108010. [Google Scholar]
- Li, J.; Zhu, J.; Wu, H.; Li, W. Synthesis, in vitro, and in silico studies of fisetin and quercetin and their metal complexes as inhibitors of α-glucosidase and thrombin. J. Mol. Liq. 2022, 349, 118164. [Google Scholar] [CrossRef]
- Kong, F.; Qin, Y.; Su, Z.; Ning, Z.; Yu, S. Optimization of extraction of hypoglycemic ingredients from grape seeds and evaluation of α-glucosidase and α-amylase inhibitory effects in vitro. J. Food Sci. 2018, 83, 1422–1429. [Google Scholar] [CrossRef]
- Xu, P.; Wu, J.; Zhang, Y.; Chen, H.; Wang, Y. Physicochemical characterization of puerh tea polysaccharides and their antioxidant and α-glycosidase inhibition. J. Funct. Foods 2014, 6, 545–554. [Google Scholar] [CrossRef]
- Roy, A.S.; Tripathy, D.R.; Chatterjee, A.; Dasgupta, S. A spectroscopic study of the interaction of the antioxidant naringin with bovine serum albumin. J. Biophys. Chem. 2010, 1, 141–152. [Google Scholar] [CrossRef]
- Sun, L.; Warren, F.J.; Gidley, M.J.; Guo, Y.; Miao, M. Mechanism of binding interactions between young apple polyphenols and porcine pancreatic α-amylase. Food Chem. 2019, 283, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, X.; Qi, W.; Su, R.; He, Z. Affinity of rosmarinic acid to human serum albumin and its effect on protein conformation stability. Food Chem. 2016, 192, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, G.; Pan, J.; Gong, D. Galangin competitively inhibits xanthine oxidase by a ping-pong mechanism. Food Res. Int. 2016, 89, 152–160. [Google Scholar] [CrossRef]
- Shi, W.; Han, W.; Liao, Y.; Wen, J.; Zhang, G. Inhibition mechanism of fisetin on acetylcholinesterase and its synergistic effect with galantamine. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 305, 123452. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Karrar, E.; Jin, Q.; Zhang, H.; Wu, G.; Wang, X. Mechanisms of sesamol and sesamin inhibiting α-glucosidase activity by spectroscopy and molecular docking. Food Biosci. 2023, 53, 102680. [Google Scholar] [CrossRef]
- Saha, I.; Bhattacharyya, J.; Kumar, G.S. Thermodynamic investigations of ligand–protein interactions: Binding of the phenazinium dyes phenosafranin and safranin O with human serum albumin. J. Chem. Thermodyn. 2013, 56, 114–122. [Google Scholar] [CrossRef]
- Shi, J.-h.; Zhu, Y.-Y.; Wang, J.; Chen, J.; Shen, Y.-J. Intermolecular interaction of prednisolone with bovine serum albumin: Spectroscopic and molecular docking methods. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2013, 103, 287–294. [Google Scholar] [CrossRef]
- Ren, G.; Sun, H.; Guo, J.; Fan, J.; Li, G.; Xu, S. Molecular mechanism of the interaction between resveratrol and trypsin via spectroscopy and molecular docking. Food Funct. 2019, 10, 3291–3302. [Google Scholar] [CrossRef]
- Wang, M.; Shi, J.; Wang, L.; Hu, Y.; Ye, X.; Liu, D.; Chen, J. Inhibitory kinetics and mechanism of flavonoids from lotus (Nelumbo nucifera Gaertn.) leaf against pancreatic α-amylase. Int. J. Biol. Macromol. 2018, 120, 2589–2596. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Li, K.; Liu, Z. Research on the influences of five food-borne polyphenols on in vitro slow starch digestion and the mechanism of action. J. Agric. Food Chem. 2019, 67, 8617–8625. [Google Scholar] [CrossRef] [PubMed]
- Poureshghi, F.; Ghandforoushan, P.; Safarnejad, A.; Soltani, S. Interaction of an antiepileptic drug, lamotrigine with human serum albumin (HSA): Application of spectroscopic techniques and molecular modeling methods. J. Photochem. Photobiol. B Biol. 2017, 166, 187–192. [Google Scholar] [CrossRef]
- Sun, C.-P.; Yan, J.-K.; Yi, J.; Zhang, X.-Y.; Yu, Z.-L.; Huo, X.-K.; Liang, J.-H.; Ning, J.; Feng, L.; Wang, C. The study of inhibitory effect of natural flavonoids toward β-glucuronidase and interaction of flavonoids with β-glucuronidase. Int. J. Biol. Macromol. 2020, 143, 349–358. [Google Scholar] [CrossRef]
- Azad, I.; Nasibullah, M.; Khan, T.; Hassan, F.; Akhter, Y. Exploring the novel heterocyclic derivatives as lead molecules for design and development of potent anticancer agents. J. Mol. Graph. Model. 2018, 81, 211–228. [Google Scholar] [CrossRef]
- Vijaya, P.; Sundaraselvan, G. Synthesis, characterization, PASS analysis and ADMET properties of oxazolone ring containing hydrazone derivatives. Mater. Today: Proc. 2022, 48, 502–507. [Google Scholar] [CrossRef]

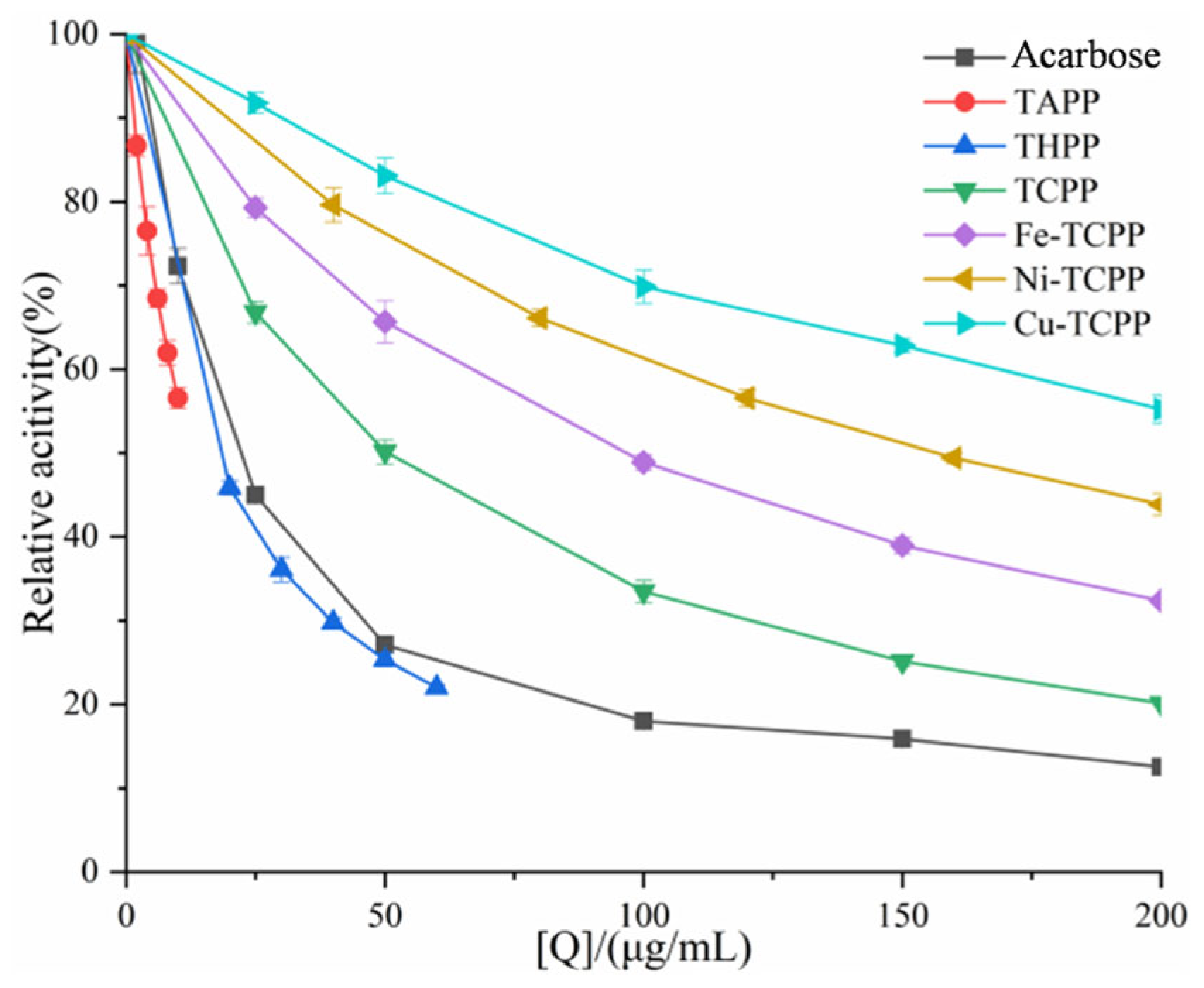
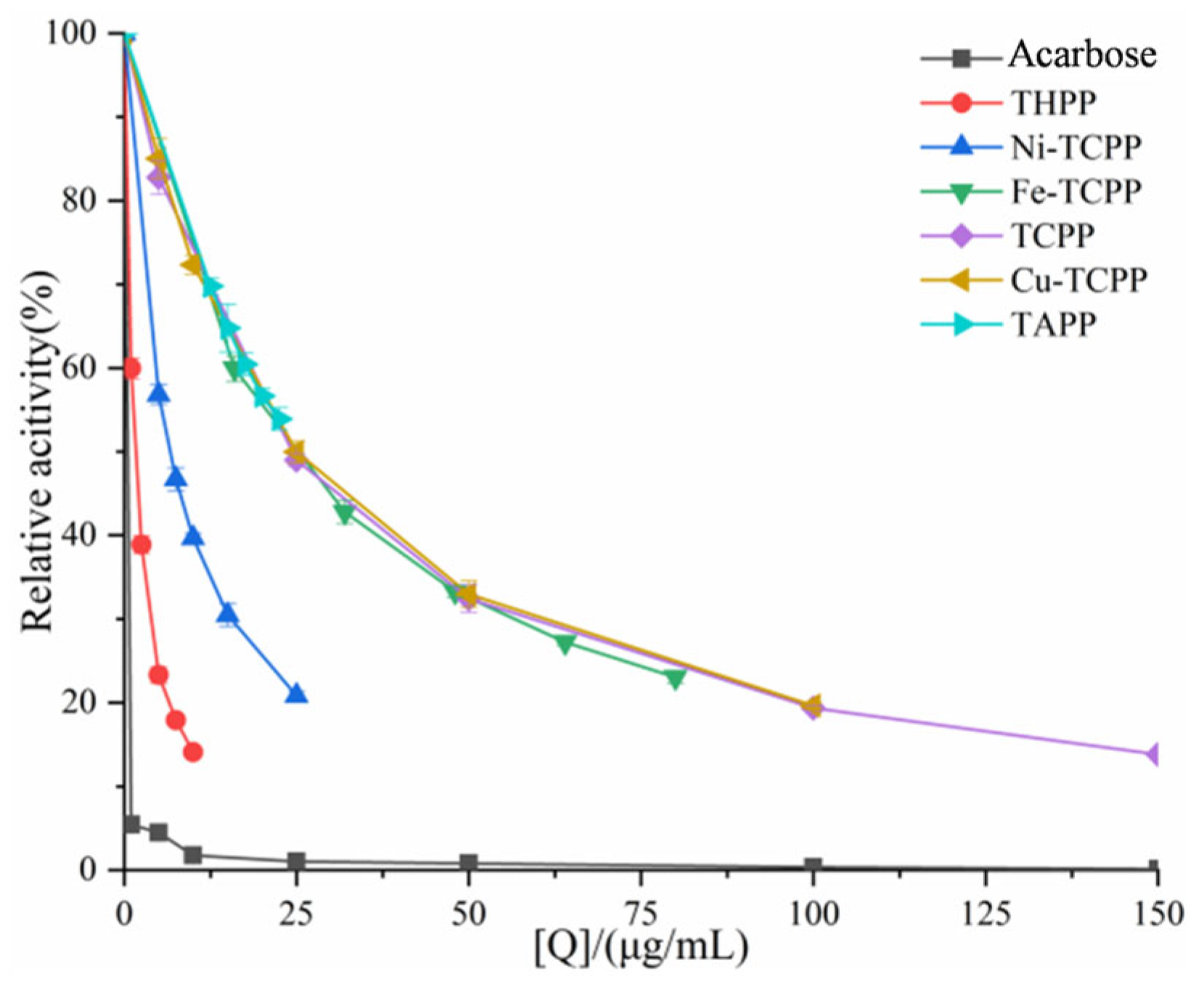
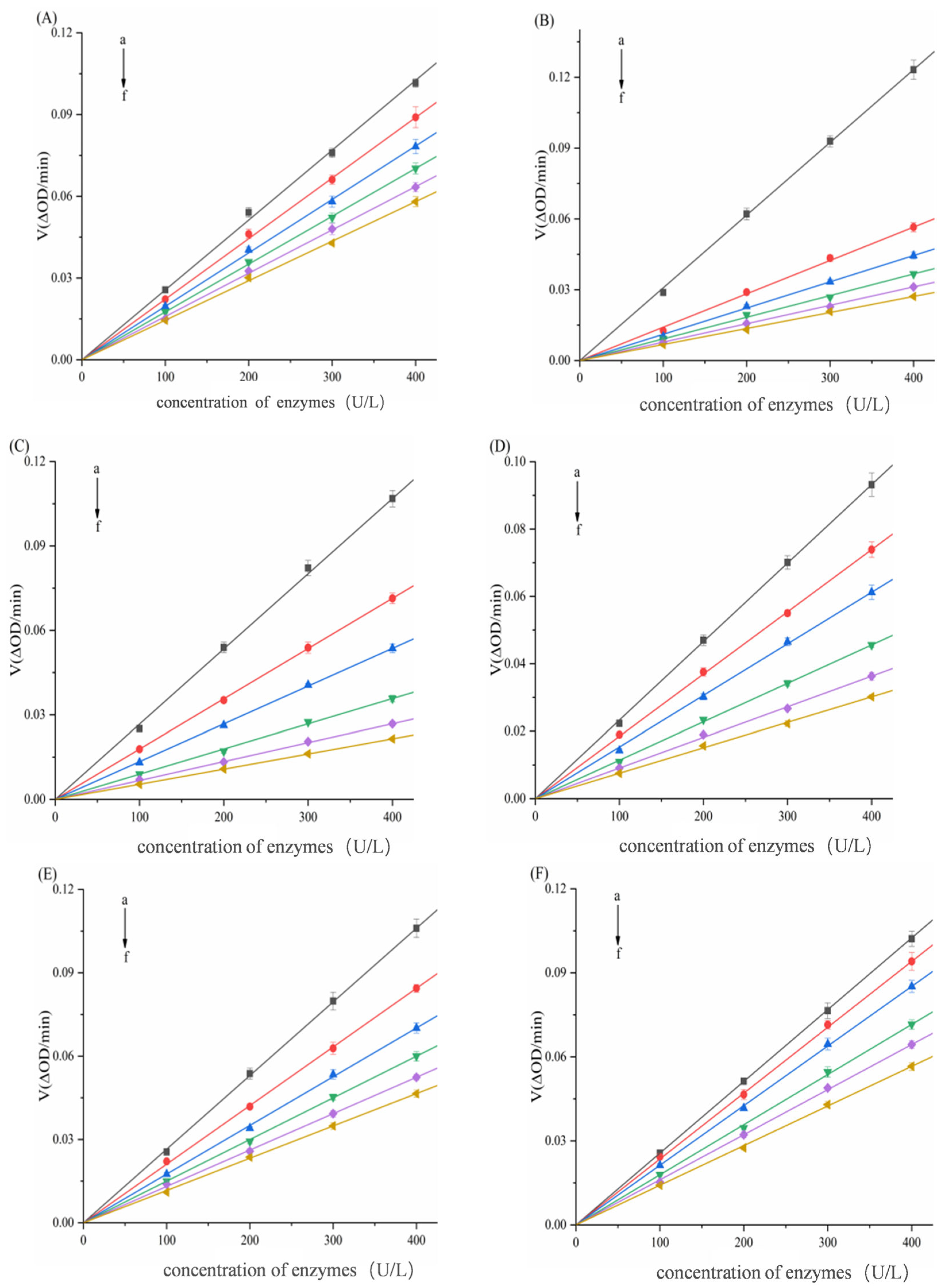
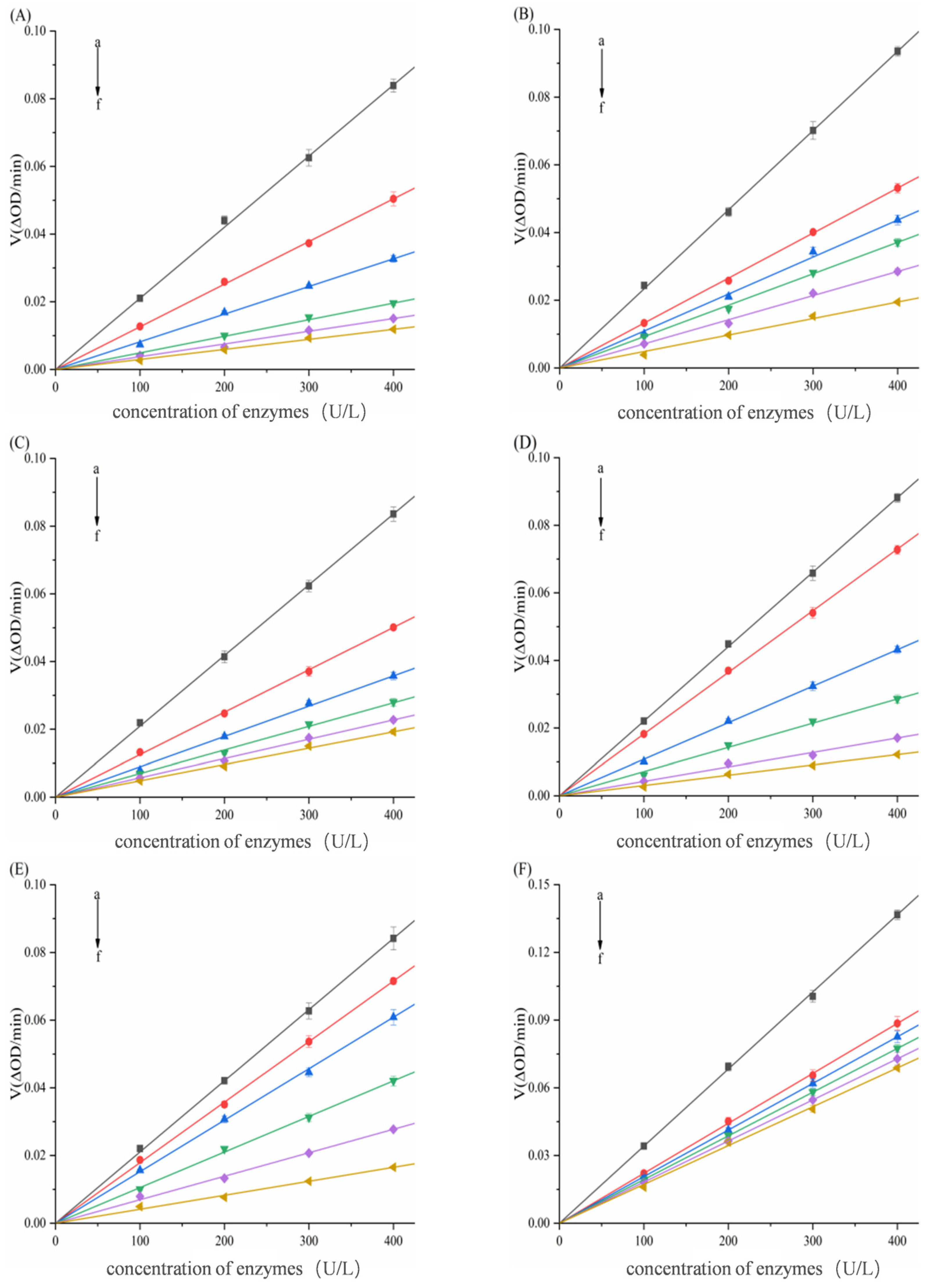

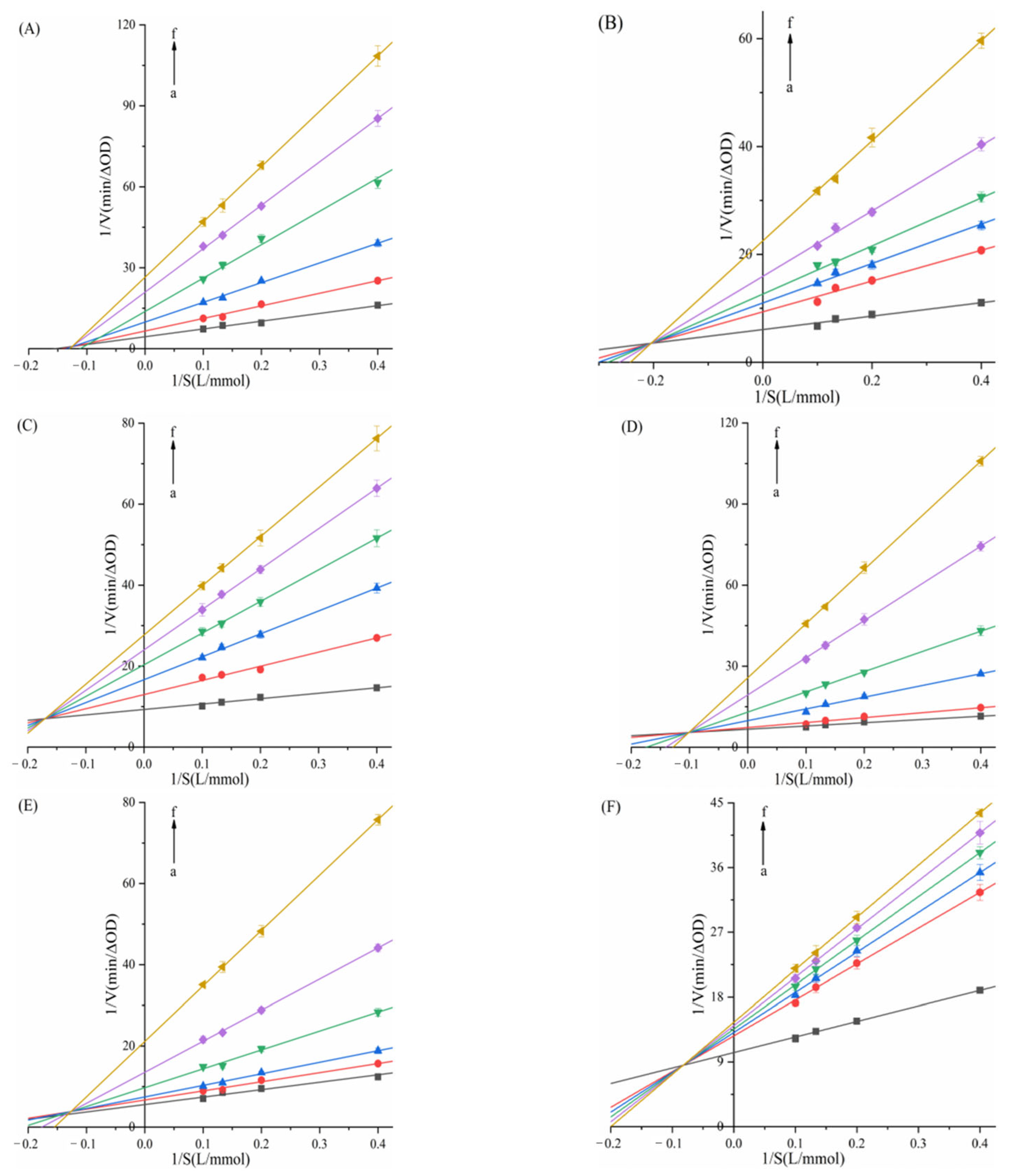






| System | C (μg/mL) | Km (μg/mL) | Vmax (ΔOD/min) | Ki (μg/mL) | Kis (μg/mL) |
|---|---|---|---|---|---|
| α-AMY–TAPP | 0 | 3.9047 ± 0.1139 | 0.1826 ± 0.0053 | 8.7057 ± 0.0325 | 21.2847 ± 0.0677 |
| 2 | 4.3893 ± 0.0776 | 0.1670 ± 0.0030 | |||
| 4 | 4.8719 ± 0.1211 | 0.1562 ± 0.0039 | |||
| 6 | 5.1449 ± 0.1158 | 0.1425 ± 0.0032 | |||
| 8 | 5.4460 ± 0.1386 | 0.1328 ± 0.0034 | |||
| 10 | 5.7081 ± 0.1284 | 0.1243 ± 0.0028 | |||
| α-AMY–THPP | 0 | 3.6644 ± 0.0843 | 0.1390 ± 0.0032 | 9.2445 ± 0.0411 | 23.1012 ± 0.1027 |
| 20 | 7.8640 ± 0.2930 | 0.0740 ± 0.0028 | |||
| 30 | 8.0378 ± 0.1371 | 0.0580 ± 0.0010 | |||
| 40 | 8.3899 ± 0.2135 | 0.0491 ± 0.0012 | |||
| 50 | 8.6487 ± 0.2634 | 0.0426 ± 0.0013 | |||
| 60 | 8.8467 ± 0.2802 | 0.0376 ± 0.0012 | |||
| α-AMY–TCPP | 0 | 2.5848 ± 0.0724 | 0.1244 ± 0.0035 | 19.9216 ± 0.0524 | 132.4382 ± 0.1403 |
| 25 | 4.9029 ± 0.1157 | 0.1046 ± 0.0025 | |||
| 50 | 6.3562 ± 0.1007 | 0.0872 ± 0.0014 | |||
| 100 | 8.8655 ± 0.2311 | 0.0709 ± 0.0018 | |||
| 150 | 10.3380 ± 0.3732 | 0.0583 ± 0.0021 | |||
| 200 | 11.3676 ± 0.1523 | 0.0496 ± 0.0007 | |||
| α-AMY–Fe–TCPP | 0 | 2.4394 ± 0.0636 | 0.1387 ± 0.0036 | 69.5865 ± 0.1284 | 117.2152 ± 0.1432 |
| 25 | 2.7329 ± 0.0882 | 0.1143 ± 0.0037 | |||
| 50 | 2.9384 ± 0.0447 | 0.0972 ± 0.0015 | |||
| 100 | 3.2079 ± 0.1016 | 0.0748 ± 0.0024 | |||
| 150 | 3.3360 ± 0.0508 | 0.0601 ± 0.0009 | |||
| 200 | 3.4920 ± 0.0491 | 0.0512 ± 0.0007 | |||
| α-AMY–Ni–TCPP | 0 | 3.6368 ± 0.0487 | 0.1831 ± 0.0025 | 157.9542 ± 0.1375 | |
| 40 | 3.6229 ± 0.1082 | 0.1456 ± 0.0043 | |||
| 80 | 3.6134 ± 0.1167 | 0.1238 ± 0.0040 | |||
| 120 | 3.6072 ± 0.0593 | 0.1032 ± 0.0017 | |||
| 160 | 3.6027 ± 0.1141 | 0.0901 ± 0.0029 | |||
| 200 | 3.5987 ± 0.0938 | 0.0799 ± 0.0021 | |||
| α-AMY–Cu–TCPP | 0 | 2.5129 ± 0.0827 | 0.1155 ± 0.0038 | 135.8043 ± 0.1608 | 420.9420 ± 0.2189 |
| 25 | 2.8021 ± 0.0410 | 0.1101 ± 0.0016 | |||
| 50 | 3.1685 ± 0.0729 | 0.1050 ± 0.0024 | |||
| 100 | 3.5949 ± 0.0569 | 0.0923 ± 0.0015 | |||
| 150 | 3.9016 ± 0.1355 | 0.0860 ± 0.0031 | |||
| 200 | 4.2470 ± 0.0673 | 0.0785 ± 0.0012 |
| System | C (μg/mL) | Km (μg/mL) | Vmax (ΔOD/min) | Ki (μg/mL) | Kis (μg/mL) |
|---|---|---|---|---|---|
| α-Glucosidase–THPP | 0 | 6.7070 ± 0.1543 | 0.2310 ± 0.0053 | 1.6508 ± 0.0135 | |
| 1 | 7.1219 ± 0.0954 | 0.1528 ± 0.0020 | |||
| 2.5 | 7.3949 ± 0.2523 | 0.1013 ± 0.0035 | |||
| 5 | 7.5880 ± 0.2213 | 0.0649 ± 0.0019 | |||
| 7.5 | 7.6790 ± 0.1357 | 0.0489 ± 0.0009 | |||
| 10 | 7.7319 ± 0.1921 | 0.0377 ± 0.0009 | |||
| α-Glucosidase–Ni–TCPP | 0 | 2.0514 ± 0.0462 | 0.1649 ± 0.0037 | 3.8703 ± 0.0201 | 9.2257 ± 0.0312 |
| 5 | 3.0491 ± 0.0776 | 0.1036 ± 0.0026 | |||
| 7.5 | 3.3243 ± 0.0748 | 0.0909 ± 0.0020 | |||
| 10 | 3.5279 ± 0.0812 | 0.0791 ± 0.0018 | |||
| 15 | 3.8090 ± 0.1419 | 0.0628 ± 0.0023 | |||
| 25 | 4.1248 ± 0.0704 | 0.0444 ± 0.0008 | |||
| α-Glucosidase–Fe–TCPP | 0 | 1.4377 ± 0.0366 | 0.1076 ± 0.0027 | 9.8917 ± 0.0514 | 40.4085 ± 0.1084 |
| 16 | 2.6958 ± 0.0821 | 0.0771 ± 0.0023 | |||
| 32 | 3.3979 ± 0.1076 | 0.0601 ± 0.0019 | |||
| 48 | 3.9231 ± 0.1098 | 0.0502 ± 0.0014 | |||
| 64 | 4.1566 ± 0.0981 | 0.0416 ± 0.0010 | |||
| 80 | 4.3847 ± 0.0694 | 0.0361 ± 0.0006 | |||
| α-Glucosidase–TCPP | 0 | 1.8019 ± 0.0470 | 0.1499 ± 0.0039 | 9.5746 ± 0.0611 | 52.5334 ± 0.1408 |
| 5 | 2.5045 ± 0.0904 | 0.1369 ± 0.0049 | |||
| 25 | 4.4087 ± 0.0591 | 0.1016 ± 0.0014 | |||
| 50 | 5.5731 ± 0.1452 | 0.0745 ± 0.0019 | |||
| 100 | 7.1021 ± 0.2293 | 0.0516 ± 0.0017 | |||
| 150 | 7.7894 ± 0.1186 | 0.0389 ± 0.0006 | |||
| α-Glucosidase–Cu–TCPP | 0 | 2.7842 ± 0.0882 | 0.1689 ± 0.0053 | 13.7332 ± 0.0823 | 38.9429 ± 0.1021 |
| 5 | 3.3657 ± 0.0431 | 0.1496 ± 0.0019 | |||
| 10 | 3.8284 ± 0.1475 | 0.1344 ± 0.0052 | |||
| 25 | 4.7824 ± 0.1009 | 0.1028 ± 0.0022 | |||
| 50 | 5.4948 ± 0.1841 | 0.0718 ± 0.0024 | |||
| 100 | 6.4626 ± 0.1103 | 0.0473 ± 0.0008 | |||
| α-Glucosidase–TAPP | 0 | 2.0896 ± 0.0305 | 0.0969 ± 0.0014 | 9.4635 ± 0.0572 | 55.1227 ± 0.1247 |
| 12.5 | 3.9347 ± 0.1025 | 0.0791 ± 0.0021 | |||
| 15 | 4.2527 ± 0.0701 | 0.0767 ± 0.0013 | |||
| 17.5 | 4.5048 ± 0.0928 | 0.0736 ± 0.0015 | |||
| 20 | 4.7611 ± 0.0754 | 0.0711 ± 0.0011 | |||
| 22.5 | 5.0007 ± 0.0856 | 0.0688 ± 0.0012 |
| System | T | Ksv (×104 L/mol) | Kq (×1012 L/mol/s) | n | Ka (×105 L/mol) |
|---|---|---|---|---|---|
| α-Amylase–TAPP | 298 | 4.07 ± 0.05 | 4.07 ± 0.05 | 1.48 | 52.90 ± 0.07 |
| 304 | 3.37 ± 0.02 | 3.37 ± 0.02 | 1.40 | 21.79 ± 0.07 | |
| 310 | 3.07 ± 0.09 | 3.07 ± 0.09 | 1.31 | 7.46 ± 0.02 | |
| α-Amylase–THPP | 298 | 8.90 ± 0.06 | 8.90 ± 0.06 | 1.19 | 5.65 ± 0.06 |
| 304 | 8.20 ± 0.02 | 8.20 ± 0.02 | 1.16 | 3.65 ± 0.03 | |
| 310 | 7.64 ± 0.07 | 7.64 ± 0.07 | 1.12 | 2.46 ± 0.01 | |
| α-Amylase–TCPP | 298 | 4.04 ± 0.01 | 4.04 ± 0.01 | 1.36 | 12.54 ± 0.05 |
| 304 | 3.55 ± 0.08 | 3.55 ± 0.08 | 1.28 | 5.01 ± 0.03 | |
| 310 | 3.01 ± 0.07 | 3.01 ± 0.07 | 1.20 | 2.18 ± 0.02 | |
| α-Amylase–Fe–TCPP | 298 | 4.32 ± 0.06 | 4.32 ± 0.06 | 1.65 | 944.30 ± 0.18 |
| 304 | 3.36 ± 0.06 | 3.36 ± 0.06 | 1.38 | 40.38 ± 0.09 | |
| 310 | 3.29 ± 0.08 | 3.29 ± 0.08 | 1.05 | 2.13 ± 0.05 | |
| α-Amylase–Ni–TCPP | 298 | 9.78 ± 0.03 | 9.78 ± 0.03 | 1.44 | 99.31 ± 0.12 |
| 304 | 8.47 ± 0.07 | 8.47 ± 0.07 | 1.19 | 5.75 ± 0.04 | |
| 310 | 7.11 ± 0.05 | 7.11 ± 0.05 | 0.99 | 0.61 ± 0.02 | |
| α-Amylase–Cu–TCPP | 298 | 4.37 ± 0.07 | 4.37 ± 0.07 | 0.93 | 0.20 ± 0.01 |
| 304 | 4.33 ± 0.03 | 4.33 ± 0.03 | 0.84 | 0.10 ± 0.01 | |
| 310 | 4.32 ± 0.02 | 4.32 ± 0.02 | 0.78 | 0.06 ± 0.01 |
| System | T | Ksv (×104 L/mol) | Kq (×1012 L/mol/s) | n | Ka (×105 L/mol) |
|---|---|---|---|---|---|
| α-Glucosidase–THPP | 298 | 7.40 ± 0.06 | 7.40 ± 0.06 | 1.29 | 15.49 ± 0.08 |
| 304 | 6.97 ± 0.06 | 6.97 ± 0.06 | 1.23 | 8.01 ± 0.06 | |
| 310 | 6.69 ± 0.04 | 6.69 ± 0.04 | 1.16 | 3.39 ± 0.03 | |
| α-Glucosidase–Ni–TCPP | 298 | 17.73 ± 0.15 | 17.73 ± 0.15 | 1.03 | 2.34 ± 0.04 |
| 304 | 14.23 ± 0.09 | 14.23 ± 0.09 | 1.04 | 2.09 ± 0.02 | |
| 310 | 12.31 ± 0.08 | 12.31 ± 0.08 | 1.03 | 1.83 ± 0.01 | |
| α-Glucosidase–Fe–TCPP | 298 | 14.52 ± 0.07 | 14.52 ± 0.07 | 1.01 | 1.65 ± 0.03 |
| 304 | 13.18 ± 0.05 | 13.18 ± 0.05 | 1.00 | 1.26 ± 0.03 | |
| 310 | 12.20 ± 0.07 | 12.20 ± 0.07 | 0.98 | 1.02 ± 0.02 | |
| α-Glucosidase–TCPP | 298 | 7.45 ± 0.04 | 7.45 ± 0.04 | 1.30 | 13.73 ± 0.06 |
| 304 | 4.90 ± 0.02 | 4.90 ± 0.02 | 1.19 | 3.17 ± 0.03 | |
| 310 | 4.22 ± 0.09 | 4.22 ± 0.09 | 1.07 | 0.97 ± 0.02 | |
| α-Glucosidase–Cu–TCPP | 298 | 7.35 ± 0.09 | 7.35 ± 0.09 | 1.44 | 55.21 ± 0.11 |
| 304 | 6.74 ± 0.03 | 6.74 ± 0.03 | 1.25 | 7.38 ± 0.04 | |
| 310 | 5.05 ± 0.02 | 5.05 ± 0.02 | 1.03 | 0.69 ± 0.01 | |
| α-Glucosidase–TAPP | 298 | 6.25 ± 0.04 | 6.25 ± 0.04 | 1.14 | 2.78 ± 0.01 |
| 304 | 5.71 ± 0.01 | 5.71 ± 0.01 | 1.09 | 1.42 ± 0.03 | |
| 310 | 5.66 ± 0.02 | 5.66 ± 0.02 | 1.01 | 0.65 ± 0.02 |
| System | T | ΔH0 (kJ/mol) | ΔG0 (kJ/mol) | ΔS0 (J/mol/k) | R2 |
|---|---|---|---|---|---|
| α-AMY– TAPP | 298 | −125.25 ± 0.12 | −38.36 ± 0.11 | −291.29 ± 0.12 | 0.99 |
| 304 | −36.89 ± 0.12 | ||||
| 310 | −34.85 ± 0.10 | ||||
| α-AMY–THPP | 298 | −53.25 ± 0.05 | −32.82 ± 0.08 | −68.61 ± 0.06 | 0.99 |
| 304 | −32.37 ± 0.08 | ||||
| 310 | −31.99 ± 0.07 | ||||
| α-AMY–TCPP | 298 | −112.04 ± 0.06 | −34.79 ± 0.09 | −259.31 ± 0.09 | 0.99 |
| 304 | −33.17 ± 0.07 | ||||
| 310 | −31.68 ± 0.07 | ||||
| α-AMY–Fe–TCPP | 298 | −390.15 ± 0.17 | −45.50 ± 0.07 | −1156.68 ± 0.13 | 0.99 |
| 304 | −38.45 ± 0.06 | ||||
| 310 | −31.62 ± 0.08 | ||||
| α-AMY–Ni–TCPP | 298 | −326.36 ± 0.18 | −39.92 ± 0.08 | −961.90 ± 0.08 | 0.99 |
| 304 | −33.52 ± 0.06 | ||||
| 310 | −28.39 ± 0.07 | ||||
| α-AMY–Cu–TCPP | 298 | −79.68 ± 0.07 | −24.55 ± 0.05 | −185.16 ± 0.05 | 0.99 |
| 304 | −23.29 ± 0.03 | ||||
| 310 | −22.33 ± 0.05 |
| System | T | ΔH0 (kJ/mol) | ΔG0 (kJ/mol) | ΔS0 (J/mol/k) | R2 |
|---|---|---|---|---|---|
| α-GLU–THPP | 298 | −97.17 ± 0.09 | −35.31 ± 0.10 | −207.25 ± 0.13 | 0.98 |
| 304 | −34.36 ± 0.09 | ||||
| 310 | −32.82 ± 0.06 | ||||
| α-GLU–Ni–TCPP | 298 | −15.84 ± 0.05 | −30.63 ± 0.13 | 49.67 ± 0.08 | 0.99 |
| 304 | −30.96 ± 0.06 | ||||
| 310 | −31.23 ± 0.07 | ||||
| α-GLU–Fe–TCPP | 298 | −30.90 ± 0.06 | −29.76 ± 0.05 | −3.88 ± 0.01 | 0.99 |
| 304 | −29.68 ± 0.07 | ||||
| 310 | −29.72 ± 0.06 | ||||
| α-GLU–TCPP | 298 | −169.47 ± 0.10 | −35.01 ± 0.06 | −451.52 ± 0.12 | 0.99 |
| 304 | −32.01 ± 0.07 | ||||
| 310 | −29.60 ± 0.05 | ||||
| α-GLU–Cu–TCPP | 298 | −280.76 ± 0.13 | −38.46 ± 0.08 | −812.45 ± 0.17 | 0.99 |
| 304 | −34.15 ± 0.05 | ||||
| 310 | −28.70 ± 0.04 | ||||
| α-GLU–TAPP | 298 | −93.02 ± 0.08 | −31.06 ± 0.08 | −207.76 ± 0.14 | 0.99 |
| 304 | −29.98 ± 0.05 | ||||
| 310 | −28.56 ± 0.05 |
| System | Peak 1 Ex/Em (nm) | Fluorescence Intensity | Peak 2 Ex/Em (nm) | Fluorescence Intensity |
|---|---|---|---|---|
| α-Amylase (A) | 280.0/356.0 | 364.8 | 236.0/352.0 | 8.688 |
| α-Amylase–TAPP(A′) | 280.0/348.0 | 317.2 | 236.0/352.0 | 7.013 |
| α-Amylase(B/C) | 280.0/352.0 | 608.4 | 236.0/352.0 | 85.34 |
| α-Amylase–THPP(B′) | 280.0/352.0 | 297.5 | 236.0/352.0 | 32.69 |
| α-Amylase–TCPP(C′) | 280.0/352.0 | 461.6 | 236.0/352.0 | 58.06 |
| α-Amylase (D) | 280.0/356.0 | 373.4 | 236.0/356.0 | 48.99 |
| α-Amylase–Fe–TCPP(D′) | 280.0/352.0 | 270.8 | 236.0/356.0 | 28.26 |
| α-Amylase (E/F) | 280.0/356.0 | 376.4 | 236.0/356.0 | 23.03 |
| α-Amylase–Ni–TCPP(E′) | 280.0/356.0 | 225.2 | 236.0/360.0 | 9.012 |
| α-Amylase–Cu–TCPP(F′) | 280.0/352.0 | 313.1 | 236.0/356.0 | 16.48 |
| System | Peak 1 Ex/Em (nm) | Fluorescence Intensity | Peak 2 Ex/Em (nm) | Fluorescence Intensity |
|---|---|---|---|---|
| α-Glucosidase(A/C/D) | 280.0/348.0 | 207.4 | 236.0/356.0 | 33.69 |
| α-Glucosidase–THPP(A′) | 280.0/348.0 | 122.3 | 236.0/352.0 | 13.28 |
| α-Glucosidase–Fe–TCPP(C′) | 280.0/344.0 | 168.0 | 236.0/352.0 | 16.79 |
| α-Glucosidase–TCPP(D′) | 280.0/348.0 | 160.0 | 236.0/352.0 | 22.17 |
| α-Glucosidase (B/E) | 280.0/348.0 | 205.7 | 236.0/356.0 | 22.99 |
| α-Glucosidase–Ni–TCPP(B′) | 280.0/352.0 | 101.6 | 236.0/352.0 | 6.157 |
| α-Glucosidase–Cu–TCPP(E′) | 280.0/348.0 | 139.0 | 236.0/356.0 | 6.458 |
| α-glucosidase (F) | 280.0/352.0 | 216.1 | 236.0/352.0 | 8.943 |
| α-Glucosidase–TAPP(F′) | 280.0/352.0 | 159.0 | 236.0/348.0 | 0.576 |
| System | α-Helix (%) | β-Sheet (%) | β-Turn (%) | Random Coil (%) | β-Antiparallel (%) |
|---|---|---|---|---|---|
| α-AMY | 24.58 ± 0.03 | 23.64 ± 0.07 | 27.34 ± 0.06 | 14.10 ± 0.04 | 10.34 ± 0.02 |
| α-AMY–TAPP | 22.45 ± 0.07 | 14.62 ± 0.03 | 34.00 ± 0.09 | 9.94 ± 0.02 | 18.98 ± 0.04 |
| α-AMY–THPP | 22.24 ± 0.06 | 12.92 ± 0.05 | 33.76 ± 0.08 | 17.71 ± 0.07 | 13.37 ± 0.03 |
| α-AMY–TCPP | 10.24 ± 0.02 | 18.51 ± 0.05 | 39.66 ± 0.09 | 19.84 ± 0.06 | 11.75 ± 0.03 |
| α-AMY–Fe–TCPP | 21.77 ± 0.07 | 12.86 ± 0.04 | 34.02 ± 0.08 | 22.59 ± 0.03 | 8.76 ± 0.02 |
| α-AMY–Ni–TCPP | 20.76 ± 0.06 | 11.20 ± 0.02 | 36.33 ± 0.13 | 21.40 ± 0.05 | 10.30 ± 0.03 |
| α-AMY–Cu–TCPP | 20.76 ± 0.05 | 10.94 ± 0.01 | 35.19 ± 0.06 | 20.94 ± 0.06 | 12.17 ± 0.02 |
| System | α-Helix (%) | β-Sheet (%) | β-Turn (%) | Random Coil (%) | β-Antiparallel (%) |
|---|---|---|---|---|---|
| α-GLU | 24.74 ± 0.03 | 14.72 ± 0.04 | 32.91 ± 0.09 | 19.42 ± 0.07 | 8.21 ± 0.03 |
| α-GLU–THPP | 21.19 ± 0.05 | 12.79 ± 0.03 | 35.20 ± 0.11 | 23.74 ± 0.06 | 7.07 ± 0.01 |
| α-GLU–Ni–TCPP | 19.75 ± 0.07 | 13.68 ± 0.04 | 37.94 ± 0.10 | 26.76 ± 0.07 | 1.88 ± 0.01 |
| α-GLU–Fe–TCPP | 22.48 ± 0.05 | 15.77 ± 0.04 | 36.97 ± 0.08 | 16.75 ± 0.03 | 8.02 ± 0.02 |
| α-GLU–TCPP | 22.71 ± 0.05 | 12.03 ± 0.02 | 33.54 ± 0.07 | 22.15 ± 0.04 | 9.57 ± 0.02 |
| α-GLU–Cu–TCPP | 23.65 ± 0.03 | 10.93 ± 0.05 | 37.78 ± 0.09 | 25.05 ± 0.07 | 2.59 ± 0.01 |
| α-GLU–TAPP | 22.82 ± 0.08 | 12.99 ± 0.13 | 38.30 ± 0.08 | 23.93 ± 0.05 | 1.96 ± 0.01 |
| System | XP GScore (kcal/mol) | Key Residues | Hydrogen Bonds |
|---|---|---|---|
| α-amylase–TAPP | −5.19 | LEU162, VAL163, LEU165, TYR62, TRP59, TRP58, PRO54, TRP357, ASP356, VAL354, GLU352, ALA307, TYR151 | |
| α-amylase–THPP | −6.82 | LEU162, VAL163, LEU165, TYR62, TRP59, TRP58, PRO54, TRP357, ASP356, VAL354, GLU352, ALA307, TYR151, ILE235 | TRP59 |
| α-amylase–TCPP | −2.90 | LEU162, VAL163, LEU165, TYR62, TRP59, TRP58, ALA107, HIE299, ASH300, ARG195, ASH197, VAL51 | HIE299, ASH300, ASH197 |
| System | XP GScore (kcal/mol) | Key Residues | Hydrogen Bonds |
|---|---|---|---|
| α-Glucosidas–THPP | −6.39 | PRO223, PHE225, ILE143, GLU141, LYS290, TRP288, LEU287, ASN258, MET229, PHE282, GLU300 | GLU141, ASN258, GLU300 |
| α-Glucosidase–TCPP | −4.30 | PRO223, PHE225, MET229, ARG231, TRP288, LEU287, MET285, LYS334, ILE304 | |
| α-Glucosidase–TAPP | −7.90 | PRO223, PHE225, ILE143, SER142, GLU141, LYS290, TRP288, LEU287, PHE282, ASN258, MET229 | SER142, GLU141, ASN258 |
| Compound | BBB | HIA | Oral Toxicity |
|---|---|---|---|
| THPP | 0.5000+ | 0.9951+ | (III)0.5450 |
| TAPP | 0.7799+ | 0.9955+ | (III)0.6175 |
| TCPP | 0.5357− | 0.9739+ | (III)0.5439 |
| Fe–TCPP | 0.5795+ | 0.8931+ | (III)0.5356 |
| Ni–TCPP | 0.5335+ | 0.7102+ | (III)0.5504 |
| Cu–TCPP | 0.5674+ | 0.7426+ | (III)0.5504 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Liu, Z.; Ma, Q.; Liu, Y.; Yin, S.; Zhou, Z.; Zhou, J.; Bai, H.; Li, T. Exploring the Inhibitory Potential of Six Porphyrin Compounds Against α-Amylase and α-Glucosidase Linked to Diabetes. Biomolecules 2025, 15, 1338. https://doi.org/10.3390/biom15091338
Zhang S, Liu Z, Ma Q, Liu Y, Yin S, Zhou Z, Zhou J, Bai H, Li T. Exploring the Inhibitory Potential of Six Porphyrin Compounds Against α-Amylase and α-Glucosidase Linked to Diabetes. Biomolecules. 2025; 15(9):1338. https://doi.org/10.3390/biom15091338
Chicago/Turabian StyleZhang, Shuo, Zi Liu, Qiurui Ma, Yangyuxin Liu, Shuren Yin, Zhihan Zhou, Jie Zhou, Helong Bai, and Tianjiao Li. 2025. "Exploring the Inhibitory Potential of Six Porphyrin Compounds Against α-Amylase and α-Glucosidase Linked to Diabetes" Biomolecules 15, no. 9: 1338. https://doi.org/10.3390/biom15091338
APA StyleZhang, S., Liu, Z., Ma, Q., Liu, Y., Yin, S., Zhou, Z., Zhou, J., Bai, H., & Li, T. (2025). Exploring the Inhibitory Potential of Six Porphyrin Compounds Against α-Amylase and α-Glucosidase Linked to Diabetes. Biomolecules, 15(9), 1338. https://doi.org/10.3390/biom15091338





