Isolation and Identification of Duck Intestinal Probiotics and Their Effects on the Production and Immune Performance of Pekin Ducks
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Probiotic Samples
2.2. Isolation and Identification of Probiotics from Duck Intestine
2.3. Biological Characteristics of the Isolates
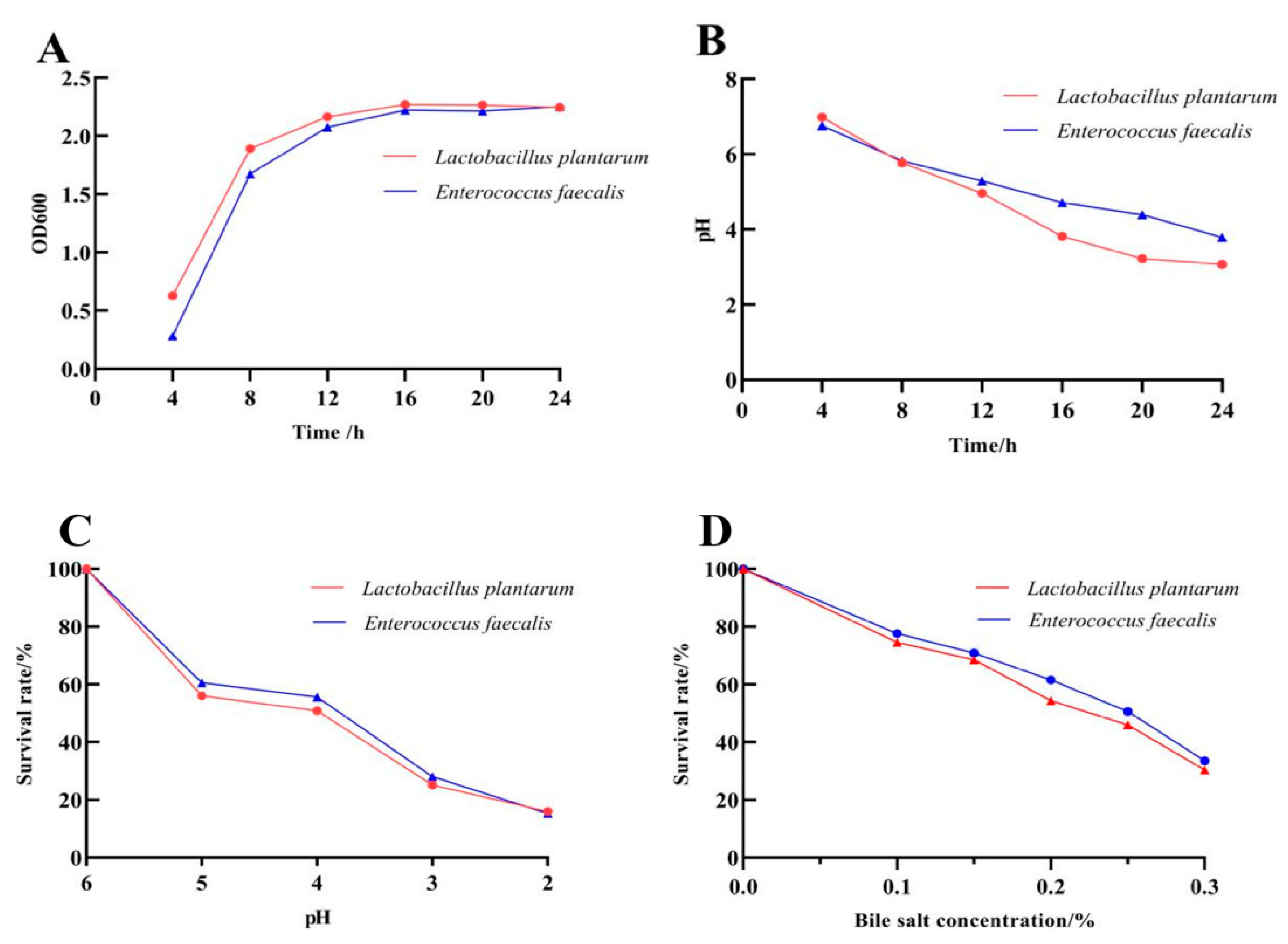
2.3.1. The Growth and Acid Production Capacity
2.3.2. Tolerance to Low pH
2.3.3. Tolerance to Bile Salt
2.3.4. Antimicrobial Activity
2.4. Effects of Isolated Probiotics on the Production and Immunity of Pekin Ducks
2.4.1. Analysis of the Growth and Slaughter Performance
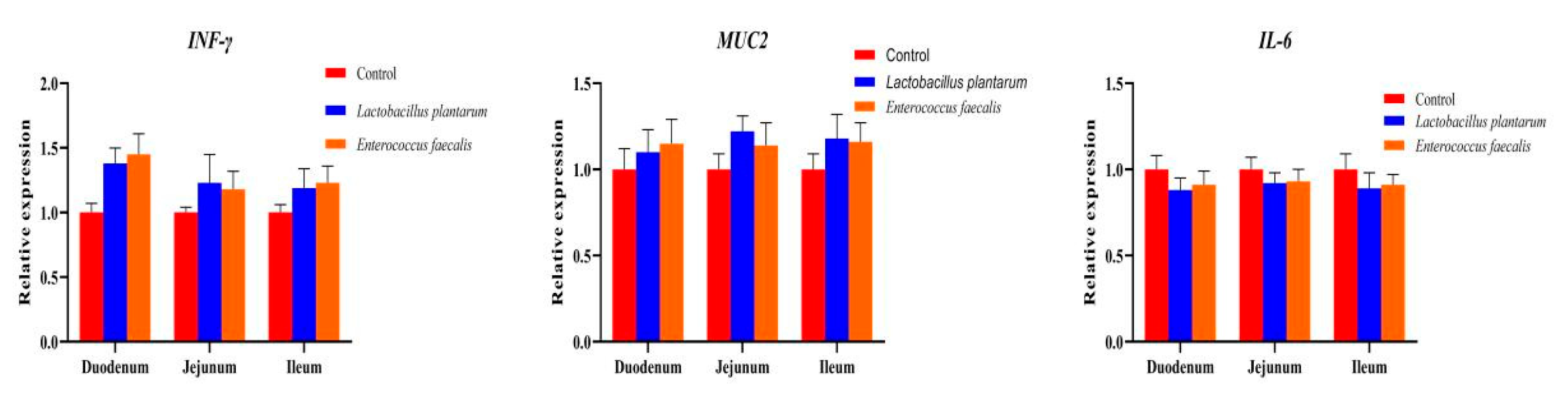
2.4.2. Histomorphological Examination and RT-PCR for Small Intestine
2.4.3. Content of Serum IgG and IgA
2.4.4. Changes in Cecal Microflora
2.5. Statistical Analysis
3. Results
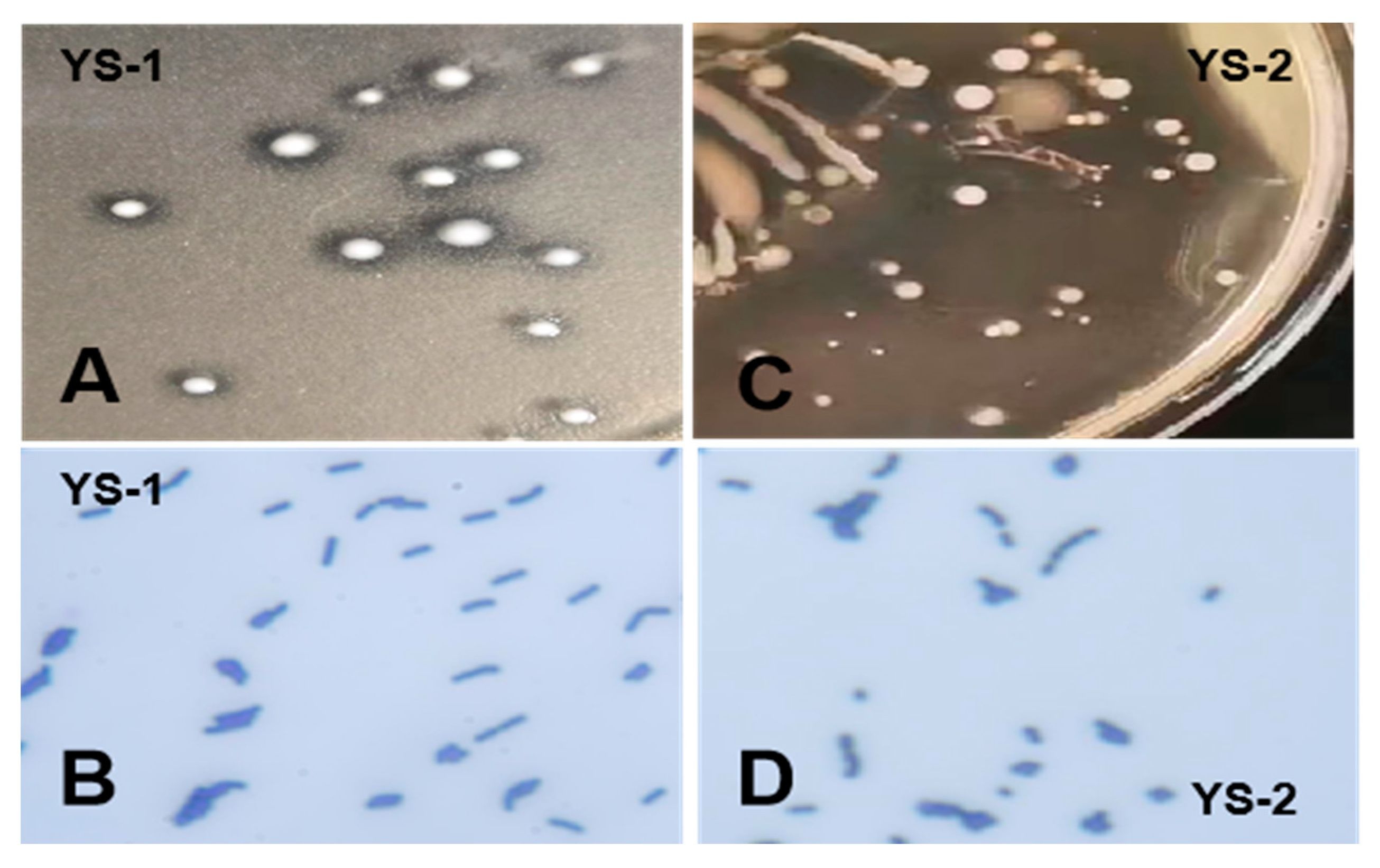
3.1. Isolation and Identification of Bacterial Strains
3.2. Biological Characteristics of the Probiotics
3.3. Effects of L. plantarum and E. faecalis on Production and Immunity of Pekin Duck
3.4. Effects of L. plantarum and E. faecalis on the Intestinal Flora of Pekin Ducks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Round, J.L.; Mazmanian, S.K. The gut microbiome shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Stegeman, J.A.; Smidt, H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018, 9, 235. [Google Scholar] [CrossRef]
- Carrasco, J.M.D.; Casanova, N.A.; Miyakawa, M.E.F. Microbiota, gut health and chicken productivity: What is the connection? Microorganisms 2019, 7, 374. [Google Scholar] [CrossRef] [PubMed]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Lamot, D.M.; Stegeman, J.A.; Smidt, H. Take care of the environment: Housing conditions affect the interplay of nutritional interventions and intestinal microbiota in broiler chickens. Anim. Microbiome 2019, 1, 10. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, V.; Flórez, M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poult. Sci. 2018, 97, 1006–1021. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.L.; Kasper, D.L. Interactions between the intestinal microbiota and innate lymphoid cells. Gut Microbes 2014, 5, 129–140. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [CrossRef]
- Modi, S.R.; Collins, J.J.; Relman, D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014, 124, 4212–4218. [Google Scholar] [CrossRef]
- Yoon, M.Y.; Yoon, S.S. Disruption of the gut ecosystem by antibiotics. Yonsei Med. J. 2018, 59, 4–12. [Google Scholar] [CrossRef]
- Echegaray, N.; Yilmaz, B.; Sharma, H.; Kumar, M.; Pateiro, M.; Ozogul, F.; Lorenzo, J.M. A novel approach to Lactiplantibacillus plantarum: From probiotic properties to the omics insights. Microbiol. Res. 2023, 268, 127289. [Google Scholar] [CrossRef]
- Mokoena, M.P. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- Troche, J.M.R.; Adame, E.C.; Díaz, M.A.V.; Escudero, O.G.; Chávez, M.E.I.; Chávez-Barrera, J.A.; Mondragón, F.Z.; Velasco, J.A.R.V.; Tavares, G.R.A.; Pedrín, M.A.L.; et al. Lactobacillus acidophilus LB: A useful pharmabiotic for the treatment of digestive disorders. Therap. Adv. Gastroenterol. 2020, 13, 1756284820971201. [Google Scholar] [CrossRef]
- De Filippis, F.; Pasolli, E.; Ercolini, D. The food-gut axis: Lactic acid bacteria and their link to food, the gut microbiome and human health. FEMS Microbiol. Rev. 2020, 44, 454–489. [Google Scholar] [CrossRef]
- Meruvu, H.; Harsa, S.T. Lactic acid bacteria: Isolation-characterization approaches and industrial applications. Crit. Rev. Food Sci. Nutr. 2023, 63, 8337–8356. [Google Scholar] [CrossRef]
- Ahmed, Z.; Vohra, M.S.; Khan, M.N.; Ahmed, A.; Khan, T.A. Antimicrobial role of Lactobacillus species as potential probiotics against enteropathogenic bacteria in chickens. J. Infect. Dev. Ctries. 2019, 13, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Suryadi, U.; Nugraheni, Y.R.; Prasetyo, A.F.; Awaludin, A. Evaluation of effects of a novel probiotic feed supplement on the quality of broiler meat. Vet. World 2019, 12, 1775–1778. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.L.; Liu, G.; Zhang, H.H.; Hu, X.F.; Zhang, X.Y.; Han, F.; Cui, H.Z.; Luo, J.J.; Guo, R.; Li, R.; et al. High-mobility group box 1 protein (HMGB1) from Cherry Valley duck mediates signaling pathways and antiviral activity. Vet. Res. 2020, 51, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Liu, C.; An, K.Y.; Gong, X.W.; Xia, Z.F. Effect of dietary Clostridium butyricum supplementation on growth performance, intestinal barrier function, immune function, and microbiota diversity of Pekin ducks. Animals 2021, 11, 2514. [Google Scholar] [CrossRef]
- Pechrkong, T.; Incharoen, T.; Hwanhlem, N.; Kaewkong, W.; Subsoontorn, P.; Tartrakoon, W.; Numthuam, S.; Jiménez, G.; Charoensook, R. Effect of Bacillus toyonensis BCT-7112T supplementation on growth performance, intestinal morphology, immune-related gene expression, and gutmicrobiome in Barbary ducks. Poult. Sci. 2023, 102, 102991. [Google Scholar] [CrossRef]
- Hu, Z.G.; Cao, J.T.; Zhang, J.Q.; Ge, L.Y.; Zhang, H.L.; Liu, X.L. Skeletal muscle transcriptome analysis of Hanzhong Ma duck at different growth stages using RNA-Seq. Biomolecules 2021, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Dong, L.; Guo, S.J.; Xu, Q.Q.; Miao, L.Z.; Xie, J.W.; Zhang, Y.; Ma, L.; Shen, Z.Q. Establishment of SYBR Green Ⅰ qPCR detection method for irp2 gene of poultry pathogenic Escherichia coli virulence island. Chin. Vet. Sci. 2017, 47, 871–876. (In Chinese) [Google Scholar]
- Pasquali, F.; Rossi, M.; Manfreda, G.; Zanoni, R. Complete nucleotide sequence of the gyrA gene of Helicobacter pullorum and identification of a point mutation leading to ciprofloxacin resistance in poultry isolates. Int. J. Antimicrob. Agents 2007, 30, 222–238. [Google Scholar] [CrossRef] [PubMed]
- Li, F.H.; Lou, L.P.; Jiang, T.T.; Liu, M.X.; Deng, Y.G.; Liu, J.C. Establishment of SYBR Green Ⅰ multiplex quantitative PCR method for Escherichia coli, Salmonella, Campylobacter jejuni and Proteus mirabilis in chicken. Mod. J. Anim. Husb. Vet. Med. 2022, 11, 1–6. (In Chinese) [Google Scholar]
- Sun, S.D.; Tian, Y.; Gao, Y.W.; Niu, L.Y.; Du, Y.Y.; Wang, W.J.; Zhong, J.H.; Tang, X.Q.; Liu, J.Q.; Zhou, W.P. Construction and application of fluorescent quantitative PCR method for avian Salmonella. Hunan J. Ani. Sci. Vet. Med. 2024, 1, 41–45. (In Chinese) [Google Scholar]
- Yang, M.; Yin, Y.; Wang, F.; Bao, X.T.; Long, L.N.; Tan, B.; Yin, Y.L.; Chen, J.S. Effects of dietary rosemary extract supplementation on growth performance, nutrient digestibility, antioxidant capacity, intestinal morphology, and microbiota of weaning pigs. J. Anim. Sci. 2021, 99, 237. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Probiotics in irritable bowel syndrome: An immunomodulatory strategy? J. Am. Coll. Nutr. 2007, 26, 684S–690S. [Google Scholar] [CrossRef]
- Power, S.E.; O’Toole, P.W.; Stanton, C.; Ross, R.P.; Fitzgerald, G.F. Intestinal microbiota, diet and health. Br. J. Nutr. 2014, 111, 387–402. [Google Scholar] [CrossRef]
- Sureshkumar, S.; Lee, H.C.; Jung, S.K.; Kim, D.J.; Oh, K.B.; Yang, H.; Jo, Y.J.; Lee, H.S.; Lee, S.; Byun, S.J.; et al. Inclusion of Lactobacillus salivarius strain revealed a positive effect on improving growth performance, fecal microbiota and immunological responses in chicken. Arch. Microbiol. 2021, 203, 847–853. [Google Scholar] [CrossRef] [PubMed]
- An, K.; Gao, W.; Li, P.; Li, L.; Xia, Z. Dietary Lactobacillus plantarum improves the growth performance and intestinal health of Pekin ducks. Poult. Sci. 2022, 101, 101844. [Google Scholar] [CrossRef] [PubMed]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus plantarum-nomad and ideal probiotic. Front. Microbiol. 2021, 12, 712236. [Google Scholar] [CrossRef] [PubMed]
- Talib, M.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Isolation and characterization of Lactobacillus spp. from kefir samples in Malaysia. Molecules 2019, 24, 2606. [Google Scholar] [CrossRef]
- Daca, A.; Jarzembowski, T. From the friend to the foe-Enterococcus faecalis diverse impact on the human immune system. Int. J. Mol. Sci. 2024, 25, 2422. [Google Scholar] [CrossRef]
- Ditu, L.M.; Chifiriuc, M.C.; Bezirtzoglou, E.; Voltsi, C.; Bleotu, C.; Pelinescu, D.; Mihaescu, G.; Lazar, V. Modulation of virulence and antibiotic susceptibility of enteropathogenic Escherichia coli strains by Enterococcus faecium probiotic strain culture fractions. Anaerobe 2011, 17, 448–451. [Google Scholar] [CrossRef]
- Kassa, G.; Alemayehu, D.; Andualem, B. Isolation, identification, and molecular characterization of probiotic bacteria from locally selected Ethiopian free range chickens gastrointestinal tract. Poult. Sci. 2024, 103, 103311. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.U.; Jahid, I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef]
- Yin, Y.Y.; Liao, Y.Y.; Li, J.; Pei, Z.; Wang, L.P.; Shi, Y.; Peng, H.Y.; Tan, Y.Z.; Li, C.T.; Bai, H.L.; et al. Lactobacillus plantarum GX17 benefits growth performance and improves functions of intestinal barrier/intestinal flora among yellow-feathered broilers. Front. Immunol. 2023, 14, 1195382. [Google Scholar] [CrossRef] [PubMed]
- Shehata, A.A.; Tarabees, R.; Basiouni, S.; ElSayed, M.S.; Gaballah, A.; Krueger, M. Effect of a potential probiotic candidate Enterococcus faecalis-1 on growth performance, intestinal microbiota, and immune response of commercial broiler chickens. Probiotics Antimicrob. Proteins 2020, 12, 451–460. [Google Scholar] [CrossRef]
- De Sousa-Pereira, P.; Woof, J.M. IgA: Structure, Function, and Developability. Antibodies 2019, 8, 57. [Google Scholar] [CrossRef]
- Damelang, T.; Brinkhaus, M.; van Osch, T.L.J.; Schuurman, J.; Labrijn, A.F.; Rispens, T.; Vidarsson, G. Impact of structural modifications of IgG antibodies on effector functions. Front. Immunol. 2024, 14, 1304365. [Google Scholar] [CrossRef]
- Quagliato, L.A.; Nardi, A.E. Cytokine alterations in panic disorder: A systematic review. J. Affect. Disord. 2018, 228, 91–96. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, Z.; Zhu, L.; Ma, S.; Luo, Y.; Liang, H.; Liu, Q.; Chen, J.; Guli, S.; Chen, X. Orchestration of MUC2-The key regulatory target of gut barrier and homeostasis: A review. Int. J. Biol. Macromol. 2023, 236, 123862. [Google Scholar] [CrossRef]
- Mancabelli, L.; Ferrario, C.; Milani, C.; Mangifesta, M.; Turroni, F.; Duranti, S.; Lugli, G.A.; Viappiani, A.; Ossiprandi, M.C.; van Sinderen, D.; et al. Insights into the biodiversity of the gut microbiota of broiler chickens. Environ. Microbiol. 2016, 18, 4727–4738. [Google Scholar] [CrossRef]
- Ha, C.W.Y.; Lam, Y.Y.; Holmes, A.J. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J. Gastroenterol. 2014, 20, 16498–16517. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.G.; Hur, K.Y.; Lee, M.S. Alterations in gut microbiota and immunity by dietary fat. Yonsei Med. J. 2017, 58, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, H.; Liu, W.; Wu, H.; Li, C.; Lin, F.; Yan, L.; Huang, C. Beneficial effects of duck-derived lactic acid bacteria on growth performance and meat quality through modulation of gut histomorphology and intestinal microflora in Muscovy ducks. Poult. Sci. 2024, 103, 104195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yu, A.; Wu, H.; Xiong, X.; Li, J.; Chen, L. Lactobacillus acidophilus and Bacillus subtilis significantly change the growth performance, serum immunity and cecal microbiota of Cherry Valley ducks during the fattening period. Anim. Sci. J. 2024, 95, e13946. [Google Scholar] [CrossRef] [PubMed]
| Ingredient | Content (%) | Nutrient | Content (%) |
|---|---|---|---|
| Corn | 56.00 | Avian metabolizable energy | 2875 Mcal·kg−1 |
| Soybean meal | 23.80 | Crude protein | 15.700 |
| Corn gluten meal | 10.00 | Calcium | 0.900 |
| Limestone | 7.00 | Total phosphorus | 0.680 |
| CaHPO4 | 1.50 | Available phosphorus | 0.450 |
| Premix | 1.00 | Salt | 0.370 |
| NaCl | 0.30 | Lysine | 0.760 |
| Lys·HCl | 0.30 | Methionine | 0.387 |
| DL-Met | 0.10 | Methionine + Cystine | 0.654 |
| Total | 100.00 | Isoleucine | 0.534 |
| Threonine | 0.579 | ||
| Tryptophan | 0.194 | ||
| Crude fiber | 4.100 | ||
| Crude fat | 3.400 | ||
| Crude ash | 5.200 |
| Name (Target Gene) | Primer (5′-3′) | Length | Reference |
|---|---|---|---|
| INF-γ | F:GCTGATGGCAATCCTGTTTT | 247 bp | [21] |
| R:GGATTTTCAAGCCAGTCAGC | |||
| MUC2 | F:AGTTCTTGCCTAATTCCTCAGTCT | 146 bp | [22] |
| R:TTGCCGTTCATATCCAGGTTCA | |||
| IL-6 | F:TTCGACGAGGAGAAATGCTT | 150 bp | [23] |
| R:CCTTATCGTCGTTGCCAGAT | |||
| β-actin | F:CCCTGTATGCCTCTGGTCG | 194 bp | [24] |
| R:CTCGGCTGTGGTGGTGAAG | |||
| Escherichia coli (irp2) | F: GCTCTGTGCCCTTTGA | 261 bp | [25] |
| R: GGCGGGAGGAGTAGTT | |||
| Helicobacter pullorum (gyrA) | F: CAAGAATCGTGGGTGATG | 351 bp | [26] |
| R: GTGGAATATTTGTCGCCA | |||
| Campylobacter sp. (aicsxii) | F: CACATTAAATCTTTATTTTCAACCCGCTGAA | 73 bp | [27] |
| R: ACAATCCATCTTCTATCATTGCCTTAGC | |||
| Salmonella sp. (invA) | F: CAATGGCGGCGAATTACGAG | 100 bp | [28] |
| R: AAGGCTGAGGAAGGTACTGC | |||
| Lactobacillus sp. | F: CGATGAGTGCTAGGTGTTGGA | 186 bp | [29] |
| R: CAAGATGTCAAGACCTGGTAAG |
| Name | Strain and Diameter of Antibacterial Zone (mm) | ||
|---|---|---|---|
| Staphylococcus aureus | Escherichia coli | Salmonella gallinarum | |
| L. plantarum | 12.20 ± 0.92 | 12.92 ± 0.74 | 13.42 ± 1.40 |
| E. faecalis | 13.92 ± 2.12 | 12.48 ± 0.23 | 14.07 ± 2.41 |
| Performance | Control Group | LP Group | EF Group |
|---|---|---|---|
| ADFI (g/d) | 251.77 ± 3.89 b | 259.34 ± 4.32 a | 257.18 ± 4.18 b |
| ADG (g/d) | 110.32 ± 2.04 b | 118.15 ± 1.84 a | 116.58 ± 1.99 a |
| FCR | 2.28 ± 0.22 | 2.20 ± 0.23 | 2.21 ± 0.26 |
| Live weight (g) | 3278.74 ± 30.76 b | 3462.15 ± 47.85 a | 3432.69 ± 40.32 a |
| Carcass weight (g) | 2866.32 ± 27.55 | 2902.38 ± 35.69 | 2887.65 ± 34.82 |
| Eviscerated weight (g) | 2401.58 ± 35.83 b | 2573.46 ± 33.28 a | 2568.14 ± 34.49 a |
| Half-eviscerated weight (g) | 2698.14 ± 40.48 b | 2832.08 ± 38.77 a | 2828.78 ± 39.21 a |
| Breast muscle weight (g) | 231.23 ± 8.54 b | 245.55 ± 9.85 a | 242.14 ± 9.42 a |
| Leg muscle weight (g) | 266.12 ± 7.15 b | 282.27 ± 8.56 a | 289.32 ± 8.58 a |
| Items | Control Group | LP Group | EF Group |
|---|---|---|---|
| Villus height in duodenum (μm) | 1007.35 ± 71.33 b | 1107.54 ± 50.05 a | 1101.21 ± 53.26 a |
| Crypt depth in duodenum (μm) | 207.78 ± 20.26 | 213.98 ± 25.45 | 208.85 ± 31.23 |
| V/C in duodenum | 4.85 ± 0.33 b | 5.18 ± 0.42 a | 5.27 ± 0.40 a |
| Villus height in jejunum (μm) | 637.65 ± 45.01 b | 656.56 ± 37.75 a | 650.50 ± 38.44 a |
| Crypt depth in jejunum (μm) | 208.42 ± 18.83 | 211.23 ± 21.36 | 210.38 ± 24.45 |
| V/C in jejunum | 3.06 ± 0.35 | 3.11 ± 0.22 | 3.09 ± 0.33 |
| Villus height in ileum (μm) | 664.83 ± 87.35 b | 679.88 ± 56.58 a | 677.67 ± 67.43 a |
| Crypt depth in ileum (μm) | 155.90 ± 20.63 | 158.54 ± 24.08 | 156.38 ± 23.15 |
| V/C in ileum | 4.26 ± 0.25 | 4.29 ± 0.38 | 4.33 ± 0.29 |
| Thymus index (%) | 1.71 ± 0.21 | 1.78 ± 0.16 | 1.75 ± 0.16 |
| Bursa index (%) | 0.62 ± 0.11 b | 0.70 ± 0.09 a | 0.72 ± 0.10 a |
| Spleen index (%) | 0.57 ± 0.07 b | 0.64 ± 0.05 a | 0.63 ± 0.08 a |
| Serum IgG content (μg/mL) | 752.79 ± 42.34 a | 679.08 ± 48.25 b | 684.23 ± 36.12 b |
| Serum IgA content (μg/mL) | 123.28 ± 12.48 b | 136.78 ± 15.87 a | 137.74 ± 13.75 a |
| Strains | Standard Curve | Correlation Coefficient (R2) | Amplification Efficiency (E) |
|---|---|---|---|
| Escherichia coli | y = −2.8283x + 24.113 | 0.9883 | 102.3 % |
| Helicobacterpullorum | y = −2.9817x + 28.337 | 0.9956 | 107.5 % |
| Campylobacter sp. | y = −3.0549x + 28.961 | 0.9913 | 100.1 % |
| Salmonella sp. | y = −3.1228x + 33.136 | 0.9942 | 98.8 % |
| Lactobacillus sp. | y = −3.2248x + 31.456 | 0.9938 | 100.9 % |
| Name | Control Group | LP Group | EF Group |
|---|---|---|---|
| Escherichia coli | 5.28 ± 0.13 a | 4.72 ± 0.08 b | 4.49 ± 0.11 b |
| Helicobacter pullorum | 3.73 ± 0.11 | 3.64 ± 0.08 | 3.58 ± 0.09 |
| Campylobacter sp. | 4.27 ± 0.27 a | 3.84 ± 0.15 b | 4.02 ± 0.23 b |
| Salmonella sp. | 4.79 ± 0.20 a | 3.53 ± 0.35 b | 3.77 ± 0.45 b |
| Lactobacillus sp. | 3.66 ± 0.22 B | 5.38 ± 0.29 A | 4.86 ± 0.36 A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.; Zhi, Z.; Zhang, H.; Zhou, J.; Cui, M.; Zhang, J.; Xue, D.; Liu, X. Isolation and Identification of Duck Intestinal Probiotics and Their Effects on the Production and Immune Performance of Pekin Ducks. Biomolecules 2025, 15, 1217. https://doi.org/10.3390/biom15091217
Hu Z, Zhi Z, Zhang H, Zhou J, Cui M, Zhang J, Xue D, Liu X. Isolation and Identification of Duck Intestinal Probiotics and Their Effects on the Production and Immune Performance of Pekin Ducks. Biomolecules. 2025; 15(9):1217. https://doi.org/10.3390/biom15091217
Chicago/Turabian StyleHu, Zhigang, Zhuo Zhi, Huiya Zhang, Jie Zhou, Mengmeng Cui, Jianqin Zhang, Dongfeng Xue, and Xiaolin Liu. 2025. "Isolation and Identification of Duck Intestinal Probiotics and Their Effects on the Production and Immune Performance of Pekin Ducks" Biomolecules 15, no. 9: 1217. https://doi.org/10.3390/biom15091217
APA StyleHu, Z., Zhi, Z., Zhang, H., Zhou, J., Cui, M., Zhang, J., Xue, D., & Liu, X. (2025). Isolation and Identification of Duck Intestinal Probiotics and Their Effects on the Production and Immune Performance of Pekin Ducks. Biomolecules, 15(9), 1217. https://doi.org/10.3390/biom15091217





