Exploring the Allosteric Pathways of Asciminib in the Dual Inhibition of BCR-ABL1
Abstract
1. Introduction
2. Materials and Methods
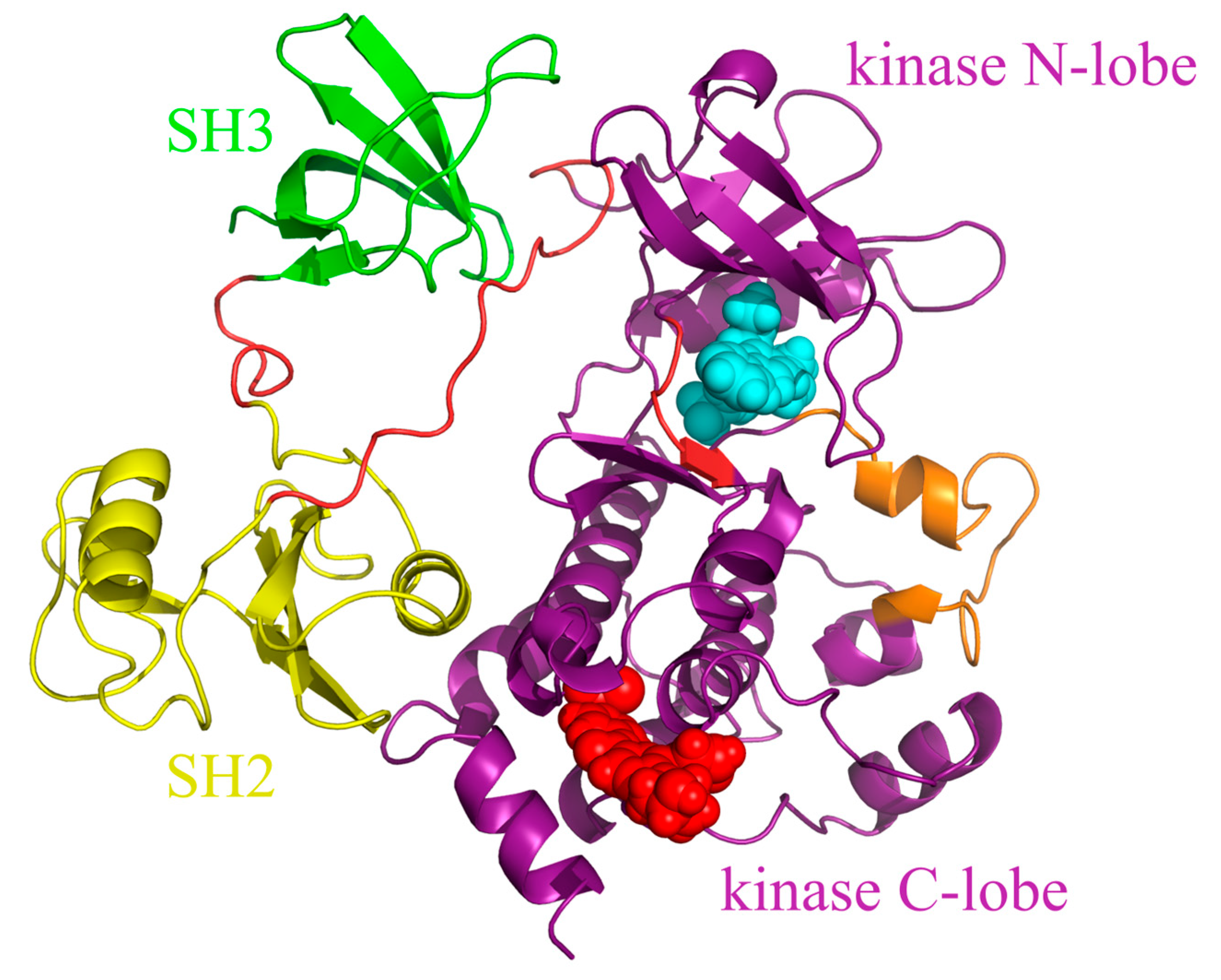
2.1. System Preparation
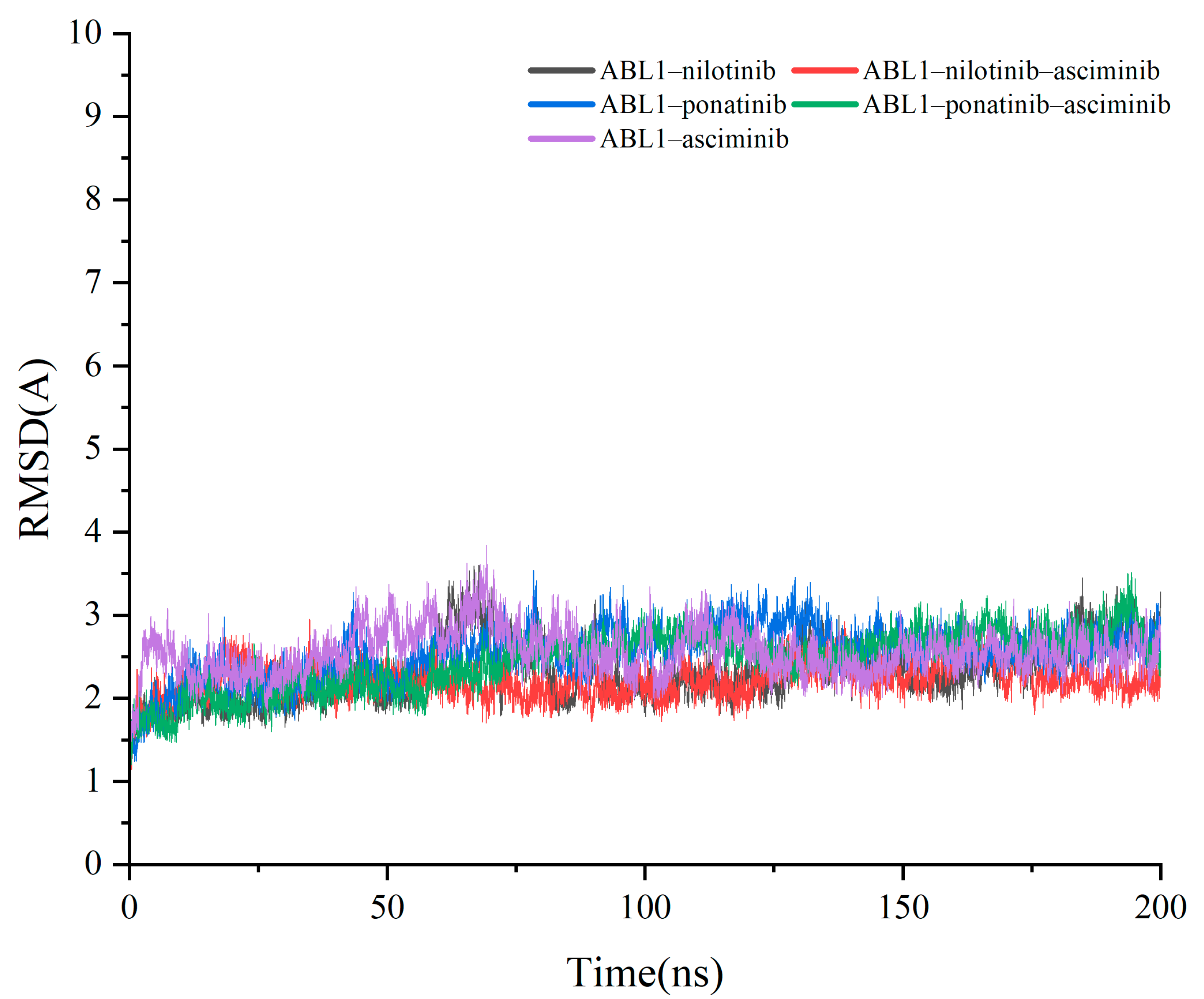
2.2. Molecular Dynamics (MD) Simulations
2.3. MM-GB/SA Calculations
2.4. Residue Interaction Network
3. Results and Discussion
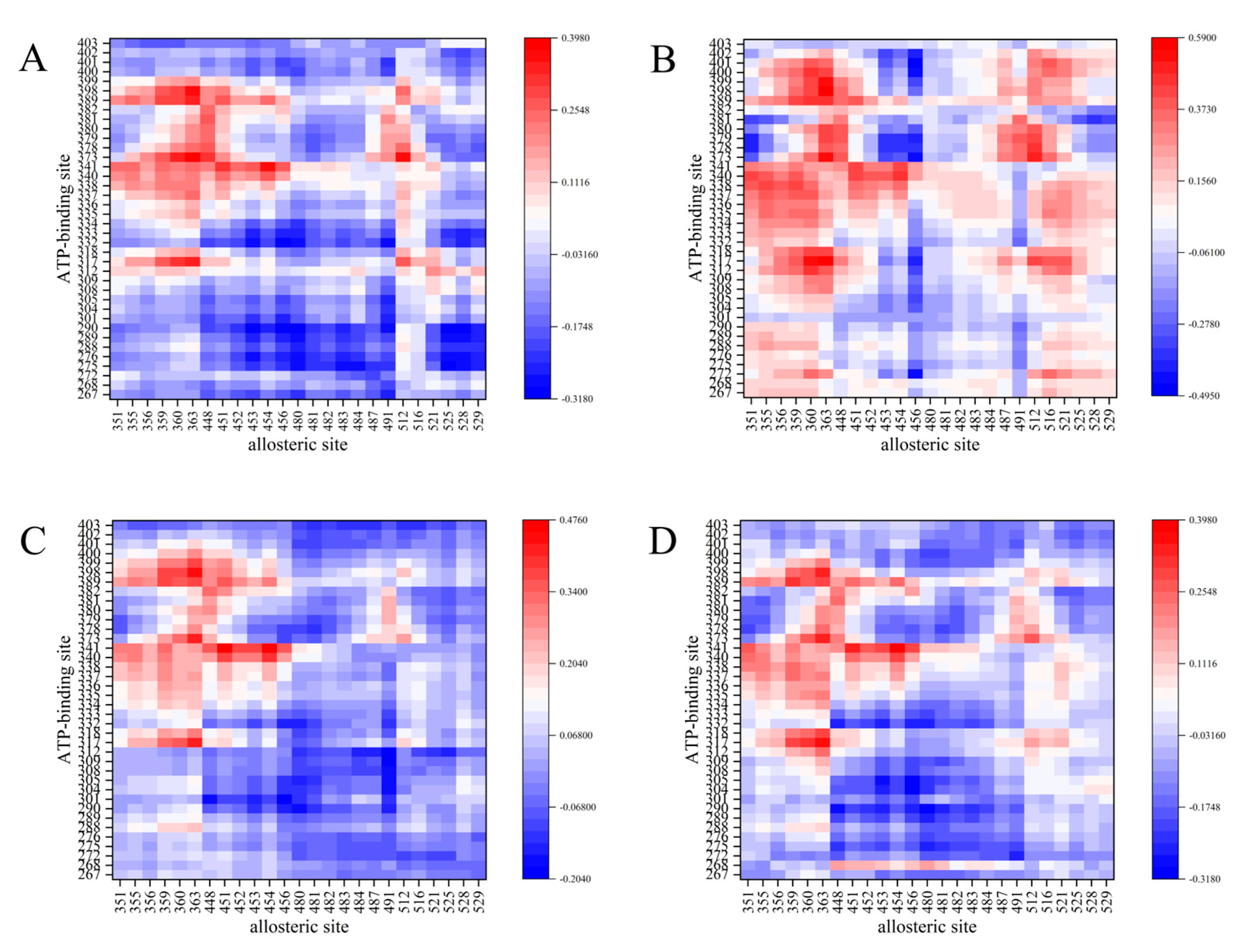
3.1. Correlation of ATP Binding Site with Allosteric Site
3.2. Binding Free Energy Calculations
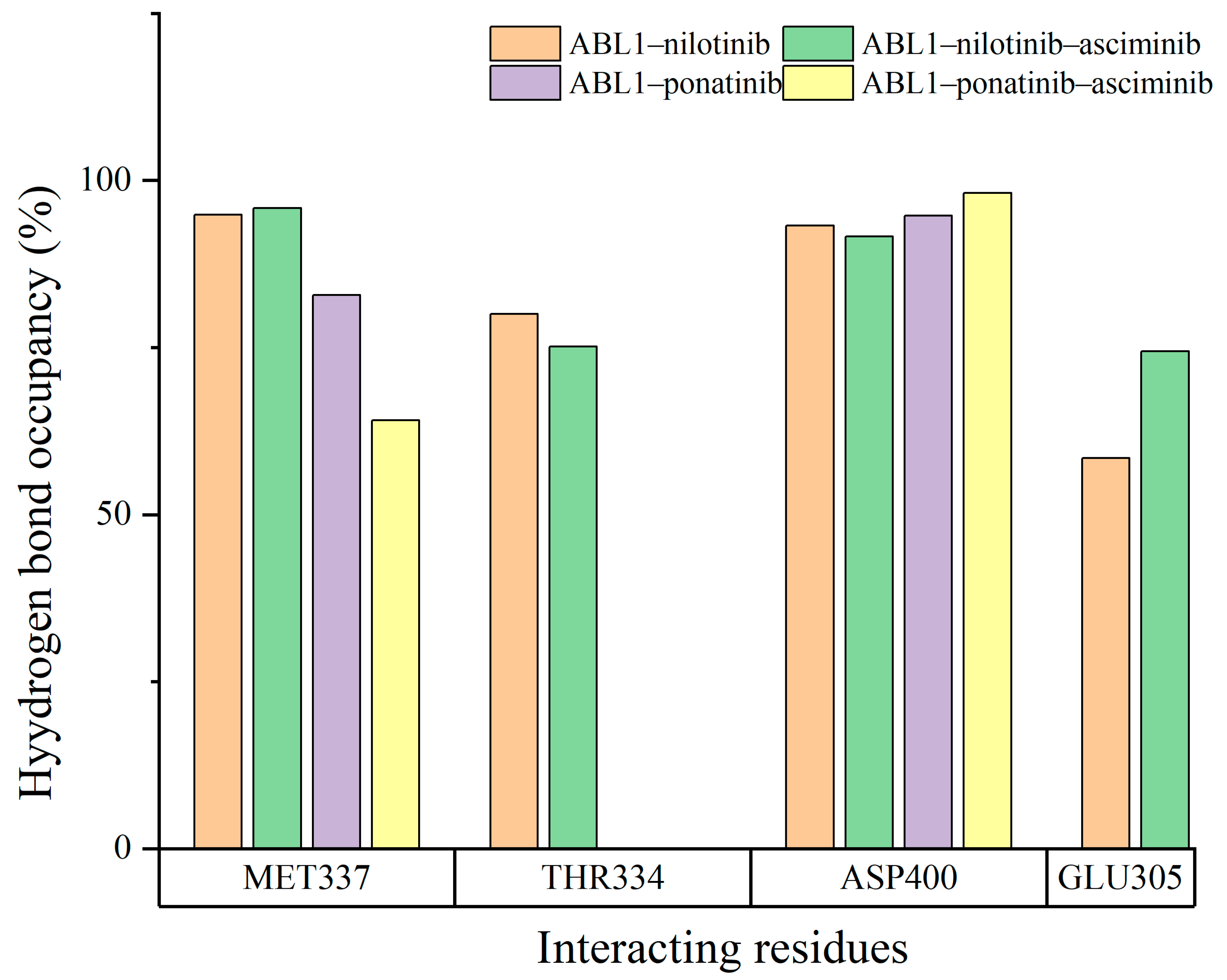
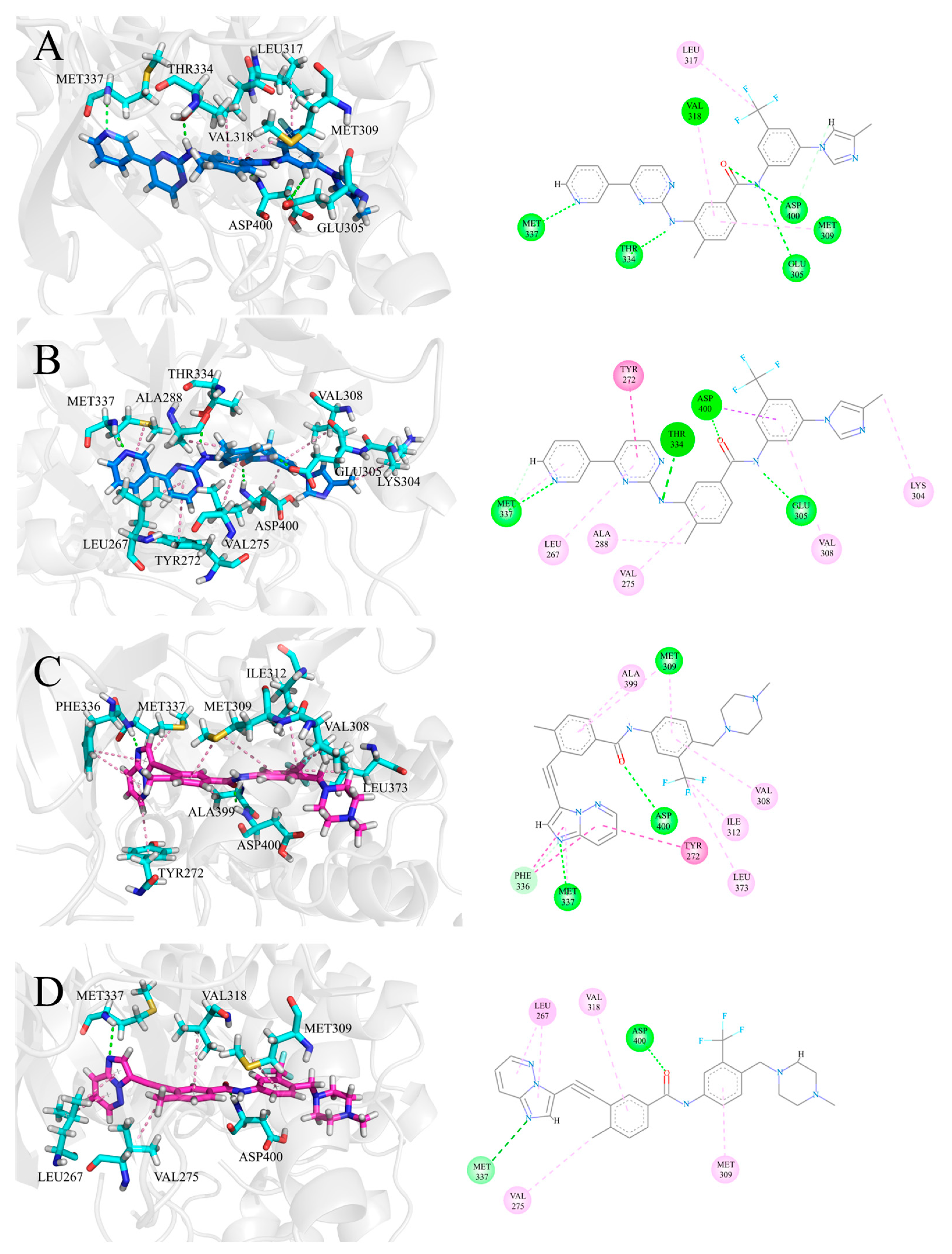
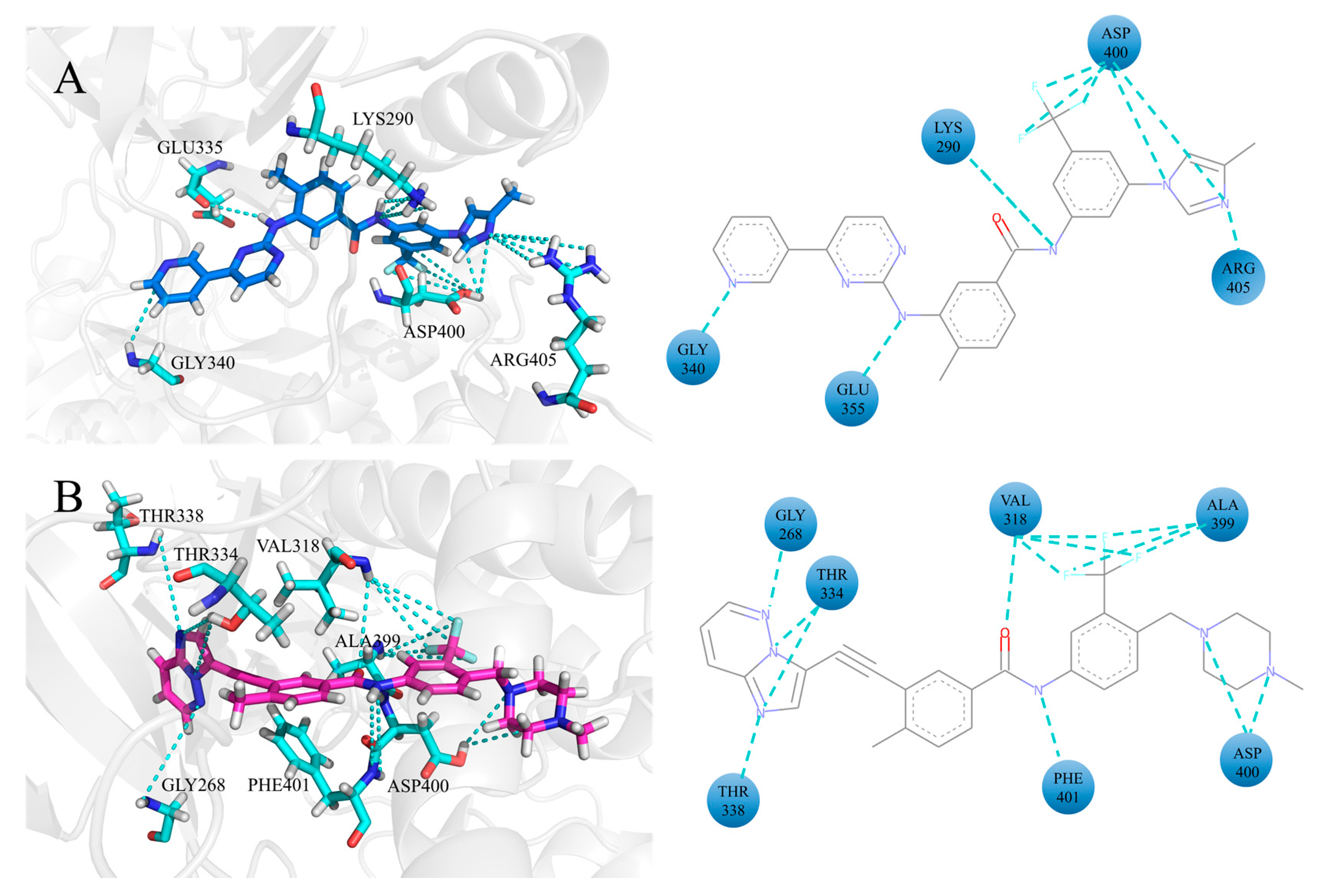
3.3. The H-Bonding Ability of Inhibitors at the ATP Binding Site
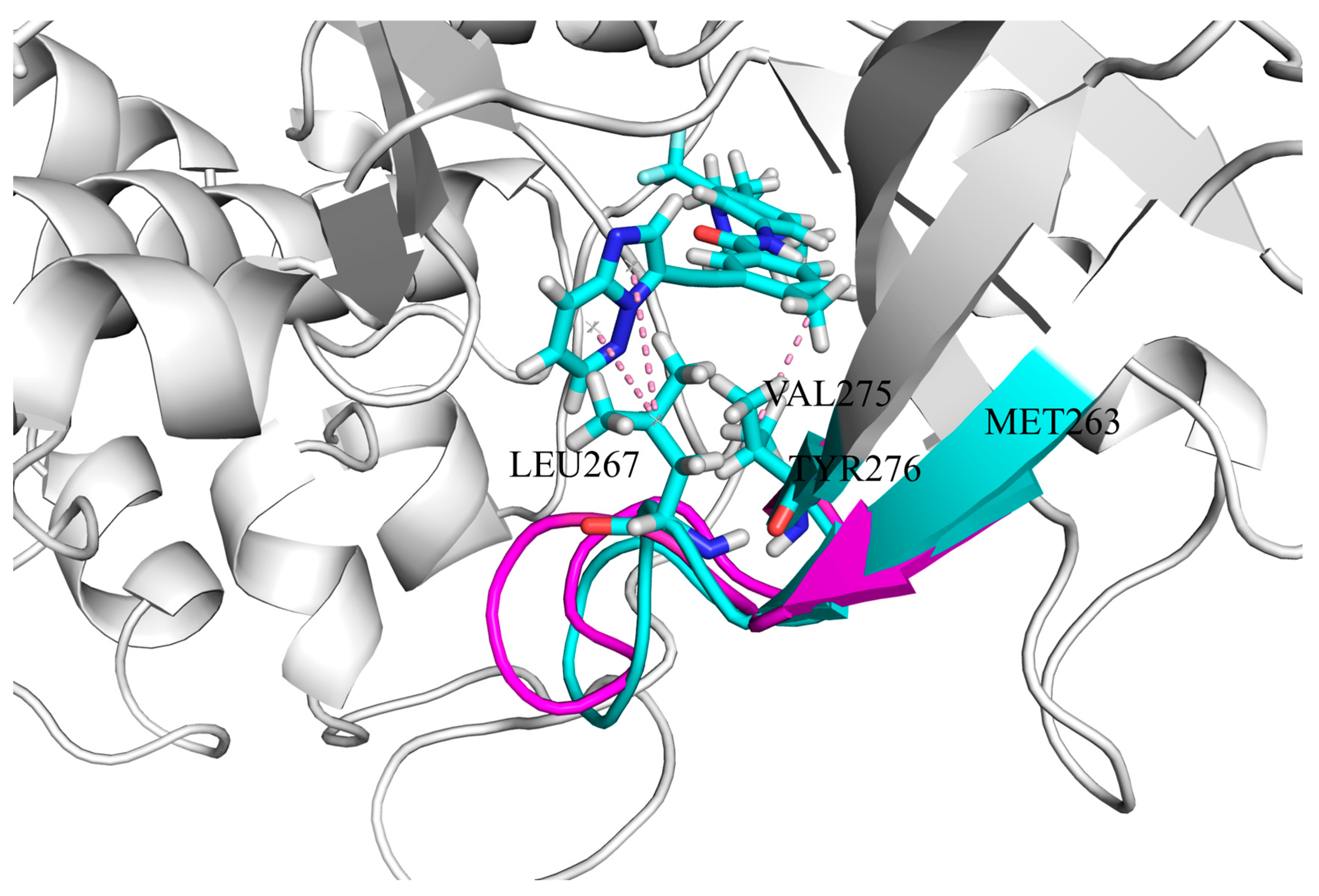
3.4. The Hydrophobic Interaction of Inhibitors at the ATP Binding Site
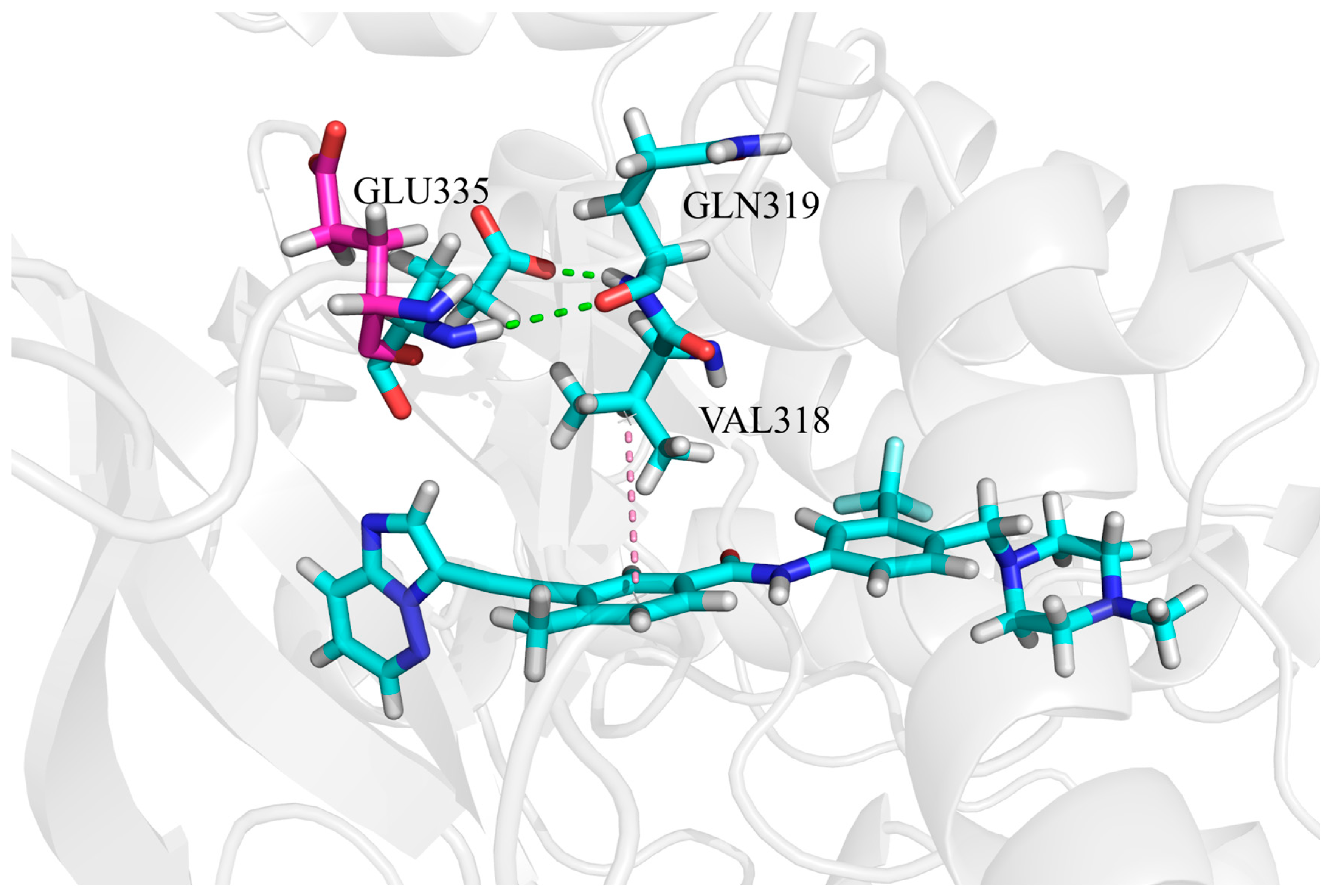
3.5. The Electrostatic Interaction of Inhibitors at the ATP Binding Site
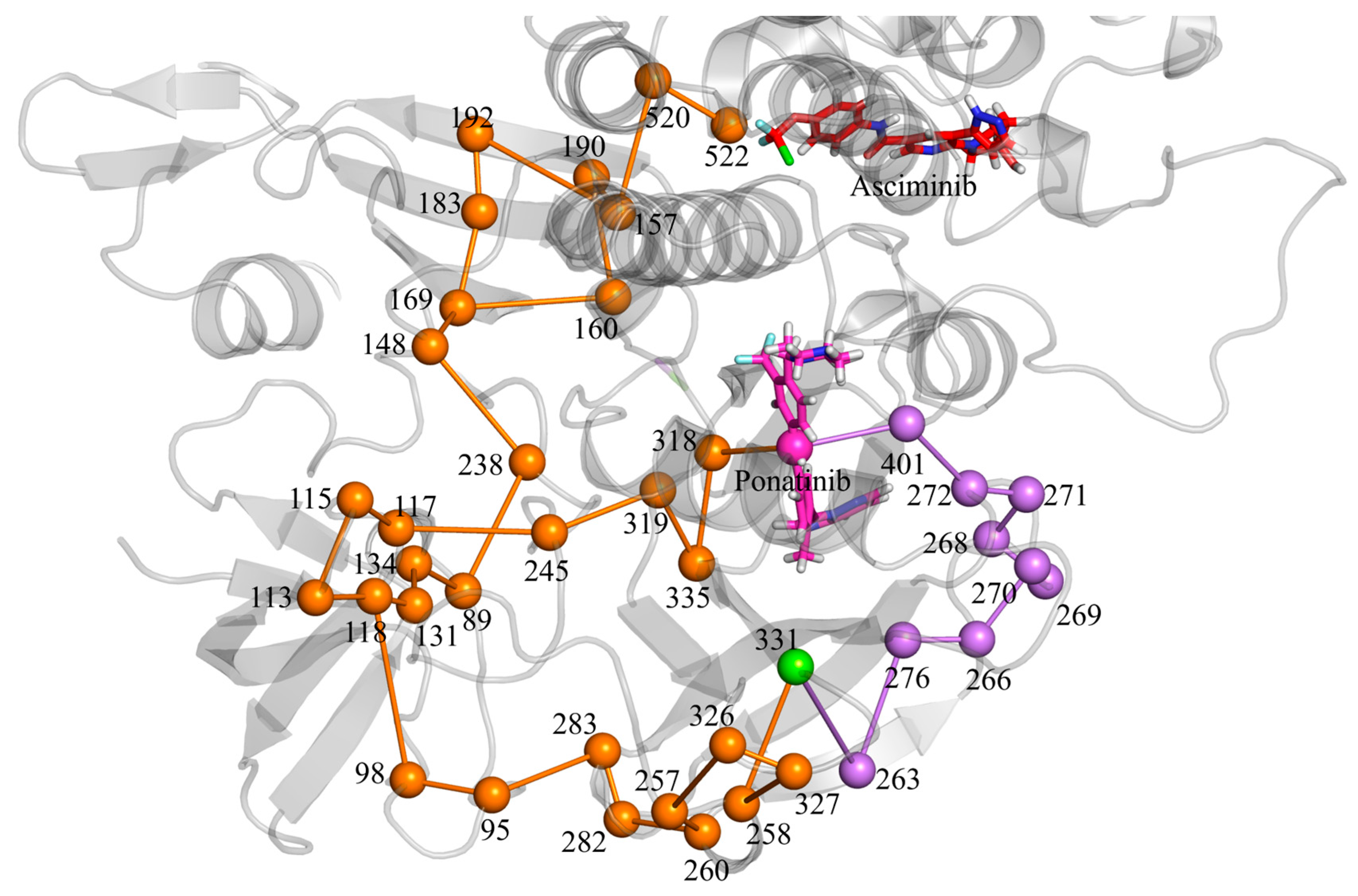
3.6. Allosteric Communications Between Asciminib and Ponatinib
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.D. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine Fluorescence and Giemsa Staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef]
- El-Tanani, M.; Nsairat, H.; Matalka, I.I.; Lee, Y.F.; Rizzo, M.; Aljabali, A.A.; Mishra, V.; Mishra, Y.; Hromić-Jahjefendić, A.; Tambuwala, M.M. The impact of the BCR-ABL oncogene in the pathology and treatment of chronic myeloid leukemia. Pathol.-Res. Pract. 2024, 254, 155161. [Google Scholar] [CrossRef]
- Han, H.-J.; Kim, J.J.; Pyne, D.; Travas, A.; Ambalavanan, A.; Kimura, S.; Deininger, M.W.; Kim, J.-W.; Kim, D.D.H. In vitro evidence of synergistic efficacy with asciminib combined with reduced dose of ATP-binding pocket tyrosine kinase inhibitors according to the ABL1 kinase domain mutation profile. Leukemia 2023, 38, 412–415. [Google Scholar] [CrossRef]
- Hunter, T. Treatment for chronic myelogenous leukemia: The long road to imatinib. J. Clin. Investig. 2007, 117, 2036–2043. [Google Scholar] [CrossRef]
- Chandrasekhar, C.; Kumar, P.S.; Sarma, P.V.G.K. Novel mutations in the kinase domain of BCR-ABL gene causing imatinib resistance in chronic myeloid leukemia patients. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Saglio, G. Second-generation TKIs: Which and when? Leuk. Suppl. 2012, 1, S40–S42. [Google Scholar] [CrossRef]
- Abruzzese, E.; Breccia, M.; Latagliata, R. Second-Generation Tyrosine Kinase Inhibitors in First-Line Treatment of Chronic Myeloid Leukaemia (CML). BioDrugs 2013, 28, 17–26. [Google Scholar] [CrossRef]
- Blay, J.-Y.; von Mehren, M. Nilotinib: A Novel, Selective Tyrosine Kinase Inhibitor. Semin. Oncol. 2011, 38, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.; Moura, S.; Parra, L.; Vasconcellos, V.; Costa, G.; Leite, D.; Dias, M.; Fernandes, T.V.A.; Hoelz, L.; Pimentel, L.; et al. Ponatinib: A Review of the History of Medicinal Chemistry behind Its Development. Pharmaceuticals 2024, 17, 1361. [Google Scholar] [CrossRef]
- Weisberg, E.; Manley, P.; Mestan, J.; Cowan-Jacob, S.; Ray, A.; Griffin, J.D. AMN107 (nilotinib): A novel and selective inhibitor of BCR-ABL. Br. J. Cancer 2006, 94, 1765–1769. [Google Scholar] [CrossRef] [PubMed]
- Lambert, G.K.; Duhme-Klair, A.-K.; Morgan, T.; Ramjee, M.K. The background, discovery and clinical development of BCR-ABL inhibitors. Drug. Discov. Today 2013, 18, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Kwarcinski, F.E.; Brandvold, K.R.; Phadke, S.; Beleh, O.M.; Johnson, T.K.; Meagher, J.L.; Seeliger, M.A.; Stuckey, J.A.; Soellner, M.B. Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity. ACS. Chem. Biol. 2016, 11, 1296–1304. [Google Scholar] [CrossRef]
- Gorre, M.E.; Mohammed, M.; Ellwood, K.; Hsu, N.; Paquette, R.; Rao, P.N.; Sawyers, C.L. Clinical Resistance to STI-571 Cancer Therapy Caused by BCR-ABL Gene Mutation or Amplification. Science 2001, 293, 876–880. [Google Scholar] [CrossRef]
- Manley, P.W.; Barys, L.; Cowan-Jacob, S.W. The specificity of asciminib, a potential treatment for chronic myeloid leukemia, as a myristate-pocket binding ABL inhibitor and analysis of its interactions with mutant forms of BCR-ABL1 kinase. Leuk. Res. 2020, 98, 106458. [Google Scholar] [CrossRef] [PubMed]
- Tamai, M.; Inukai, T.; Kojika, S.; Abe, M.; Kagami, K.; Harama, D.; Shinohara, T.; Watanabe, A.; Oshiro, H.; Akahane, K.; et al. T315I mutation of BCR-ABL1 into human Philadelphia chromosome-positive leukemia cell lines by homologous recombination using the CRISPR/Cas9 system. Sci. Rep. 2018, 8, 9966. [Google Scholar] [CrossRef]
- Mian, A.; Schüll, M.; Zhao, Z.; Oancea, C.; Hundertmark, A.; Beissert, T.; Ottmann, O.G.; Ruthardt, M. The gatekeeper mutation T315I confers resistance against small molecules by increasing or restoring the ABL-kinase activity accompanied by aberrant transphosphorylation of endogenous BCR, even in loss-of-function mutants of BCR/ABL. Leukemia 2009, 23, 1614–1621. [Google Scholar] [CrossRef]
- Zabriskie, M.S.; Eide, C.A.; Tantravahi, S.K.; Vellore, N.A.; Estrada, J.; Nicolini, F.E.; Khoury, H.J.; Larson, R.A.; Konopleva, M.; Cortes, J.E.; et al. BCR-ABL1 Compound Mutations Combining Key Kinase Domain Positions Confer Clinical Resistance to Ponatinib in Ph Chromosome-Positive Leukemia. Cancer Cell 2014, 26, 428–442. [Google Scholar] [CrossRef]
- Georgoulia, P.S.; Todde, G.; Bjelic, S.; Friedman, R. The catalytic activity of Abl1 single and compound mutations: Implications for the mechanism of drug resistance mutations in chronic myeloid leukaemia. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 732–741. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Wang, F.; Zhang, Y.; Chen, X.; Nie, D.; Li, Y.; Tan, Y.; Xu, Y.; Ma, X. Dynamic Evolution of Ponatinib Resistant BCR-ABL1 T315 and Compound Mutations. Blood 2019, 134, 3796. [Google Scholar] [CrossRef]
- Heiblig, M.; Rea, D.; Chrétien, M.-L.; Charbonnier, A.; Rousselot, P.; Coiteux, V.; Escoffre-Barbe, M.; Dubruille, V.; Huguet, F.; Cayssials, E.; et al. Ponatinib evaluation and safety in real-life chronic myelogenous leukemia patients failing more than two tyrosine kinase inhibitors: The PEARL observational study. Exp. Hematol. 2018, 67, 41–48. [Google Scholar] [CrossRef]
- Teng, M.; Luskin, M.R.; Cowan-Jacob, S.W.; Ding, Q.; Fabbro, D.; Gray, N.S. The Dawn of Allosteric BCR-ABL1 Drugs: From a Phenotypic Screening Hit to an Approved Drug. J. Med. Chem. 2022, 65, 7581–7594. [Google Scholar] [CrossRef] [PubMed]
- Assanto, G.M.; Scalzulli, E.; Breccia, M. Asciminib in chronic myeloid leukemia. Drugs Today 2022, 58, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Schuld, P.; Grzesiek, S.; Schlotte, J.; Habazettl, J.M.; Jahnke, W.; Barys, L.; Cowan-Jacob, S.W.; Loo, A.; Wiget, A.; Manley, P.W. Structural and Biochemical Studies Confirming the Mechanism of Action of Asciminib, an Agent Specifically Targeting the ABL Myristoyl Pocket (STAMP). Blood 2020, 136, 34–35. [Google Scholar] [CrossRef]
- Schoepfer, J.; Jahnke, W.; Berellini, G.; Buonamici, S.; Cotesta, S.; Cowan-Jacob, S.W.; Dodd, S.; Drueckes, P.; Fabbro, D.; Gabriel, T.; et al. Discovery of Asciminib (ABL001), an Allosteric Inhibitor of the Tyrosine Kinase Activity of BCR-ABL1. J. Med. Chem. 2018, 61, 8120–8135. [Google Scholar] [CrossRef] [PubMed]
- Beyett, T.S.; To, C.; Heppner, D.E.; Rana, J.K.; Schmoker, A.M.; Jang, J.; De Clercq, D.J.H.; Gomez, G.; Scott, D.A.; Gray, N.S.; et al. Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors. Nat. Commun. 2022, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lindström, H.J.G.; Friedman, R. The effects of combination treatments on drug resistance in chronic myeloid leukaemia: An evaluation of the tyrosine kinase inhibitors axitinib and asciminib. BMC. Cancer 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Jahnke, W.; Paladini, J.; Habazettl, J.M.; Wiget, A.; Loo, A.; Jacob, S.W.C.; Grzesiek, S.; Manley, P.W. Correspondence on “Synergy and Antagonism between Allosteric and Active-Site Inhibitors of Abl Tyrosine Kinase”. Angew. Chem. Int. Ed. 2022, 61, e202117276. [Google Scholar] [CrossRef]
- Kim, C.; Ludewig, H.; Hadzipasic, A.; Kutter, S.; Nguyen, V.; Kern, D. A biophysical framework for double-drugging kinases. Proc. Natl. Acad. Sci. USA 2023, 120, e2304611120. [Google Scholar] [CrossRef]
- Okamoto, N.; Yagi, K.; Imawaka, S.; Takaoka, M.; Aizawa, F.; Niimura, T.; Goda, M.; Miyata, K.; Kawada, K.; Izawa-Ishizawa, Y.; et al. Asciminib, a novel allosteric inhibitor of BCR-ABL1, shows synergistic effects when used in combination with imatinib with or without drug resistance. Pharmacol. Res. Perspect. 2024, 12, e1214. [Google Scholar] [CrossRef]
- Wylie, A.A.; Schoepfer, J.; Jahnke, W.; Cowan-Jacob, S.W.; Loo, A.; Furet, P.; Marzinzik, A.L.; Pelle, X.; Donovan, J.; Zhu, W.; et al. The allosteric inhibitor ABL001 enables dual targeting of BCR–ABL1. Nature 2017, 543, 733–737. [Google Scholar] [CrossRef]
- El Rashedy, A.A.; Appiah-Kubi, P.; Soliman, M.E.S. A Synergistic Combination Against Chronic Myeloid Leukemia: An Intra-molecular Mechanism of Communication in BCR–ABL1 Resistance. Protein J. 2019, 38, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Gleixner, K.V.; Filik, Y.; Berger, D.; Schewzik, C.; Stefanzl, G.; Sadovnik, I.; Degenfeld-Schonburg, L.; Eisenwort, G.; Schneeweiss-Gleixner, M.; Byrgazov, K.; et al. Asciminib and ponatinib exert synergistic anti-neoplastic effects on CML cells expressing BCR-ABL1 T315I-compound mutations. Am. J. Cancer Res. 2021, 11, 4470–4484. [Google Scholar] [PubMed]
- El Rashedy, A.A.; Olotu, F.A.; Soliman, M.E.S. Dual Drug Targeting of Mutant Bcr-Abl Induces Inactive Conformation: New Strategy for the Treatment of Chronic Myeloid Leukemia and Overcoming Monotherapy Resistance. Chem. Biodivers. 2018, 15, e1700533. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., III; D.arden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Florová, P.; Sklenovský, P.; Banáš, P.; Otyepka, M. Explicit Water Models Affect the Specific Solvation and Dynamics of Unfolded Peptides While the Conformational Behavior and Flexibility of Folded Peptides Remain Intact. J. Chem. Theory Comput. 2010, 6, 3569–3579. [Google Scholar] [CrossRef]
- Amber 2024. Available online: https://ambermd.org/CiteAmber.php (accessed on 13 July 2024).
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Accounts Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Sun, H.; Pan, P.; Zhu, F.; Chang, S.; Xu, L.; Li, Y.; Hou, T. Importance of protein flexibility in molecular recognition: A case study on Type-I1/2 inhibitors of ALK. Phys. Chem. Chem. Phys. 2018, 20, 4851–4863. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Shen, M.; Tian, S.; Xu, L.; Pan, P.; Guan, Y.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys. Chem. Chem. Phys. 2014, 16, 22035–22045. [Google Scholar] [CrossRef]
- Federico, L.B.; Silva, G.M.; Gomes, S.Q.; Francischini, I.A.G.; Barcelos, M.P.; dos Santos, C.B.R.; Costa, L.T.; Rosa, J.M.C.; Silva, C.H.T.d.P.d. Potential colchicine binding site inhibitors unraveled by virtual screening, molecular dynamics and MM/PBSA. Comput. Biol. Med. 2021, 137, 104817. [Google Scholar] [CrossRef]
- Seeber, M.; Felline, A.; Raimondi, F.; Mariani, S.; Fanelli, F. WebPSN: A web server for high-throughput investigation of structural communication in biomacromolecules. Bioinformatics 2014, 31, 779–781. [Google Scholar] [CrossRef]
- Felline, A.; Seeber, M.; Fanelli, F. PSNtools for standalone and web-based structure network analyses of conformational ensembles. Comput. Struct. Biotechnol. J. 2022, 20, 640–649. [Google Scholar] [CrossRef]
- Sun, D.; Wang, Z.; Zheng, Q.; Zhang, H. Exploring the inhibition mechanism on HIF-2 by inhibitor PT2399 and 0X3 using molecular dynamics simulations. J. Mol. Recognit. 2018, 31, e2730. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Fukuda, I.; Nakamura, H.; Agarwal, P.K. A Novel Approach of Dynamic Cross Correlation Analysis on Molecular Dynamics Simulations and Its Application to Ets1 Dimer–DNA Complex. PLoS ONE 2014, 9, e112419. [Google Scholar] [CrossRef] [PubMed]
- Curik, N.; Laznicka, A.; Polivkova, V.; Krizkova, J.; Pokorna, E.; Semerak, P.; Suchankova, P.; Burda, P.; Hochhaus, A.; Polakova, K.M. Combination therapies with ponatinib and asciminib in a preclinical model of chronic myeloid leukemia blast crisis with compound mutations. Leukemia 2024, 38, 1415–1418. [Google Scholar] [CrossRef]

| System | ||||||
| ABL1–nilotinib | −71.75 | −34.5284 | 45.075 | −9.1536 | 31.5174 | −38.8396 |
| ABL1–nilotinib–asciminib | −72.3503 | −37.3234 | 47.6184 | −9.4476 | 30.2918 | −41.2111 |
| ABL1–ponatinib | −72.3503 | −23.5641 | 29.3745 | −9.0356 | 29.3277 | −44.5496 |
| ABL1–ponatinib–asciminib | −73.4241 | −30.6382 | 34.2425 | −9.3242 | 27.285 | −51.859 |
| System | ||||||
| ABL1–ascminib | −43.7724 | −24.8216 | 33.4048 | −5.7043 | 22.6313 | −18.2622 |
| ABL1–nilotinib–asciminib | −43.9577 | −22.5531 | 35.7391 | −5.6030 | 22.0603 | −14.3145 |
| ABL1–ponatinib–asciminib | −44.9838 | −19.3029 | 33.4972 | −5.6402 | 21.3559 | −15.0738 |
| Inhibitor | Residues | Percentage Change | ||
| nilotinib | PHE378 | −1.014 | −0.695 | −31% |
| ASP400 | −2.865 | −2.305 | −20% | |
| PHE401 | −1.926 | −1.55 | −20% | |
| ponatinib | LEU267 | −0.844 | −1.575 | 87% |
| VAL275 | −0.847 | −1.573 | 86% | |
| THR334 | −1.256 | −1.76 | 40% | |
| GLU335 | −0.574 | −1.014 | 77% | |
| PHE336 | −1.669 | −0.87 | −48% |
| Value | ABL1–Ponatinib–Asciminib System | ABL1–Ponatinib System |
| Number Of Nodes in MetaPath | 73 | 45 |
| Specific Nodes in MetaPath | 48 (65.75%) | 20 (44.44%) |
| Shared Nodes in MetaPath | 25 (34.25%) | 25 (55.56%) |
| Number Of Links MetaPath | 73 | 45 |
| Specific Links in MetaPath | 53 (72.60%) | 25 (55.56%) |
| Shared Links in MetaPath | 20 (27.40%) | 20 (44.44%) |
| Number of Shortest Paths | 2039 | 2359 |
| Length Of Smallest Path | 3 | 4 |
| Average Path Length | 18.78 | 16.16 |
| Length of Longest Path | 38 | 31 |
| Minimum Path Force | 2.75 | 3.80 |
| Average Path Force | 7.34 | 8.17 |
| Maximum Path Force | 11.17 | 13.62 |
| Minimum Path Correlation | 0.80 | 0.80 |
| Average Path Correlation | 0.89 | 0.88 |
| Maximum Path Correlation | 0.94 | 0.93 |
| Minimum % Of Corr. Nodes | 2.78 | 3.45 |
| Average % Of Corr. Nodes | 9.27 | 10.51 |
| Maximum % Of Corr. Nodes | 100 | 75 |
| Minimum Path Hubs % | 10 | 0 |
| Average Path Hubs % | 42.66 | 38.64 |
| Maximum Path Hubs % | 80 | 75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ming, J.; Gao, H.; Zhan, J. Exploring the Allosteric Pathways of Asciminib in the Dual Inhibition of BCR-ABL1. Biomolecules 2025, 15, 1214. https://doi.org/10.3390/biom15091214
Ming J, Gao H, Zhan J. Exploring the Allosteric Pathways of Asciminib in the Dual Inhibition of BCR-ABL1. Biomolecules. 2025; 15(9):1214. https://doi.org/10.3390/biom15091214
Chicago/Turabian StyleMing, Jie, Hongwei Gao, and Jiuyu Zhan. 2025. "Exploring the Allosteric Pathways of Asciminib in the Dual Inhibition of BCR-ABL1" Biomolecules 15, no. 9: 1214. https://doi.org/10.3390/biom15091214
APA StyleMing, J., Gao, H., & Zhan, J. (2025). Exploring the Allosteric Pathways of Asciminib in the Dual Inhibition of BCR-ABL1. Biomolecules, 15(9), 1214. https://doi.org/10.3390/biom15091214







