The Multifaceted Role of VIRMA, a Core Component of the Methyltransferase Complex, in Cancer and Cancer Therapy
Abstract
1. Introduction
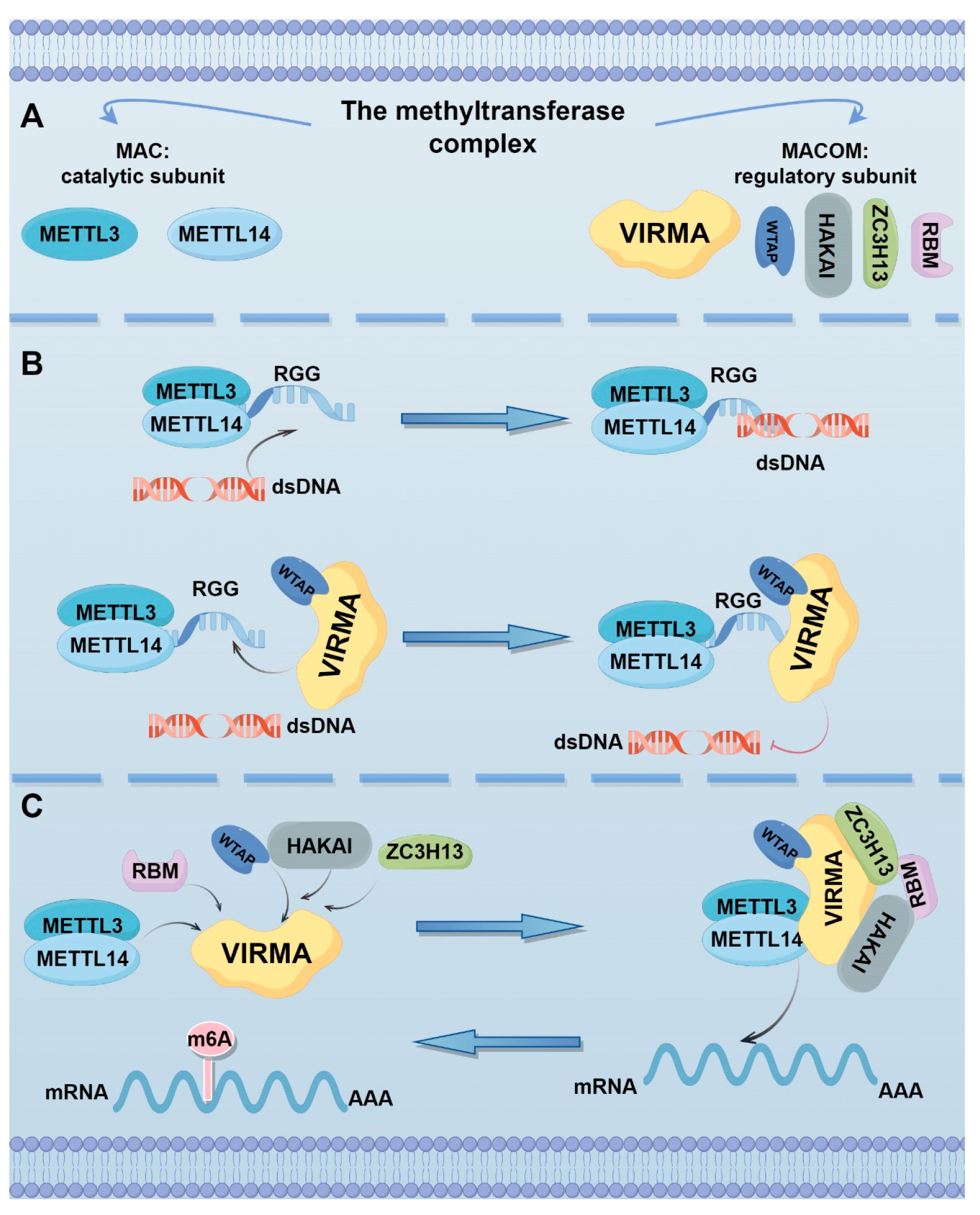
2. Localization, Structure, and Function of VIRMA
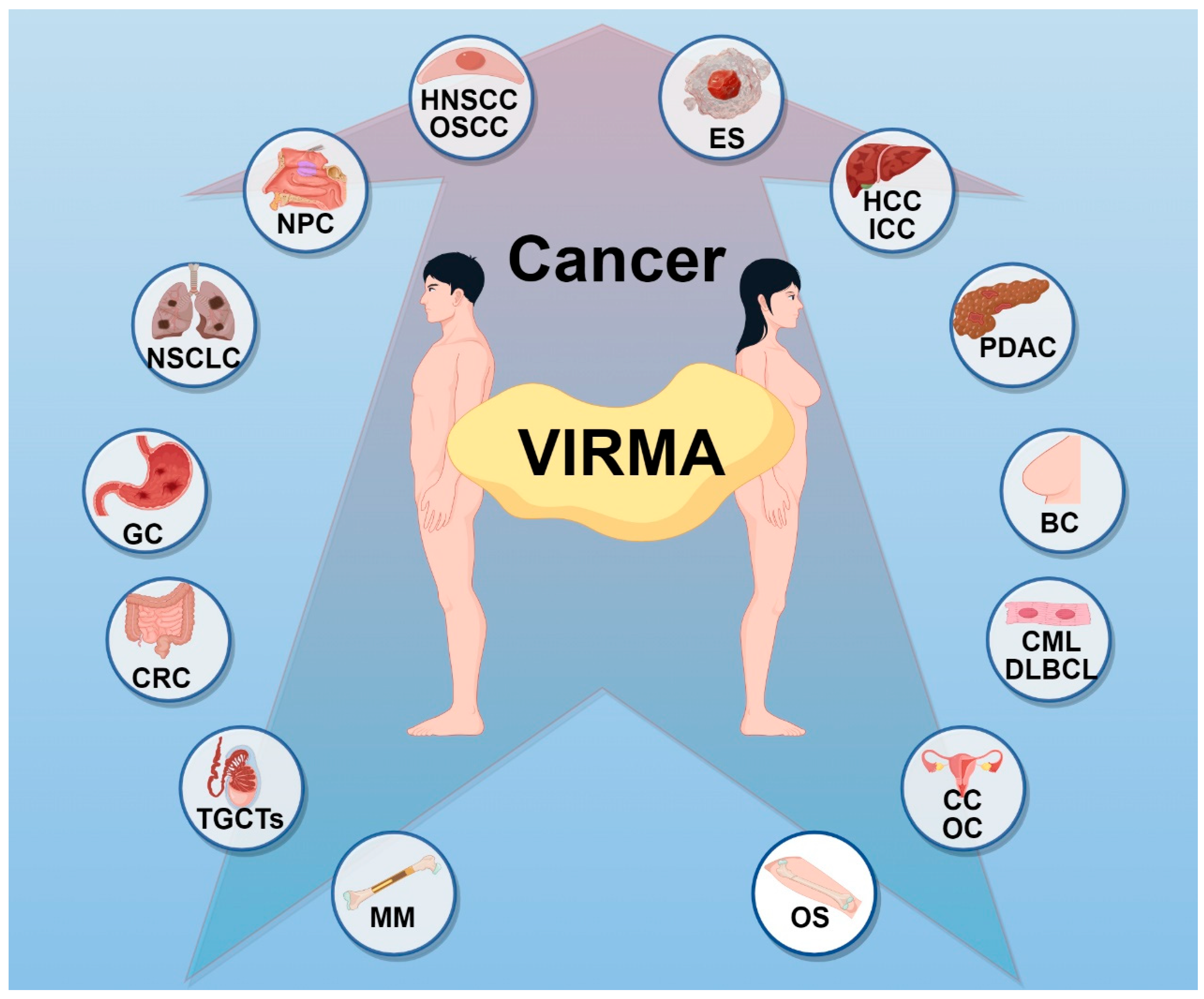
3. The Role of VIRMA in Cancer
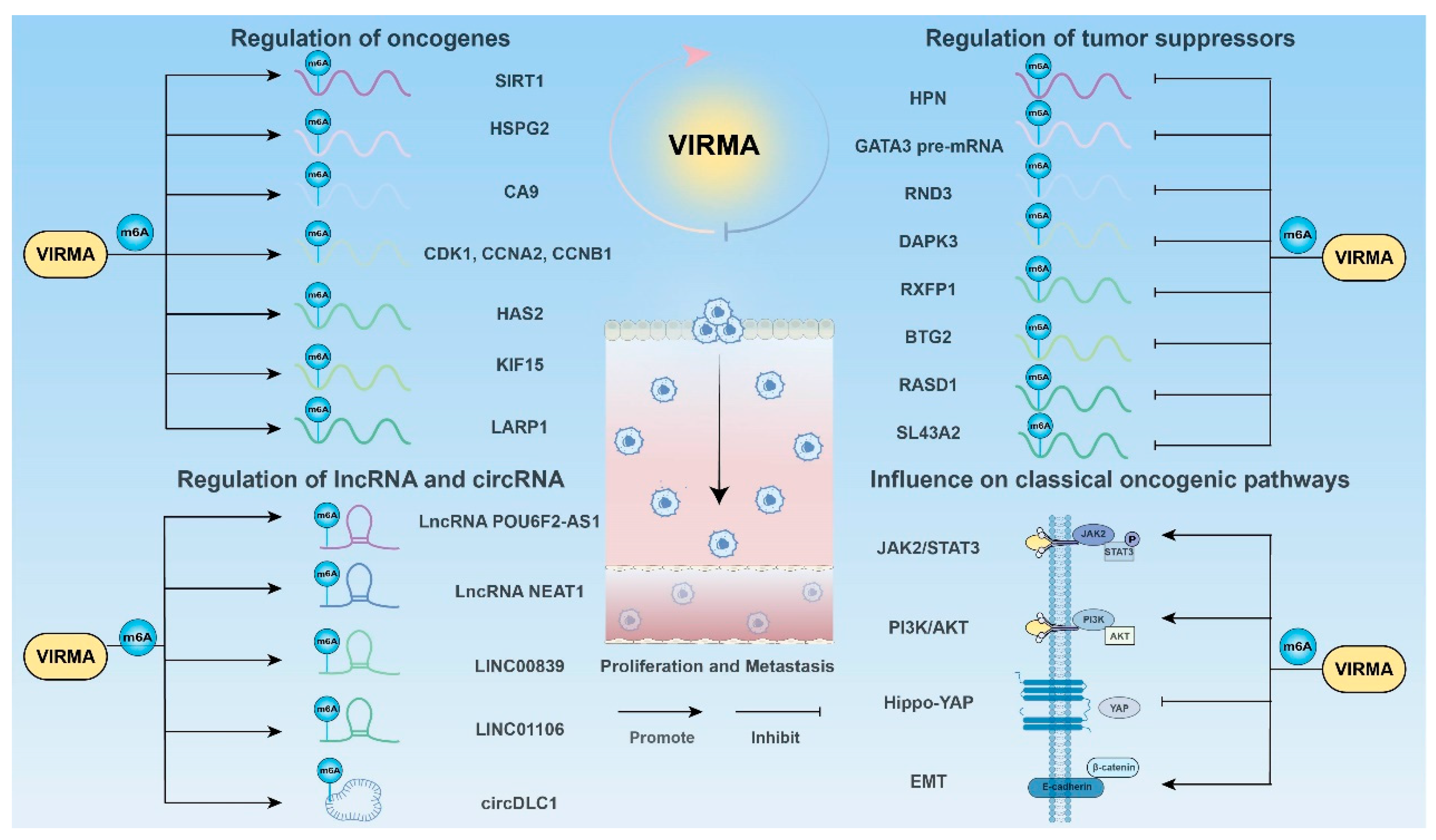
3.1. Promoting Cancer Cell Proliferation and Metastasis
3.1.1. Regulation of Oncogenes
3.1.2. Regulation of Tumor Suppressors
3.1.3. Regulation of lncRNA
3.1.4. Regulation of Circ RNA
3.1.5. Influence on Classical Oncogenic Pathways
3.2. Influencing Cell Cycle
3.3. Affecting Cancer Metabolism
3.4. Mediating Resistance to Ferroptosis
3.5. Mediating Immune Escape
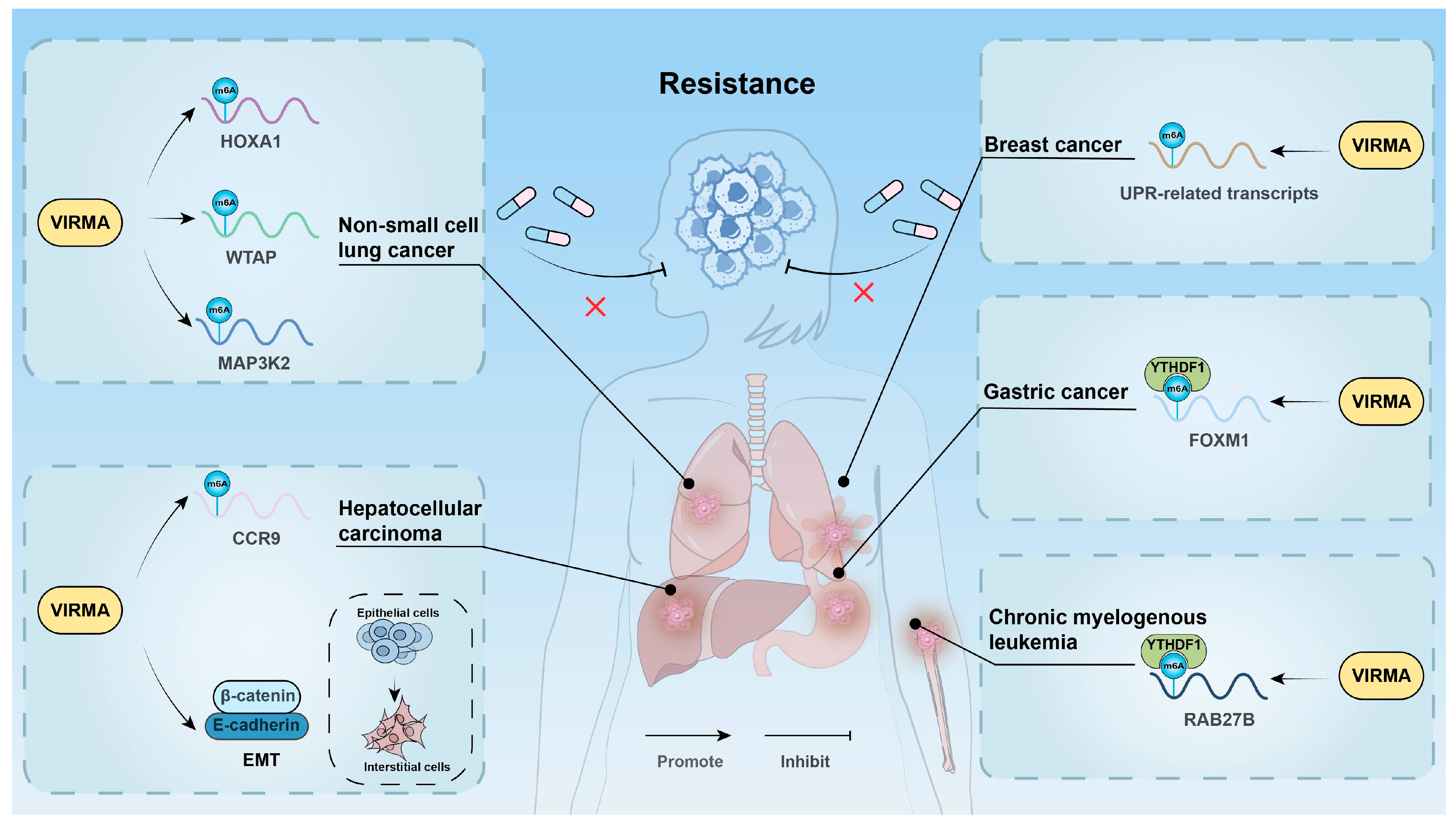
3.6. Affecting Drug Resistance
3.7. Involvement in Tumor Progression Through m6A-Independent Mechanisms
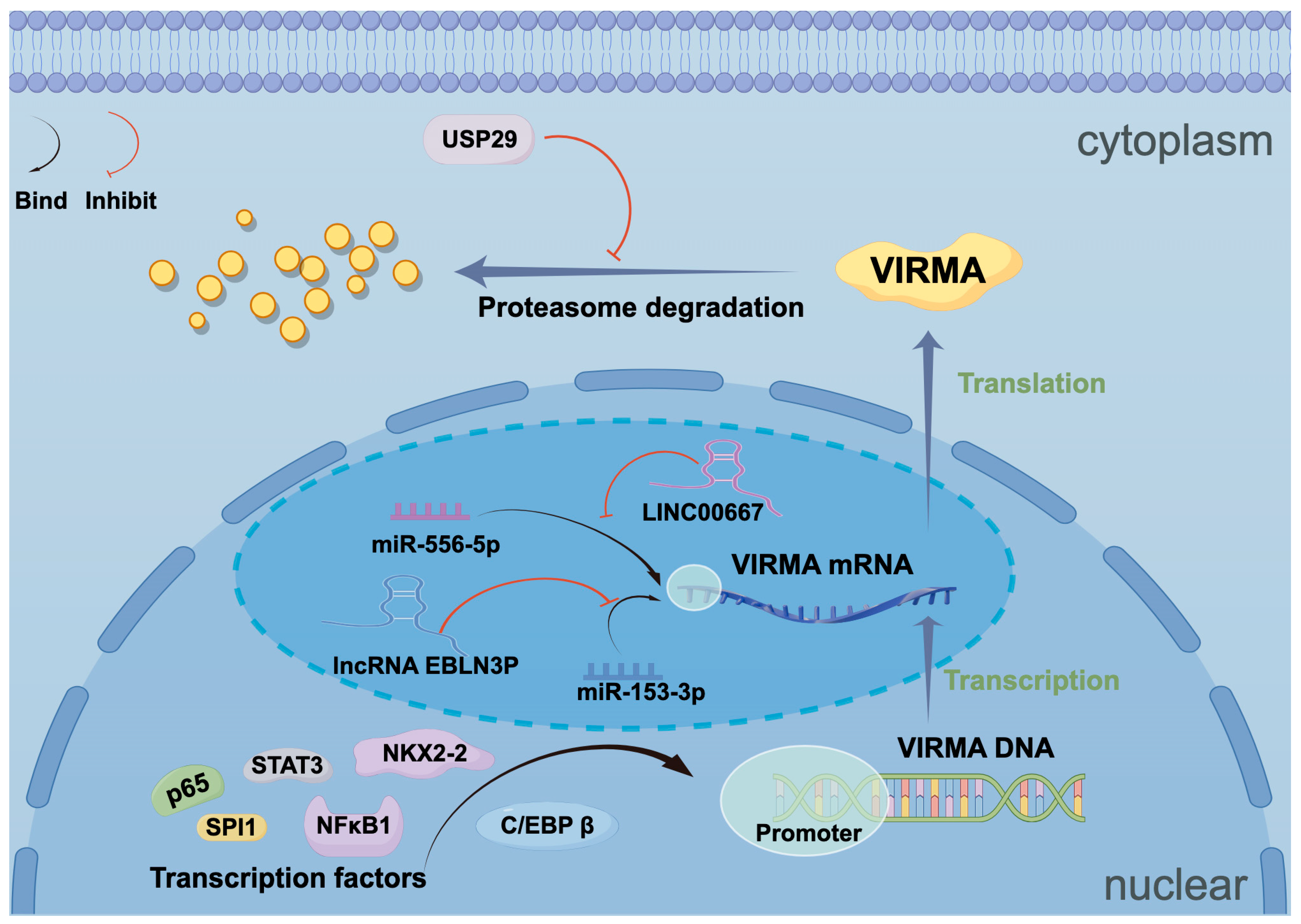
4. Upstream Regulatory Mechanisms of VIRMA
4.1. Transcription Factors
4.2. Non-Coding RNAs
4.3. Ubiquitin-Related Enzymes
5. The Potential of VIRMA in Cancer Diagnosis and Therapy
| Cancer Type | Clinical Significance | Targets | Survival Association | Ref. |
|---|---|---|---|---|
| HNSCC | Oncogene | UBR5 | Not explored | [35] |
| NPC | Oncogene | PTGS2; LINC00839; E2F7 | Worst OS | [32,46,47] |
| OSCC | Oncogene | inhibitor A; PGK1; CA9 | Worst OS | [33,48,49] |
| NSCLC | Oncogene | PXFP1; WTAP; BTG2; DAPK3; MUC3A; ARHGAP30; LINC01106; P53; KLF1; HOXA1; MAP3K2 | Worst OS | [34,50,51,52,53,54,55,56,57,58,59] |
| HCC | Oncogene | RND3; HPN; circDLC1; GATA3 pre-mRNA; ID2; HK1; SLC7A11; CCR9; HSPG2; HBx; Zeb1 | Worst OS | [18,23,60,61,62,63,64,65,66,67,85,122] |
| ICC | Oncogene | CCL3; TMED2P; ARD3B; SIRT1 | Worst OS | [68,69] |
| GC | Oncogene | RASD1; LINC00958; FOXM1; P65; c-Jun | Worst OS | [36,70,71,72,86] |
| PDAC | Oncogene | STRA6; SLC43A2; C/EBP β | WorsetOS | [24,73] |
| CRC | Oncogene | lncRNA POU6F3-AS1; USP29; SOX8; SIRT1; miR-53-3p; lncRNA EBLN3P; HIF-1; NFκB1; WEE1 | Worst OS | [20,37,38,74,75,84] |
| OS | Oncogene | CDK1; CCNA2; CCNB1; JAK-STAT | Worst OS | [40,76] |
| ES | Oncogene | NKX2-2; STAT3 | Not explored | [77] |
| MM | Oncogene | FOXM1 | Worst OS | [39] |
| CML | Oncogene | RAB27B | Not explored | [42] |
| DLBCL | Oncogene | CHST11 | Worst OS | [41] |
| BC | Oncogene | HAS2; KIF15; TFAP2A; DDR1; NEAT1; miR-556-5p; LINC00667; SMC1A; CDK1 | Worse OS | [19,21,78,79,80,81,83] |
| TGCTs | Oncogene | Not explored | Not explored | [43] |
| CC | Oncogene | BTG2; LARP1 | Worst OS | [44,82] |
| OC | Oncogene | SPI1; ENO1 | Worst OS | [45] |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| m6A | N6-methyladenosine |
| VIRMA | Virus-like m6A methyltransferase associated |
| 5′-UTR | 5′ untranslated region |
| 3′-UTR | 3′ untranslated region |
| METTL3 | Methyltransferase-like 3 |
| METTL14 | Methyltransferase-like 14 |
| WTAP | Wilms’ tumor 1-associating protein |
| RBM15 | RNA-binding motif protein 15 |
| BC | Breast cancer |
| PDAC | Pancreatic ductal adenocarcinoma |
| MAC | m6A-METTL complex |
| MACOM | m6A-METTL- associated complex |
| dsDNA | double-stranded DNA |
| M3-M14-W-V | METTL3-METTL14-WTAP-VIRMA quaternary complex |
| RBPs | RNA-binding proteins |
| IF2 | Translation initiation factor 2 |
| HNSCC | Head and neck squamous cell carcinoma |
| NPC | Nasopharyngeal carcinoma |
| OSCC | Oral squamous cell carcinoma |
| NSCLC | Non-small cell lung cancer |
| HCC | Hepatocellular carcinoma |
| ICC | Intrahepatic cholangiocarcinoma |
| GC | Gastric cancer |
| CRC | Colorectal cancer |
| MM | Multiple myeloma |
| OS | Osteosarcoma |
| DLBCL | Diffuse large B-cell lymphoma |
| CML | Chronic myeloid leukemia |
| TGCTs | Testicular germ cell tumors |
| CC | Cervical cancer |
| OC | Ovarian cancer |
| ES | Ewing’s sarcoma |
| CCL3 | Macrophage Inflammatory Protein-1 alpha, MIP-1α |
| HBV | Hepatitis B virus |
| HBx | Hepatitis B virus X protein |
| INHBA | Inhibin A |
| HAS2 | Hyaluronan synthase 2 |
| HPN | Hepsin |
| DAPK3 | Death-Associated Protein Kinase 3 |
| RXFP1 | Relaxin Family Peptide Receptor 1 |
| LUAD | Lung adenocarcinoma |
| LncRNAs | Long non-coding RNAs |
| VIRMA FL | Full-length isoform of VIRMA |
| circRNA | Circular RNA |
| EMT | Epithelial-mesenchymal transition |
| GLUT1 | Glucose transporter-1 |
| HK | Hexokinase |
| ENO1 | Alpha-enolase |
| ECAR | Extracellular acidification rate |
| OCR | Oxygen consumption rate |
| LDHA | Lactate dehydrogenase A |
| GSH | Glutathione |
| ROS | Reactive oxygen species |
| ECM | Extracellular matrix |
| RTK | Receptor tyrosine kinase |
| SHBs | Hepatitis B virus surface small antigen |
| ER | Endoplasmic reticulum |
| UPR | Unfolded protein response |
| IH | Infantile Hemangioma |
| HIF-1 | Hypoxia-inducible factor 1 |
| COAD | Colon adenocarcinoma |
| USPs | Ubiquitin-specific protease |
| GDSC | Cancer drug sensitivity genomics |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Deng, X.; Qing, Y.; Horne, D.; Huang, H.; Chen, J. The roles and implications of RNA m(6)A modification in cancer. Nat. Rev. Clin. Oncol. 2023, 20, 507–526. [Google Scholar] [CrossRef]
- Gilbert, W.V.; Nachtergaele, S. mRNA Regulation by RNA Modifications. Annu. Rev. Biochem. 2023, 92, 175–198. [Google Scholar] [CrossRef] [PubMed]
- Dierks, D.; Garcia-Campos, M.A.; Uzonyi, A.; Safra, M.; Edelheit, S.; Rossi, A.; Sideri, T.; Varier, R.A.; Brandis, A.; Stelzer, Y.; et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 2021, 18, 1060–1067. [Google Scholar] [CrossRef]
- Yue, S.W.; Liu, H.L.; Su, H.F.; Luo, C.; Liang, H.F.; Zhang, B.X.; Zhang, W. m6A-regulated tumor glycolysis: New advances in epigenetics and metabolism. Mol. Cancer 2023, 22, 137. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.B.; Wang, Z.; Yang, C. The m(6)A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 2021, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yin, K.; Zhang, Y.; Tian, J.; Wang, S. The RNA m6A writer METTL14 in cancers: Roles, structures, and applications. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188609. [Google Scholar] [CrossRef]
- Lence, T.; Paolantoni, C.; Worpenberg, L.; Roignant, J.Y. Mechanistic insights into m(6)A RNA enzymes. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 222–229. [Google Scholar] [CrossRef]
- Zhang, X.L.; Chen, X.H.; Xu, B.; Chen, M.; Zhu, S.; Meng, N.; Wang, J.Z.; Zhu, H.; Chen, D.; Liu, J.B.; et al. K235 acetylation couples with PSPC1 to regulate the m(6)A demethylation activity of ALKBH5 and tumorigenesis. Nat. Commun. 2023, 14, 3815. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Wang, T.; Kong, S.; Tao, M.; Ju, S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer 2020, 19, 88. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, C.; Chen, J.; Chen, D.; Yang, B.; He, B.; Hu, W.; Zhang, Y.; Liu, H.; Dai, L.; et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 2019, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, F.; Wang, D.; Yang, Q. Regulation of ULK1 by WTAP/IGF2BP3 axis enhances mitophagy and progression in epithelial ovarian cancer. Cell Death Dis. 2024, 15, 97. [Google Scholar] [CrossRef]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef]
- Yan, X.; Liu, F.; Yan, J.; Hou, M.; Sun, M.; Zhang, D.; Gong, Z.; Dong, X.; Tang, C.; Yin, P. WTAP-VIRMA counteracts dsDNA binding of the m(6)A writer METTL3-METTL14 complex and maintains N(6)-adenosine methylation activity. Cell Discov. 2023, 9, 100. [Google Scholar] [CrossRef]
- Shan, M.; Liu, D.; Sun, L.; Yang, M.; He, M.; Zhang, Y.; Xiang, L.; Lu, L.; He, H.; Niu, D.; et al. KIAA1429 facilitates metastasis via m6A-YTHDC1-dependent RND3 down-regulation in hepatocellular carcinoma cells. Cancer Lett. 2024, 584, 216598. [Google Scholar] [CrossRef]
- Li, N.; Zhu, Z.; Deng, Y.; Tang, R.; Hui, H.; Kang, Y.; Rana, T.M. KIAA1429/VIRMA promotes breast cancer progression by m(6) A-dependent cytosolic HAS2 stabilization. EMBO Rep. 2023, 24, e55506. [Google Scholar] [CrossRef]
- Zhou, Y.; Pei, Z.; Maimaiti, A.; Zheng, L.; Zhu, Z.; Tian, M.; Zhou, Z.; Tan, F.; Pei, Q.; Li, Y.; et al. m(6)A methyltransferase KIAA1429 acts as an oncogenic factor in colorectal cancer by regulating SIRT1 in an m(6)A-dependent manner. Cell Death Discov. 2022, 8, 83. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, X.Y.; Qian, J.Y.; Xu, F.; Wang, Z.W.; Xia, T.; Zhou, X.J.; Li, X.X.; Shi, L.; Wei, J.F.; et al. SMC1A regulated by KIAA1429 in m6A-independent manner promotes EMT progress in breast cancer. Mol. Ther. Nucleic Acids 2022, 27, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Guo, J.; Jia, R. N6-Methyladenosine Methyltransferase Component KIAA1429 Is a Potential Target of Cancer Therapy. Biomolecules 2024, 14, 1319. [Google Scholar] [CrossRef]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhong, Z.; Zou, J.; Liao, J.Y.; Chen, S.; Zhou, S.; Zhao, Y.; Li, J.; Yin, D.; Huang, K.; et al. Glycolysis and tumor progression promoted by the m(6)A writer VIRMA via m(6)A-dependent upregulation of STRA6 in pancreatic ductal adenocarcinoma. Cancer Lett. 2024, 590, 216840. [Google Scholar] [CrossRef] [PubMed]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes. Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef]
- Su, S.; Li, S.; Deng, T.; Gao, M.; Yin, Y.; Wu, B.; Peng, C.; Liu, J.; Ma, J.; Zhang, K. Cryo-EM structures of human m(6)A writer complexes. Cell Res. 2022, 32, 982–994. [Google Scholar] [CrossRef]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, D.; Sun, S.; Ren, M.; Zhou, L.; Chen, C.; Zhao, J.; Wei, H.; Zhao, Q.; Qi, Y.; et al. RBMS1 Coordinates with the m(6)A Reader YTHDF1 to Promote NSCLC Metastasis through Stimulating S100P Translation. Adv. Sci. 2024, 11, e2307122. [Google Scholar] [CrossRef]
- Niessen, M.; Schneiter, R.; Nothiger, R. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics 2001, 157, 679–688. [Google Scholar] [CrossRef]
- Destefanis, E.; Sighel, D.; Dalfovo, D.; Gilmozzi, R.; Broso, F.; Cappannini, A.; Bujnicki, J.M.; Romanel, A.; Dassi, E.; Quattrone, A. The three YTHDF paralogs and VIRMA are strong cross-histotype tumor driver candidates among m(6)A core genes. NAR Cancer 2024, 6, zcae040. [Google Scholar] [CrossRef]
- Zheng, Z.Q.; Huang, Z.H.; Liang, Y.L.; Zheng, W.H.; Xu, C.; Li, Z.X.; Liu, N.; Yang, P.Y.; Li, Y.Q.; Ma, J.; et al. VIRMA promotes nasopharyngeal carcinoma, tumorigenesis, and metastasis by upregulation of E2F7 in an m6A-dependent manner. J. Biol. Chem. 2023, 299, 104677. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Dai, X.; Yue, J. m(6)A methyltransferase KIAA1429 accelerates oral squamous cell carcinoma via regulating glycolysis and ferroptosis. Transl. Oncol. 2023, 36, 101745. [Google Scholar] [CrossRef]
- Lin, X.; Ye, R.; Li, Z.; Zhang, B.; Huang, Y.; Du, J.; Wang, B.; Meng, H.; Xian, H.; Yang, X.; et al. KIAA1429 promotes tumorigenesis and gefitinib resistance in lung adenocarcinoma by activating the JNK/MAPK pathway in an m(6)A-dependent manner. Drug Resist. Updat. 2023, 66, 100908. [Google Scholar] [CrossRef]
- Zhu, C.; Cheng, Y.; Yu, Y.; Zhang, Y.; Ren, G. VIRMA promotes the progression of head and neck squamous cell carcinoma by regulating UBR5 mRNA and m6A levels. Biomol. Biomed. 2024, 24, 1244–1257. [Google Scholar] [CrossRef]
- Ren, M.; Pan, H.; Zhou, X.; Yu, M.; Ji, F. KIAA1429 promotes gastric cancer progression by destabilizing RASD1 mRNA in an m(6)A-YTHDF2-dependent manner. J. Transl. Med. 2024, 22, 584. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, C.; Xu, G.; Xie, J.; Xie, Y.; Zhou, Y. Silencing of KIAA1429, a N6-methyladenine methyltransferase, inhibits the progression of colon adenocarcinoma via blocking the hypoxia-inducible factor 1 signalling pathway. J. Biochem. Mol. Toxicol. 2024, 38, e23829. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, J.; Chen, Z.; Liu, L.; Wang, H.; Deng, Q.; Chen, Y.; Que, Y.; Fu, Z. Promotive role of USP29-mediated deubiquitination in malignant proliferation of colorectal cancer cells via the KIAA1429/SOX8 axis. Biomol. Biomed. 2023, 23, 483–495. [Google Scholar] [CrossRef]
- Wu, Y.; Luo, Y.; Yao, X.; Shi, X.; Xu, Z.; Re, J.; Shi, M.; Li, M.; Liu, J.; He, Y.; et al. KIAA1429 increases FOXM1 expression through YTHDF1-mediated m6A modification to promote aerobic glycolysis and tumorigenesis in multiple myeloma. Cell Biol. Toxicol. 2024, 40, 58. [Google Scholar] [CrossRef]
- Luo, J.; Wang, X.; Chen, Z.; Zhou, H.; Xiao, Y. The role and mechanism of JAK2/STAT3 signaling pathway regulated by m6A methyltransferase KIAA1429 in osteosarcoma. J. Bone Oncol. 2023, 39, 100471. [Google Scholar] [CrossRef]
- Chen, X.; Lu, T.; Cai, Y.; Han, Y.; Ding, M.; Chu, Y.; Zhou, X.; Wang, X. KIAA1429-mediated m6A modification of CHST11 promotes progression of diffuse large B-cell lymphoma by regulating Hippo-YAP pathway. Cell Mol. Biol. Lett. 2023, 28, 32. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Zhong, F.; Jiang, J.; Cheng, Y.; Xu, S.; Liu, J.; Lin, J.; Zhang, J.; Li, S.; Li, M.; et al. The m(6)A regulator KIAA1429 stabilizes RAB27B mRNA and promotes the progression of chronic myeloid leukemia and resistance to targeted therapy. Genes Dis. 2024, 11, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Gonçalves, V.; Lobo, J.; Guimarães-Teixeira, C.; Barros-Silva, D.; Guimarães, R.; Cantante, M.; Braga, I.; Maurício, J.; Oing, C.; Honecker, F.; et al. The component of the m(6)A writer complex VIRMA is implicated in aggressive tumor phenotype, DNA damage response and cisplatin resistance in germ cell tumors. J. Exp. Clin. Cancer Res. 2021, 40, 268. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Z.; Zou, X.; Yang, D.; Xu, K. The regulatory role of KIAA1429 in epithelial-mesenchymal transition in cervical cancer via mediating m6A modification of BTG2. Cytotechnology 2025, 77, 34. [Google Scholar] [CrossRef]
- Gan, L.; Zhao, S.; Gao, Y.; Qi, Y.; Su, M.; Wang, A.; Cai, H. N6-methyladenosine methyltransferase KIAA1429 promoted ovarian cancer aerobic glycolysis and progression through enhancing ENO1 expression. Biol. Direct 2023, 18, 64. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, Y.; Fu, J. KIAA1429 Promotes Nasopharyngeal Carcinoma Progression by Mediating m6A Modification of PTGS2. Crit. Rev. Immunol. 2023, 43, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.H.; Long, Z.Q.; Zheng, Z.Q.; Zhang, L.L.; Liang, Y.L.; Li, Z.X.; Lv, J.W.; Kou, J.; Hong, X.H.; He, S.W.; et al. m6A-enriched lncRNA LINC00839 promotes tumor progression by enhancing TAF15-mediated transcription of amine oxidase AOC1 in nasopharyngeal carcinoma. J. Biol. Chem. 2023, 299, 104873. [Google Scholar] [CrossRef]
- Chen, X.; Fan, R. Inhibin A contributes to the tumorigenesis of oral squamous cell carcinoma by KIAA1429-mediated m6A modification. J. Oral. Pathol. Med. 2024, 53, 266–274. [Google Scholar] [CrossRef]
- Tu, J.; Feng, X.; Cao, Q.; Guan, Y. KIAA1429 promotes the malignancy of oral squamous cell carcinoma by regulating CA9 m6A methylation. Cytotechnology 2024, 76, 585–594. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, J.; Gong, C.; Wu, S.; Sun, Y. KIAA1429-mediated RXFP1 attenuates non-small cell lung cancer tumorigenesis via N6-methyladenosine modification. Cancer Biomark. 2024. Online ahead of print. [Google Scholar] [CrossRef]
- Ma, B.; Xiu, L.; Ding, L. The m6 RNA methylation regulator KIAA1429 is associated with autophagy-mediated drug resistance in lung cancer. FASEB Bioadv 2024, 6, 105–117. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, Q.; Zhang, X.; Qin, N.; Pu, Z.; Gu, Y.; Yan, C.; Zhu, M.; Dai, J.; Wang, C.; et al. Gene amplification-driven RNA methyltransferase KIAA1429 promotes tumorigenesis by regulating BTG2 via m6A-YTHDF2-dependent in lung adenocarcinoma. Cancer Commun. 2022, 42, 609–626. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, Y.; Yao, Y.; Xie, H.; Lu, G.; Du, C.; Cheng, J.; Zhou, J. VIRMA contributes to non-small cell lung cancer progression via N(6)-methyladenosine-dependent DAPK3 post-transcriptional modification. Cancer Lett. 2021, 522, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Xie, Y. KIAA1429 promotes the progression of lung adenocarcinoma by regulating the m6A level of MUC3A. Pathol. Res. Pract. 2021, 217, 153284. [Google Scholar] [CrossRef]
- Guo, W.; Wang, T.; Huai, Q.; Guo, L.; Wang, X.; He, J. KIAA1429 regulates lung adenocarcinoma proliferation and metastasis through the PI3K/AKT pathway by modulating ARHGAP30 expression. Thorac. Cancer 2024, 15, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, Z.; Li, F. KIAA1429 Induces m6A Modification of LINC01106 to Enhance the Malignancy of Lung Adenocarcinoma Cells via the JAK/STAT3 Pathway. Crit. Rev. Immunol. 2024, 44, 49–61. [Google Scholar] [CrossRef]
- Wu, Y.; Li, H.; Huang, Y.; Chen, Q. Silencing of m(6)A methyltransferase KIAA1429 suppresses the progression of non-small cell lung cancer by promoting the p53 signaling pathway and ferroptosis. Am. J. Cancer Res. 2023, 13, 5320–5333. [Google Scholar] [PubMed]
- Geng, R.; Ren, M.; Ma, Y.; Su, W. Mechanism of the KIAA1429/KLF1/PD-L1 Axis in Regulating Immune Escape in Non-small Cell Lung Cancer. Cell Biochem. Biophys. 2024, 82, 1835–1845. [Google Scholar] [CrossRef]
- Tang, J.; Han, T.; Tong, W.; Zhao, J.; Wang, W. N(6)-methyladenosine (m(6)A) methyltransferase KIAA1429 accelerates the gefitinib resistance of non-small-cell lung cancer. Cell Death Discov. 2021, 7, 108. [Google Scholar] [CrossRef]
- Meng, Y.; Yang, W.; Li, J.; Chai, W. KIAA1429 facilitates progression of hepatocellular carcinoma by modulating m6A levels in HPN. Heliyon 2023, 9, e22084. [Google Scholar] [CrossRef]
- Liu, H.; Lan, T.; Li, H.; Xu, L.; Chen, X.; Liao, H.; Chen, X.; Du, J.; Cai, Y.; Wang, J.; et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics 2021, 11, 1396–1411. [Google Scholar] [CrossRef]
- Cheng, X.; Li, M.; Rao, X.; Zhang, W.; Li, X.; Wang, L.; Huang, G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther. 2019, 12, 3421–3428. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shan, M.; Zeng, R.; He, M.; Dai, X.; Lu, L.; Yang, M.; He, H.; Zhang, Y.; Xiang, L.; et al. Inhibition of KIAA1429/HK1 axis enhances the sensitivity of liver cancer cells to sorafenib by regulating the Warburg effect. Biochem. Pharmacol. 2024, 227, 116419. [Google Scholar] [CrossRef]
- Wang, H.; Chen, W.; Cui, Y.; Gong, H.; Li, H. KIAA1429 protects hepatocellular carcinoma cells from ferroptotic cell death with a m(6) A-dependent posttranscriptional modification of SLC7A11. J. Cell Mol. Med. 2023, 27, 4118–4132. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Liu, L.; You, J.; Zhou, P.; Su, Y.; Zhao, K.; Zhang, J.; Zhu, F. Small HBV surface antigen drives regorafenib resistance in HCC via KIAA1429-dependent m6A modification of CCR9. J. Med. Virol. 2024, 96, e29894. [Google Scholar] [CrossRef]
- Sivasudhan, E.; Zhou, J.; Ma, J.; Wang, Y.; Liu, S.; Khan, F.I.; Lu, Z.; Meng, J.; Blake, N.; Rong, R. Hepatitis B Virus X Protein Contributes to Hepatocellular Carcinoma via Upregulation of KIAA1429 Methyltransferase and mRNA m6A Hypermethylation of HSPG2/Perlecan. Mol. Carcinog. 2025, 64, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Cheng, Y.; Wang, J.; Li, H.; Cao, X.; Wang, Y. KIAA1429 mediates epithelial mesenchymal transition in sorafenib-resistant hepatocellular carcinoma through m6A methylation modification. Cancer Med. 2023, 12, 7222–7233. [Google Scholar] [CrossRef]
- Xu, H.; Lin, X.; Li, Z.; He, X.; Li, Y.; Qiu, L.; Lu, L.; Liu, B.; Zhan, M.; He, K. VIRMA facilitates intrahepatic cholangiocarcinoma progression through epigenetic augmentation of TMED2 and PARD3B mRNA stabilization. J. Gastroenterol. 2023, 58, 925–944. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, K.; Chen, S.; Lian, G.; Huang, Y.; Yao, H.; Zhao, Y.; Huang, K.; Yin, D.; Lin, H.; et al. CCL3 secreted by hepatocytes promotes the metastasis of intrahepatic cholangiocarcinoma by VIRMA-mediated N6-methyladenosine (m(6)A) modification. J. Transl. Med. 2023, 21, 43. [Google Scholar] [CrossRef]
- Yang, D.; Chang, S.; Li, F.; Ma, M.; Yang, J.; Lv, X.; Huangfu, L.; Jia, C. m(6) A transferase KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic glycolysis through targeting GLUT1. IUBMB Life 2021, 73, 1325–1333. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhou, Y.; Chen, Y.; Zhou, Z.; Liu, W.; Zheng, L.; Pei, Q.; Tan, F.; Pei, H.; Li, Y. m(6)A Methyltransferase KIAA1429 Regulates the Cisplatin Sensitivity of Gastric Cancer Cells via Stabilizing FOXM1 mRNA. Cancers 2022, 14, 5025. [Google Scholar] [CrossRef]
- Tang, B.; Li, M.; Xu, Y.; Li, X. N(6)-methyladenosine (m(6)A) writer KIAA1429 accelerates gastric cancer oxaliplatin chemoresistance by targeting FOXM1. J. Cancer Res. Clin. Oncol. 2023, 149, 5037–5045. [Google Scholar] [CrossRef]
- Gao, L.; Lv, G.; Liu, Z.; Tian, Y.; Han, F.; Li, L.; Wang, G.; Zhang, Y. Alcohol-induced C/EBP β-driven VIRMA decreases oxidative stress and promotes pancreatic ductal adenocarcinoma growth and metastasis via the m6A/YTHDF2/SLC43A2 pathway. Oncogene 2025, 44, 1118–1132. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Chen, A. lncRNA POU6F2-AS1 Regulated by KIAA1429 Contributes to Colorectal Cancer Progression in an m(6)A Modification Manner. Mol. Biotechnol. 2025, 67, 115–122. [Google Scholar] [CrossRef]
- Chen, H.; Zhu, P.; Zhu, D.; Jin, J.; Yang, Q.; Han, X. Role and mechanism of KIAA1429 in regulating cellular ferroptosis and radioresistance in colorectal cancer. Biomol. Biomed. 2024, 24, 1669–1681. [Google Scholar] [CrossRef]
- Sun, Y.; Lei, Y.W.; Zeng, J.X.; Zhong, L.Y.; Liu, J.W.; Man, Y.N.; He, M.L. Clinical Significance and Potential Mechanisms of the RNA Methyltransferase KIAA1429 in Osteosarcoma. J. Cancer 2024, 15, 126–139. [Google Scholar] [CrossRef]
- Tan, K.; Lu, W.; Chen, F.; Shi, H.; Ma, Y.; Chen, Z.; Wu, W.; Lv, Z.; Mo, J. CRISPR-Cas9 knockout screening identifies KIAA1429 as an essential gene in Ewing sarcoma. J. Exp. Clin. Cancer Res. 2023, 42, 250. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Y.; Li, Y.; Zhang, C. VIRMA Facilitates Triple-Negative Breast Cancer Progression via Increasing m6A-Dependent KIF15 Expression. Discov. Med. 2023, 35, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Lian, B.; Yan, S.; Li, J.; Bai, Z.; Li, J. HNRNPC promotes collagen fiber alignment and immune evasion in breast cancer via activation of the VIRMA-mediated TFAP2A/DDR1 axis. Mol. Med. 2023, 29, 103. [Google Scholar] [CrossRef]
- Lee, Q.; Song, R.; Phan, D.A.V.; Pinello, N.; Tieng, J.; Su, A.; Halstead, J.M.; Wong, A.C.H.; van Geldermalsen, M.; Lee, B.S.; et al. Overexpression of VIRMA confers vulnerability to breast cancers via the m(6)A-dependent regulation of unfolded protein response. Cell Mol. Life Sci. 2023, 80, 157. [Google Scholar] [CrossRef]
- Ren, S.; Zhang, Y.; Yang, X.; Li, X.; Zheng, Y.; Liu, Y.; Zhang, X. N6-methyladenine- induced LINC00667 promoted breast cancer progression through m6A/KIAA1429 positive feedback loop. Bioengineered 2022, 13, 13462–13473. [Google Scholar] [CrossRef]
- Feng, X.; Shu, L. The methyltransferase KIAA1429 potentiates cervical cancer tumorigenesis via modulating LARP1 mRNA m6A modification and stability. Histol. Histopathol. 2024, 18843. [Google Scholar] [CrossRef]
- Qian, J.Y.; Gao, J.; Sun, X.; Cao, M.D.; Shi, L.; Xia, T.S.; Zhou, W.B.; Wang, S.; Ding, Q.; Wei, J.F. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 2019, 38, 6123–6141. [Google Scholar] [CrossRef]
- Ma, L.; Lin, Y.; Sun, S.W.; Xu, J.; Yu, T.; Chen, W.L.; Zhang, L.H.; Guo, Y.C.; Wang, Y.W.; Chen, T.; et al. KIAA1429 is a potential prognostic marker in colorectal cancer by promoting the proliferation via downregulating WEE1 expression in an m6A-independent manner. Oncogene 2022, 41, 692–703. [Google Scholar] [CrossRef]
- Wang, M.; Yang, Y.; Yang, J.; Yang, J.; Han, S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 2020, 257, 118082. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Dai, C.C.; Mei, L.; Xu, J.; Sun, S.W.; Xing, Y.L.; Wu, L.S.; Wang, M.H.; Wei, J.F. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J. Cell Physiol. 2020, 235, 7420–7432. [Google Scholar] [CrossRef]
- Gerstberger, S.; Jiang, Q.; Ganesh, K. Metastasis. Cell 2023, 186, 1564–1579. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 8421. [Google Scholar] [CrossRef]
- Singh, P.; Jenkins, L.M.; Horst, B.; Alers, V.; Pradhan, S.; Kaur, P.; Srivastava, T.; Hempel, N.; Győrffy, B.; Broude, E.V.; et al. Inhibin Is a Novel Paracrine Factor for Tumor Angiogenesis and Metastasis. Cancer Res. 2018, 78, 2978–2989. [Google Scholar] [CrossRef]
- Du, F.; Yuan, P.; Wang, T.; Zhao, J.; Zhao, Z.; Luo, Y.; Xu, B. The Significance and Therapeutic Potential of GATA3 Expression and Mutation in Breast Cancer: A Systematic Review. Med. Res. Rev. 2015, 35, 1300–1315. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef] [PubMed]
- Knutsen, E.; Harris, A.L.; Perander, M. Expression and functions of long non-coding RNA NEAT1 and isoforms in breast cancer. Br. J. Cancer 2022, 126, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.T.; Wang, Y.; Ji, F.; Chen, J.N.; Wang, T.J.; Liu, Y.; Hou, M.X.; Guo, Z.G. YTHDF1’s grip on CRC vasculature: Insights into LINC01106 and miR-449b-5p-VEGFA axis. Cancer Cell Int. 2024, 24, 195. [Google Scholar] [CrossRef]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Jin, W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial-Mesenchymal Transition. Cells 2020, 9, 217. [Google Scholar] [CrossRef]
- Riggi, N.; Suvà, M.L.; Stamenkovic, I. Ewing’s Sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef]
- Liu, J.; Gu, X.; Guan, Z.; Huang, D.; Xing, H.; Zheng, L. Role of m6A modification in regulating the PI3K/AKT signaling pathway in cancer. J. Transl. Med. 2023, 21, 774. [Google Scholar] [CrossRef]
- Lu, H.; Cao, L.L.; Ballout, F.; Belkhiri, A.; Peng, D.; Chen, L.; Chen, Z.; Soutto, M.; Wang, T.C.; Que, J.; et al. Reflux conditions induce E-cadherin cleavage and EMT via APE1 redox function in oesophageal adenocarcinoma. Gut 2023, 73, 47–62. [Google Scholar] [CrossRef]
- Williams, G.H.; Stoeber, K. The cell cycle and cancer. J. Pathol. 2012, 226, 352–364. [Google Scholar] [CrossRef]
- You, Q.; Wang, J.; Yu, Y.; Li, F.; Meng, L.; Chen, M.; Yang, Q.; Xu, Z.; Sun, J.; Zhuo, W.; et al. The histone deacetylase SIRT6 promotes glycolysis through the HIF-1α/HK2 signaling axis and induces erlotinib resistance in non-small cell lung cancer. Apoptosis 2022, 27, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yongzhi, H.; Chen, S.; Luo, X.; Lin, Y.; Zhou, Y.; Jin, H.; Hou, B.; Deng, Y.; Tu, L.; et al. The prognostic value of GLUT1 in cancers: A systematic review and meta-analysis. Oncotarget 2017, 8, 43356–43367. [Google Scholar] [CrossRef]
- Guo, D.; Tong, Y.; Jiang, X.; Meng, Y.; Jiang, H.; Du, L.; Wu, Q.; Li, S.; Luo, S.; Li, M.; et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab. 2022, 34, 1312–1324.e6. [Google Scholar] [CrossRef]
- Li, Y.; He, L.; Wang, Y.; Tan, Y.; Zhang, F. N(6)-methyladenosine methyltransferase KIAA1429 elevates colorectal cancer aerobic glycolysis via HK2-dependent manner. Bioengineered 2022, 13, 11923–11932. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Lei, G.; Zhuang, L.; Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 2022, 22, 381–396. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Roh, J.L. Epigenetic modulation of ferroptosis in cancer: Identifying epigenetic targets for novel anticancer therapy. Cell Oncol. 2023, 46, 1605–1623. [Google Scholar] [CrossRef]
- Lee, J.; Roh, J.L. SLC7A11 as a Gateway of Metabolic Perturbation and Ferroptosis Vulnerability in Cancer. Antioxidants 2022, 11, 2444. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.; Ren, Z.; Li, Y.; Zou, W.; Chen, J.; Wang, H. Overcoming cancer chemotherapy resistance by the induction of ferroptosis. Drug Resist. Updat. 2023, 66, 100916. [Google Scholar] [CrossRef]
- Tharp, K.M.; Kersten, K.; Maller, O.; Timblin, G.A.; Stashko, C.; Canale, F.P.; Menjivar, R.E.; Hayward, M.K.; Berestjuk, I.; Ten Hoeve, J.; et al. Tumor-associated macrophages restrict CD8(+) T cell function through collagen deposition and metabolic reprogramming of the breast cancer microenvironment. Nat. Cancer 2024, 5, 1045–1062. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Ojha, R.; Amaravadi, R.K. Targeting the unfolded protein response in cancer. Pharmacol. Res. 2017, 120, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Iurlaro, R.; Muñoz-Pinedo, C. Cell death induced by endoplasmic reticulum stress. Febs J. 2016, 283, 2640–2652. [Google Scholar] [CrossRef]
- Huang, L.; Nakayama, H.; Klagsbrun, M.; Mulliken, J.B.; Bischoff, J. Glucose transporter 1-positive endothelial cells in infantile hemangioma exhibit features of facultative stem cells. Stem Cells 2015, 33, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zou, Y.; Huang, Z.; Wang, W.; Li, J.; Bi, J.; Huo, R. KIAA1429 promotes infantile hemangioma regression by facilitating the stemness of hemangioma endothelial cells. Cancer Sci. 2023, 114, 1569–1581. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Li, W.; Wang, Y.; Zhang, Z.; Zhang, L.; Zhang, W.; Xing, Y.; Zhou, C. Inhibition of CDK1 Overcomes Oxaliplatin Resistance by Regulating ACSL4-mediated Ferroptosis in Colorectal Cancer. Adv. Sci. 2023, 10, e2301088. [Google Scholar] [CrossRef]
- Li, Y.; Ren, B.X.; Li, H.M.; Lu, T.; Fu, R.; Wu, Z.Q. Omeprazole suppresses aggressive cancer growth and metastasis in mice through promoting Snail degradation. Acta Pharmacol. Sin. 2022, 43, 1816–1828. [Google Scholar] [CrossRef]
- Patel, M.; Horgan, P.G.; McMillan, D.C.; Edwards, J. NF-κB pathways in the development and progression of colorectal cancer. Transl. Res. 2018, 197, 43–56. [Google Scholar] [CrossRef]
- Zhang, C.P.; Huang, X.Y. Circular RNA circ_KIAA1429 accelerates hepatocellular carcinoma progression via the miR-133a-3p/high mobility group AT-hook 2 (HMGA2) axis in an m6A-dependent manner. Hum. Cell 2023, 36, 1741–1754. [Google Scholar] [CrossRef] [PubMed]
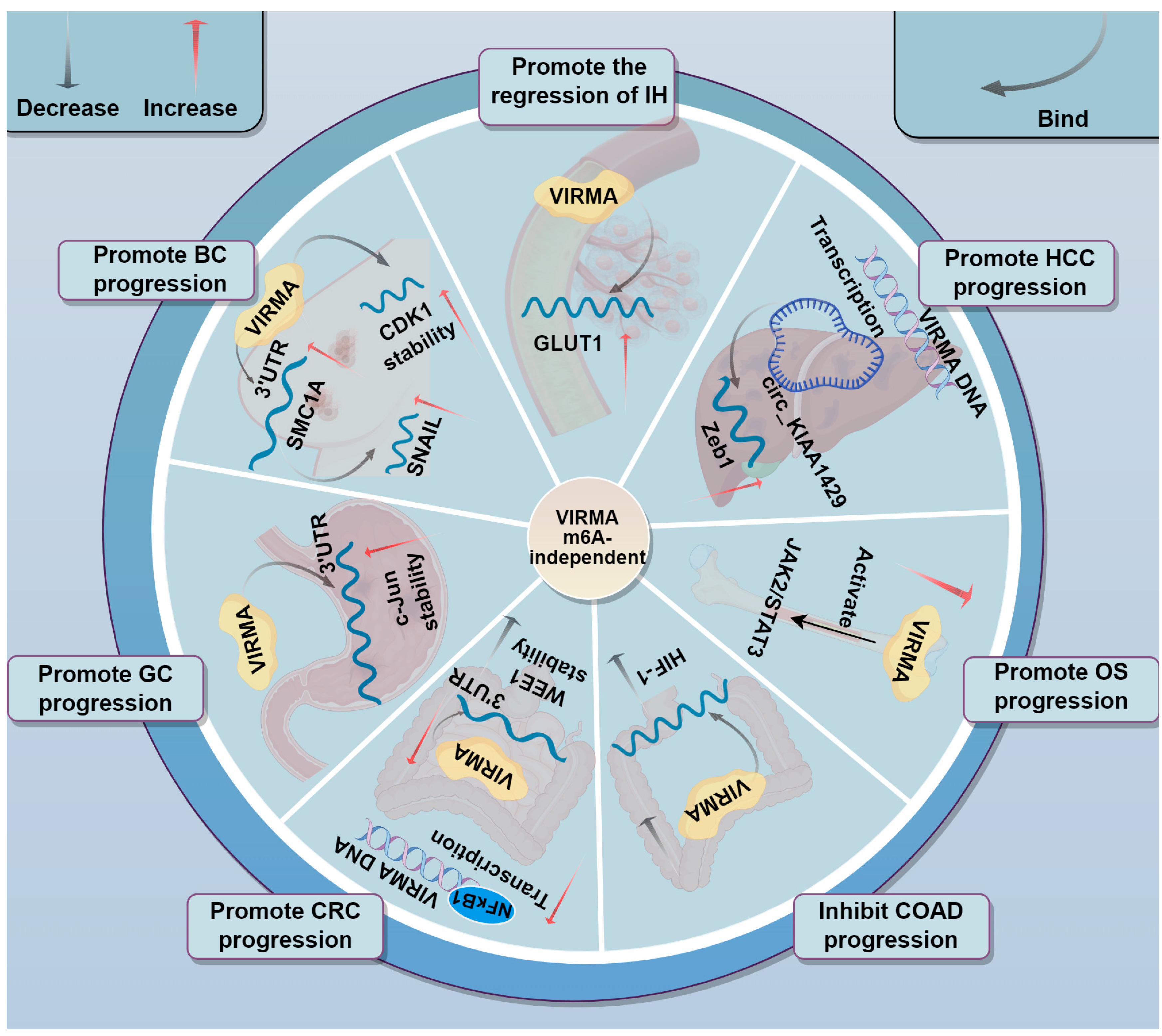
| Cancer Type | Expression of VIRMA | m6A-Dependent Regulation | Upstream Regulation | Downstream Targets | Function | In Vivo Study | Ref. |
|---|---|---|---|---|---|---|---|
| HNSCC | Up | Yes. | / | UBR5 | Promote cancer progression; influence mRNA stability | / | [35] |
| NPC | Up | Yes. | / | PTGS2 | Promote cancer progression; influence mRNA stability | Yes | [46] |
| Up | Yes | / | LINC00839 | Promote cancer progression | Yes | [47] | |
| Up | Yes | / | E2F7 | Promote cancer progression | Yes | [32] | |
| OSCC | Up | Yes | / | inhibitor A | Promote cancer progression | Yes | [48] |
| Up | Yes | / | PGK1 | Promote cancer progression; mediate resistance to ferroptosis; affect cancer metabolism | Yes | [33] | |
| Up | Yes | / | CA9 | Promote cancer progression | Yes | [49] | |
| NSCLC | Up | Yes | / | PXFP1 | Promote cancer progression | / | [50] |
| Up | Yes | / | WTAP | Promote cancer progression, affecting drug resistance. | Yes | [51] | |
| Up | Yes | / | BTG2 | Promote cancer progression; influence mRNA stability | Yes | [52] | |
| Up | Yes | / | DAPK3 | Promote cancer progression | Yes | [53] | |
| Up | Yes | / | MUC3A | Promote cancer progression by influencing the cell cycle. | / | [54] | |
| Up | Yes | / | ARHGAP30 | Promote cancer progression | / | [55] | |
| Up | Yes | / | LINC01106 | Promote cancer progression | Yes | [56] | |
| Up | Yes | / | P53 | Promote cancer progression; mediating resistance to ferroptosis | Yes | [57] | |
| Up | Yes | / | KLF1 | promoting cancer cell proliferation and metastasis; mediating immune escape | / | [58] | |
| Up | Yes | / | HOXA1 | Promote cancer progression, affecting drug resistance. | / | [59] | |
| Up | Yes | / | MAP3K2 | Promote cancer progression, affecting drug resistance. | Yes | [34] | |
| HCC | Up | Yes | / | RND3 | Promote cancer progression; influence mRNA stability | Yes | [18] |
| Up | Yes | / | HPN | Promote cancer progression | / | [60] | |
| Up | Yes | / | circDLC1 | Promote cancer progression | Yes | [61] | |
| Up | Yes | / | GATA3 pre-mRNA | Promote cancer progression; influence mRNA stability | Yes | [23] | |
| Up | Yes | / | ID2 | Promote cancer progression | / | [62] | |
| Up | Yes | / | HK1 | Promote cancer progression, affecting drug resistance and affecting cancer metabolism. | Yes | [63] | |
| Up | Yes | / | SLC7A11 | Promote cancer progression; mediating resistance to ferroptosis | Yes | [64] | |
| Up | Yes | / | CCR9 | Promote cancer progression, affecting drug resistance. | Yes | [65] | |
| Up | Yes | HBx | HSPG2 | Promote cancer progression; influence mRNA stability | / | [66] | |
| Up | Yes | / | / | Promote cancer progression, affecting drug resistance. | Yes | [67] | |
| ICC | Up | Yes | / | TMED2P; ARD3B | Promote cancer progression | Yes | [68] |
| Up | Yes | CCL3 | SIRT1 | Promote cancer progression; influence mRNA stability and influencing cell cycle. | Yes | [69] | |
| GC | Up | Yes | / | RASD1 | Promote cancer progression; influence mRNA stability; influence cell cycle | Yes | [36] |
| Up | Yes | / | LINC00958 | Promote cancer progression, affecting cancer metabolism. | Yes | [70] | |
| Up | Yes | P65 | FOXM1 | Promote cancer progression, affecting drug resistance. | Yes | [71] | |
| Up | Yes | / | FOXM1 | Promote cancer progression, affecting drug resistance. | / | [72] | |
| PDAC | Up | Yes | / | STRA6 | Promote cancer progression, affecting cancer metabolism. | Yes | [24] |
| Up | Yes | C/EBP β | SLC43A2 | Promote cancer progression; influence mRNA stability | Yes | [73] | |
| CRC | Up | Yes | / | lncRNA POU6F3-AS1 | Promote cancer progression | / | [74] |
| Up | Yes | USP29 | SOX8 | Promote cancer progression | Yes | [38] | |
| Up | Yes | / | SIRT1 | Promote cancer progression; influence mRNA stability | Yes | [20] | |
| Up | Yes | miR-153-3p | lncRNA EBLN3P | Promote cancer progression; mediating resistance to ferroptosis | / | [75] | |
| OS | Up | Yes | / | CDK1; CCNA2; CCNB1 | Promote cancer progression | Yes | [76] |
| ES | Up | Yes | NKX2-2 STAT3 | STAT3 | Promote cancer progression | Yes | [77] |
| MM | Up | Yes | / | FOXM1 | Promote cancer progression, affecting cancer metabolism. | Yes | [39] |
| CML | Up | Yes | / | RAB27B | Promote cancer progression, affecting drug resistance. | Yes | [42] |
| DLBCL | Up | Yes | / | CHST11 | Promote cancer progression by influencing the cell cycle. | Yes | [41] |
| BC | Up | Yes | / | HAS2 | Promote cancer progression; influence mRNA stability | Yes | [19] |
| Up | Yes | / | KIF15 | Promote cancer progression; influence mRNA stability | Yes | [78] | |
| Up | Yes | / | TFAP2A; DDR1 | Promote cancer progression, mediating immune escape | Yes | [79] | |
| Up | Yes | / | NEAT1 | Promote cancer progression | Yes | [80] | |
| Up | Yes | miR-556-5p | LINC00667 | Promote cancer progression | / | [81] | |
| TGCTs | Up | Yes | / | / | Promote cancer progression | Yes | [43] |
| CC | Up | Yes | / | BTG2 | Promote cancer progression; influence mRNA stability | / | [44] |
| Up | Yes | / | LARP1 | Promote cancer progression; influence mRNA stability. | Yes | [82] | |
| OC | Up | Yes | SPI1 | ENO1 | Promote cancer progression, affecting cancer metabolism. | Yes | [45] |
| OS | Up | Not explored | / | JAK-STAT | Promote cancer progression | Yes | [40] |
| CRC | Up | Not explored | / | HIF-1 | Promote cancer progression | Yes | [83] |
| Up | No | NFκB1 | WEE1 | Promote cancer progression | Yes | [84] | |
| BC | Up | No | / | SMC1A | Promote cancer progression | Yes | [21] |
| Up | No | / | CDK1 | Promote cancer progression, by influencing the cell cycle. | Yes | [83] | |
| HCC | Up | No | / | Zeb1 | Promote cancer progression | / | [85] |
| GC | Up | No | / | c-Jun | Promote cancer progression, by influencing the cell cycle. | Yes | [86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Zhang, C.; Yin, M.; You, H.; Xiong, C.; Wu, J.; Gong, Y.; Xiao, Z.; Shen, J. The Multifaceted Role of VIRMA, a Core Component of the Methyltransferase Complex, in Cancer and Cancer Therapy. Biomolecules 2025, 15, 912. https://doi.org/10.3390/biom15070912
Lu J, Zhang C, Yin M, You H, Xiong C, Wu J, Gong Y, Xiao Z, Shen J. The Multifaceted Role of VIRMA, a Core Component of the Methyltransferase Complex, in Cancer and Cancer Therapy. Biomolecules. 2025; 15(7):912. https://doi.org/10.3390/biom15070912
Chicago/Turabian StyleLu, Jinmeng, Chengyu Zhang, Mengshuang Yin, Huili You, Chao Xiong, Jing Wu, Ying Gong, Zhangang Xiao, and Jing Shen. 2025. "The Multifaceted Role of VIRMA, a Core Component of the Methyltransferase Complex, in Cancer and Cancer Therapy" Biomolecules 15, no. 7: 912. https://doi.org/10.3390/biom15070912
APA StyleLu, J., Zhang, C., Yin, M., You, H., Xiong, C., Wu, J., Gong, Y., Xiao, Z., & Shen, J. (2025). The Multifaceted Role of VIRMA, a Core Component of the Methyltransferase Complex, in Cancer and Cancer Therapy. Biomolecules, 15(7), 912. https://doi.org/10.3390/biom15070912






