Dynamic 3D-Network Coating Composite Enables Global Isolation of Phosphopeptides, Stepwise Separation of Mono- and Multi-Phosphopeptides, and Phosphoproteomics of Human Lung Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Characterization and Measurement
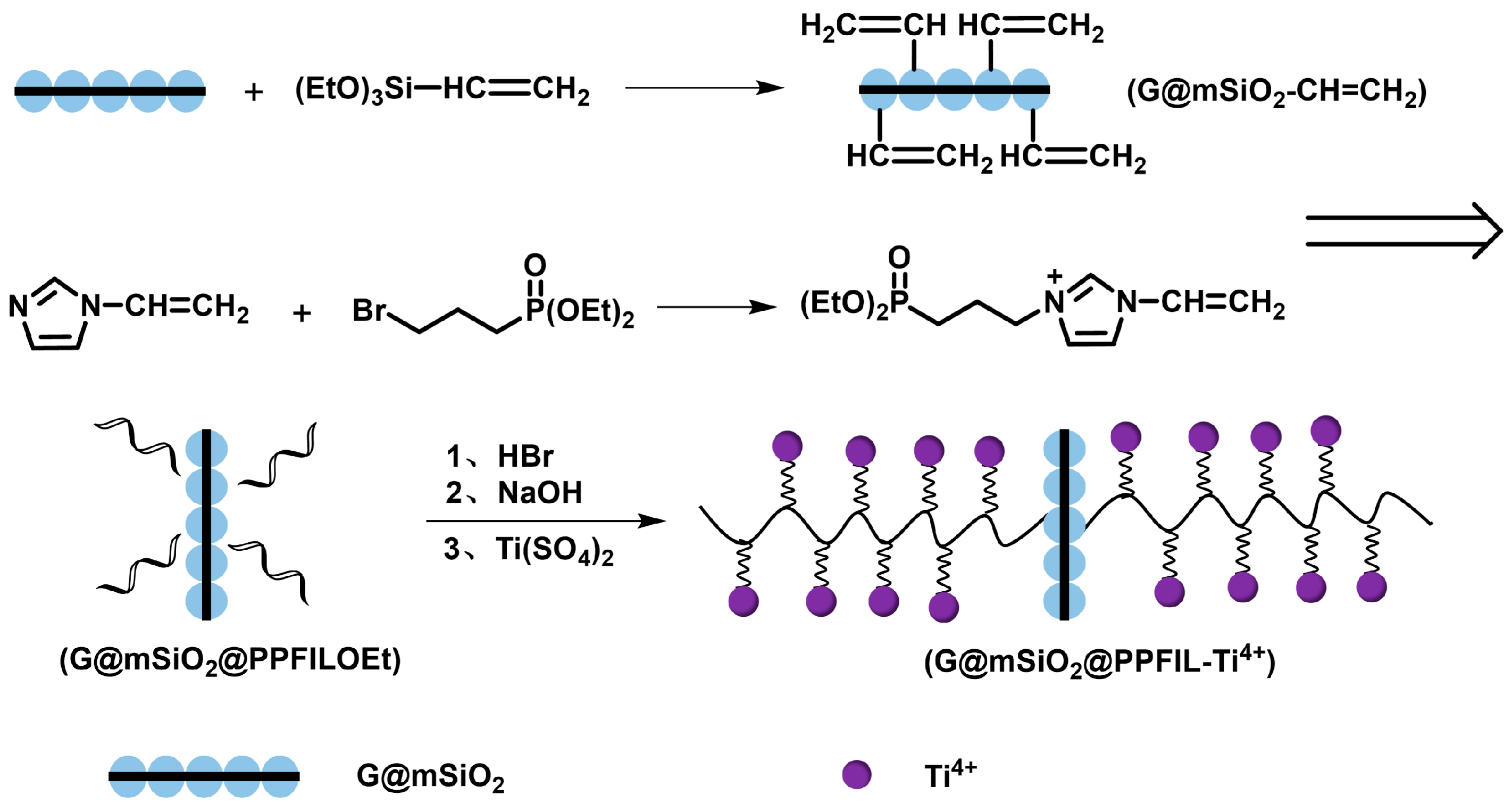
2.3. Preparation of G@mSiO2@PPFILOEt
2.4. Preparation of G@mSiO2@PPFIL-Ti4+
2.5. Sample Preparation
2.6. Enrichment of Phosphopeptides
2.7. MALDI-TOF MS Analysis
2.8. LC-MS/MS Analysis and Data Search
3. Results and Discussion
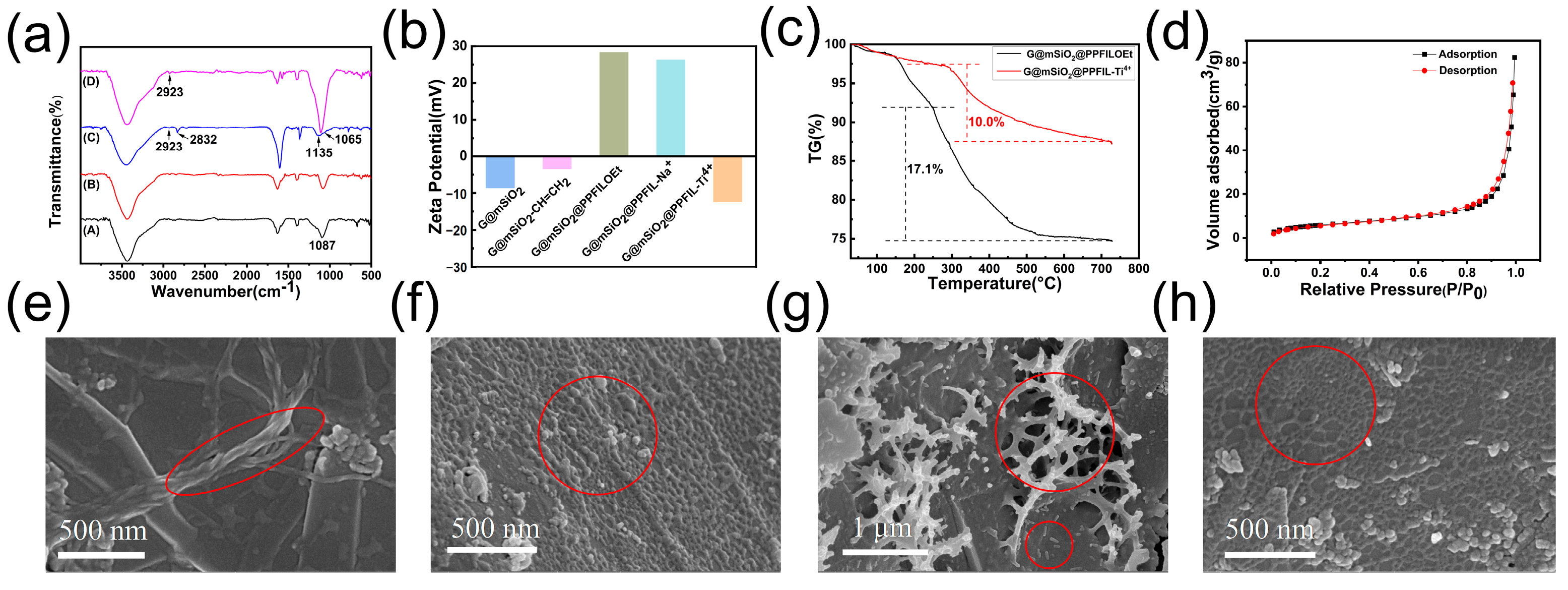
3.1. Preparation and Characterization of G@mSiO2@PPFIL-Ti4+ Composite
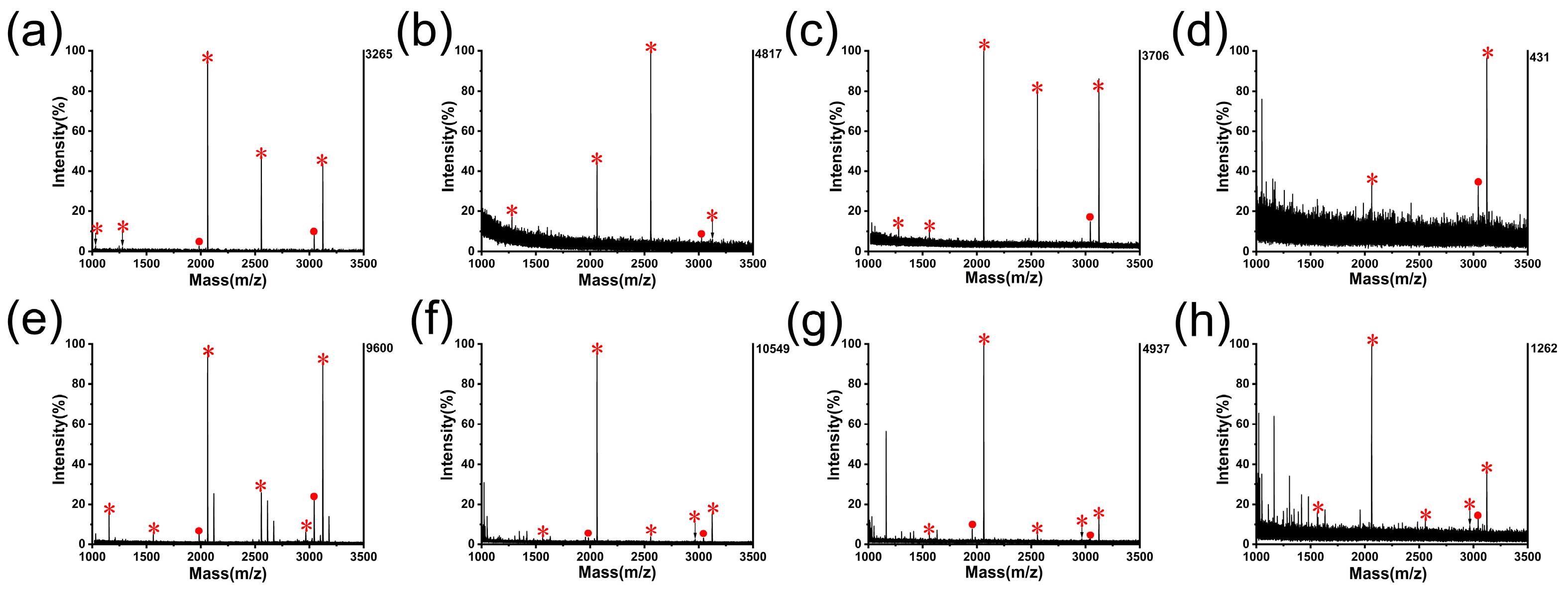
3.2. Application of G@mSiO2@PPFIL-Ti4+ Composite in Phosphopeptide Enrichment from Standard Protein Digests
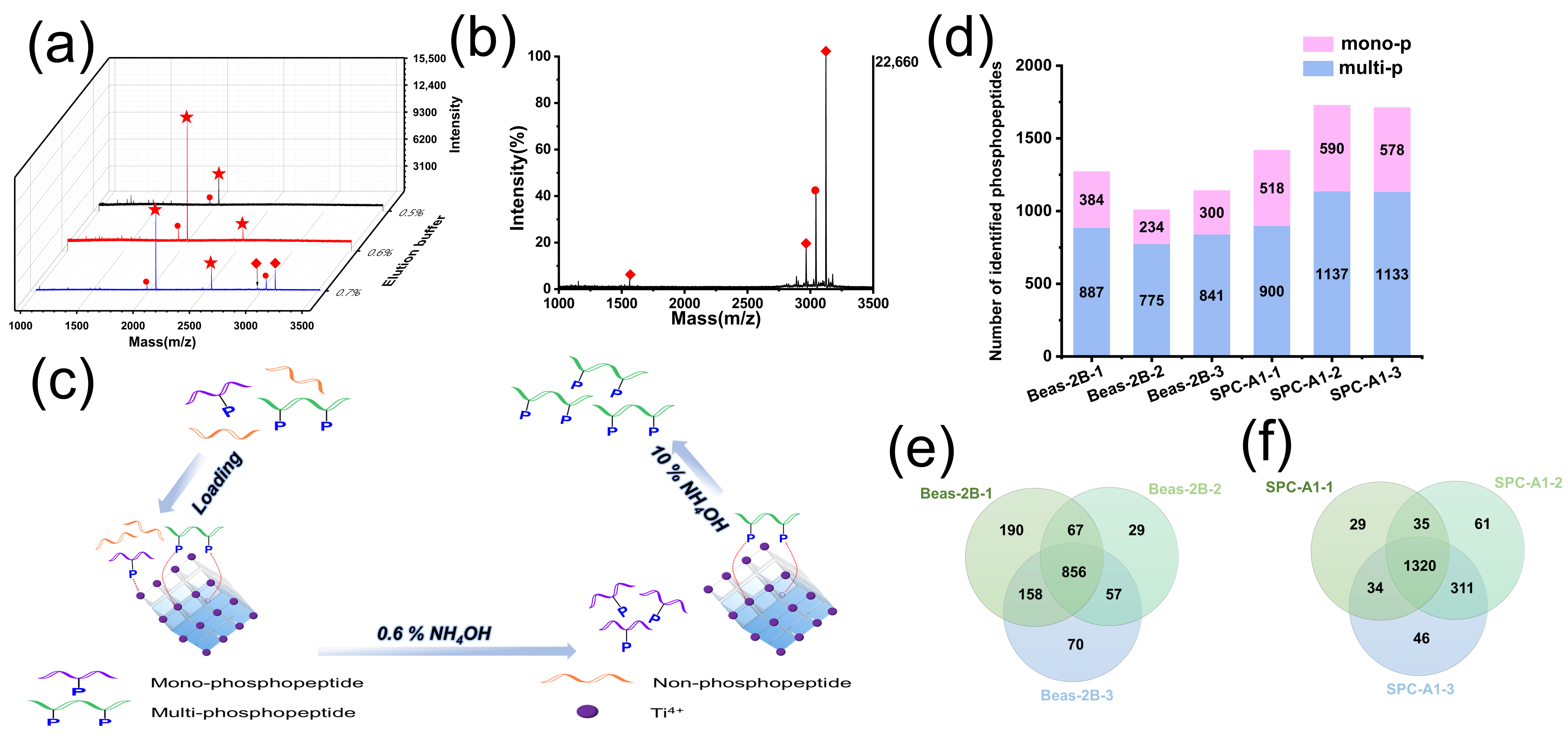
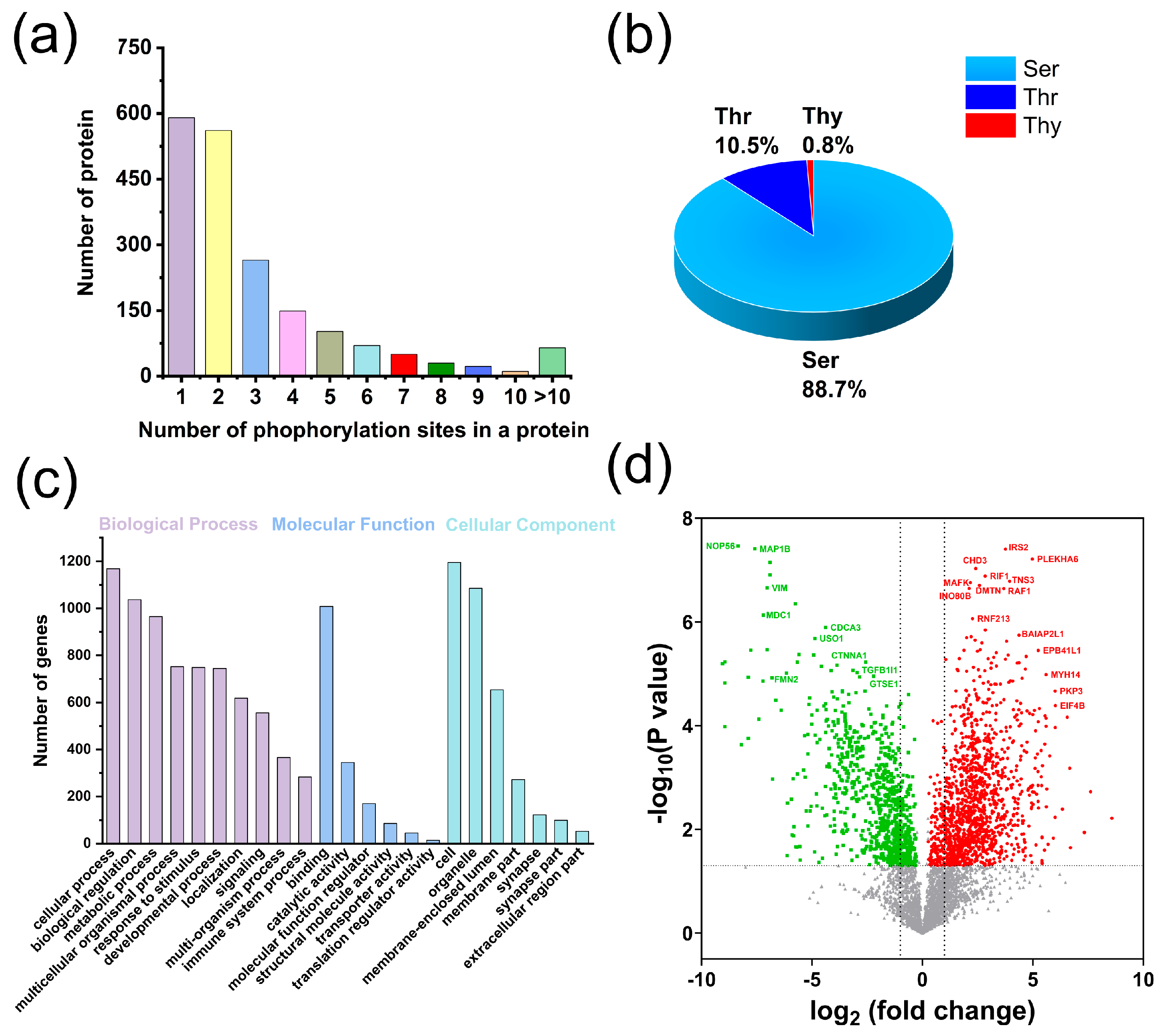
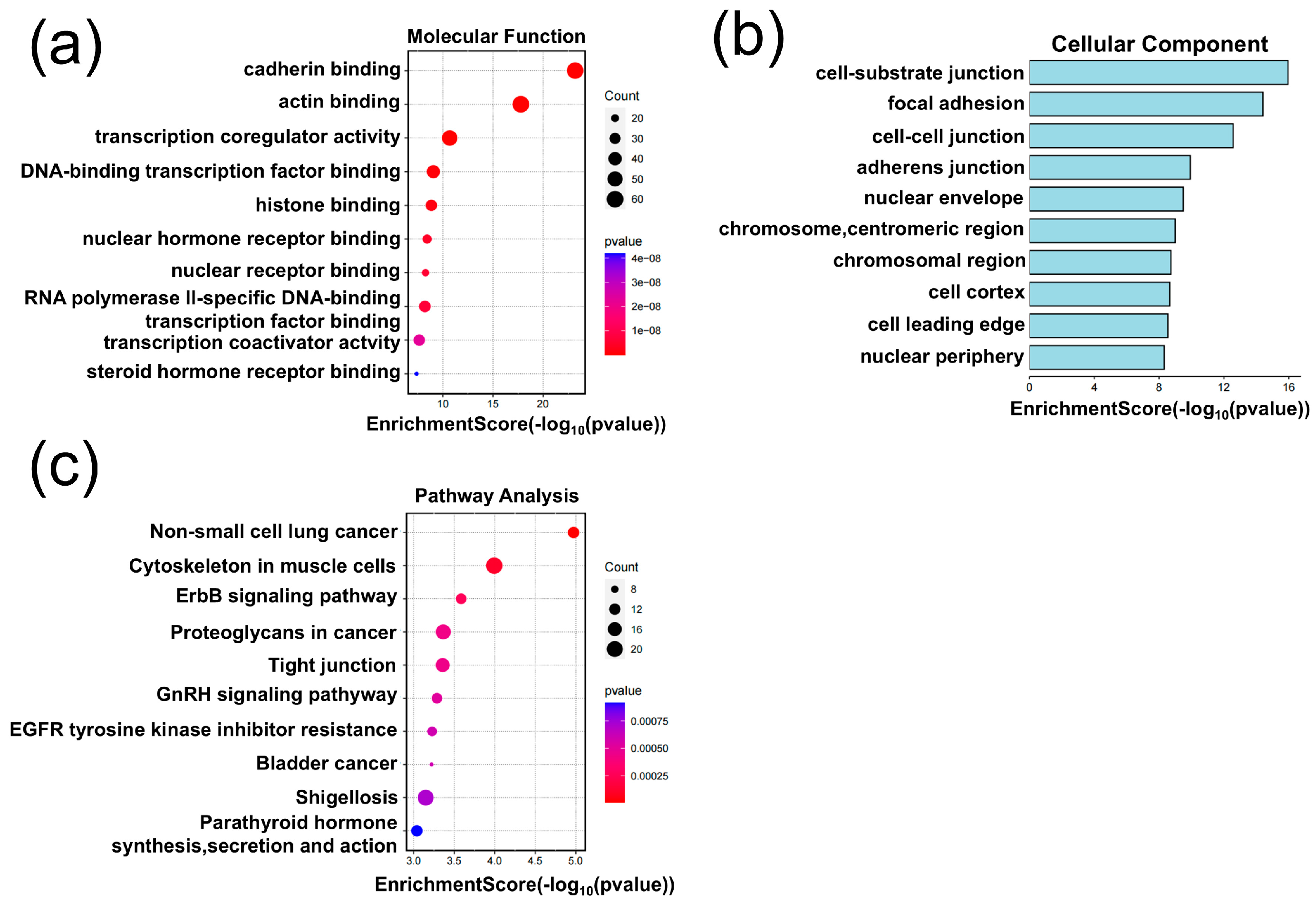
3.3. Application of the G@mSiO2@PPFIL-Ti4+ Nanocomposite in Phosphopeptide Enrichment from Human Serum, Saliva, and Lung Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Higgins, L.; Gerdes, H.; Cutillas, P.R. Principles of phosphoproteomics and applications in cancer research. Biochem. J. 2023, 480, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Urban, J. A review on recent trends in the phosphoproteomics workflow. From sample preparation to data analysis. Anal. Chim. Acta 2022, 1199, 338857. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.Z.; Bai, Q.; Ma, S.J.; Ou, J.J. Materials, workflows and applications of IMAC for phosphoproteome profiling in the recent decade: A review. TrAC-Trend Anal. Chem. 2023, 158, 116862. [Google Scholar] [CrossRef]
- Wang, B.C.; Xie, Z.H.; Ding, C.F.; Deng, C.H.; Yan, Y.H. Recent advances in metal oxide affinity chromatography materials for phosphoproteomics. TrAC-Trend Anal. Chem. 2023, 158, 116881. [Google Scholar] [CrossRef]
- Zittlau, K.; Nashier, P.; Cavarischia-Rega, C.; Macek, B.; Spät, P.; Nalpas, N. Recent progress in quantitative phosphoproteomics. Expert Rev. Proteom. 2023, 20, 469–482. [Google Scholar] [CrossRef]
- Low, T.Y.; Mohtar, M.A.; Lee, P.Y.; Omar, N.; Zhou, H.J.; Ye, M.L. Widening the Bottleneck of Phosphoproteomics: Evolving Strategies for Phosphopeptide Enrichment. Mass Spectrom. Rev. 2021, 40, 309–333. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Zhan, X.Q. Mass spectrometry analysis of phosphotyrosine-containing proteins. Mass Spectrom. Rev. 2024, 43, 857–887. [Google Scholar] [CrossRef]
- Potel, C.M.; Lin, M.H.; Heck, A.J.R.; Lemeer, S. Widespread bacterial protein histidine phosphorylation revealed by mass spectrometry-based proteomics. Nat. Methods 2018, 15, 187–190. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Jiang, H.C.; Yang, P.Y.; Lu, H.J. Fishing the PTM proteome with chemical approaches using functional solid phases. Chem. Soc. Rev. 2015, 44, 8260–8287. [Google Scholar] [CrossRef]
- Sun, N.R.; Wu, H.; Shen, X.Z.; Deng, C.H. Nanomaterials in Proteomics. Adv. Funct. Mater. 2019, 29, 1900253. [Google Scholar] [CrossRef]
- Qing, G.Y.; Lu, Q.; Xiong, Y.T.; Zhang, L.; Wang, H.X.; Li, X.L.; Liang, X.M.; Sun, T.L. New Opportunities and Challenges of Smart Polymers in Post-Translational Modification Proteomics. Adv. Mater. 2017, 29, 1604670. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.Y.; Han, G.H.; Sun, S.T.; Jiang, X.N.; Chen, R.; Wang, F.J.; Wu, R.A.; Ye, M.L.; Zou, H.F. Preparation of monodisperse immobilized Ti affinity chromatography microspheres for specific enrichment of phosphopeptides. Anal. Chim. Acta 2009, 636, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.H.; Zheng, Z.F.; Deng, C.H.; Li, Y.; Zhang, X.M.; Yang, P.Y. Hydrophilic Polydopamine-Coated Graphene for Metal Ion Immobilization as a Novel Immobilized Metal Ion Affinity Chromatography Platform for Phosphoproteome Analysis. Anal. Chem. 2013, 85, 8483–8487. [Google Scholar] [CrossRef]
- Ma, W.F.; Zhang, Y.; Li, L.L.; Zhang, Y.T.; Yu, M.; Guo, J.; Lu, H.J.; Wang, C.C. Ti4+-Immobilized Magnetic Composite Microspheres for Highly Selective Enrichment of Phosphopeptides. Adv. Funct. Mater. 2013, 23, 107–115. [Google Scholar] [CrossRef]
- Salimi, K.; Usta, D.D.; Çelikbiçak, Ö.; Pinar, A.; Salih, B.; Tuncel, A. Highly selective enrichment of phosphopeptides by titanium (IV) attached monodisperse-porous poly(vinylphosphonic acid-ethylene dimethacrylate) microspheres. J. Chromatogr. A 2017, 1496, 9–19. [Google Scholar] [CrossRef]
- Huang, Y.L.; Wang, J.; Jian, Y.H.; Yang, P.Y.; Wang, G.W.; Liu, F. Development of amphiphile 4-armed PEO-based Ti-complex for highly selective enrichment of phosphopeptides. Talanta 2019, 204, 670–676. [Google Scholar] [CrossRef]
- Wei, X.Z.; Wen, X.; Zheng, H.J.; Zhang, Y.; Jia, Q. Facile synthesis of Fe3+immobilized magnetic polydopamine polyethyleneimine composites for phosphopeptide enrichment. J. Chromatogr. A 2024, 1719, 464752. [Google Scholar] [CrossRef]
- Meng, L.Y.; Wang, B.; Zhang, S.J.; Zhang, S.; Cai, T.; Ding, C.F.; Yan, Y.H. One-step polymerization of functionalized ionic liquid as polymer microspheres for highly specific enrichment of phosphopeptides from cell lysate and serum. Microchem. J. 2024, 201, 110511. [Google Scholar] [CrossRef]
- Tao, W.A.; Wollscheid, B.; O’Brien, R.; Eng, J.K.; Li, X.J.; Bodenmiller, B.; Watts, J.D.; Hood, L.; Aebersold, R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat. Methods 2005, 2, 591–598. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Martin, V.A.; Alicie, B.M.; Geahlen, R.L.; Tao, W.A. In-depth Analyses of Kinase-dependent Tyrosine Phosphoproteomes Based on Metal Ion-functionalized Soluble Nanopolymers. Mol. Cell. Proteom. 2010, 9, 2162–2172. [Google Scholar] [CrossRef]
- Wang, B.B.; Liang, W.D.; Liang, H.Z.; Liu, B.; Wang, Q.Y.; Yan, Y.H.; Wang, T.T.; Jiang, Y.F.; Jiang, Y.L.; Qiu, H.D.; et al. Phosphonate-Functionalized Ionic Liquid: A New Surface Modifier Contributing to the Enhanced Enrichment of Phosphorylated Peptides. ACS Sustain. Chem. Eng. 2021, 9, 7930–7940. [Google Scholar] [CrossRef]
- Xia, C.L.; Wang, Q.Y.; Liang, W.D.; Wang, B.B.; Feng, Q.S.; Zhou, C.Y.; Xie, Y.S.; Yan, Y.H.; Zhao, L.L.; Jiang, B.; et al. Superhydrophilic nanocomposite adsorbents modified nitrogen-rich phosphonate-functionalized ionic liquid linkers: Enhanced phosphopeptide enrichment and phosphoproteome analysis of tau phosphorylation in the hippocampal lysate of Alzheimer’s transgenic mice. J. Mater. Chem. B 2022, 10, 7967–7978. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Xia, C.L.; Zhu, A.; Bao, Y.J.; Lu, J.N.; Chen, Y.; Xu, J.Y.; Wang, B.B.; Naman, C.B.; Li, L.P.; et al. Discrepancy of synaptic and microtubular protein phosphorylation in the hippocampus of APP/PS1 and MAPTxP301S transgenic mice at the early stage of Alzheimer’s disease. Metab. Brain Dis. 2023, 38, 1983–1997. [Google Scholar] [CrossRef] [PubMed]
- Le, J.Y.; Xia, C.L.; Xu, J.Y.; Cai, J.H.; Hu, C.W.; Bai, Y.; Chen, H.Y.; Rong, W.N.; Jiang, Y.J.; Wu, X.M.; et al. 9-Methylfascaplysin Prevents Neuroinflammation and Synaptic Damage via Cell-Specific Inhibition of Kinases in APP/PS1 Transgenic Mice. CNS Neurosci. Ther. 2024, 30, e70100. [Google Scholar] [CrossRef]
- Mel’nik, O.A.; Shaplov, A.S.; Lozinskaya, E.I.; Popova, N.A.; Makarov, M.V.; Odinets, I.L.; Lysenko, K.A.; Timofeeva, G.I.; Malyshkina, I.A.; Vygodskii, Y.S. Polymers based on ionic monomers with side phosphonate groups. Polym. Sci. Ser. B 2010, 52, 316–326. [Google Scholar] [CrossRef]
- Wang, D.Q.; Huang, J.F.; Zhang, H.R.; Ma, M.; Xu, M.; Cui, Y.S.; Shi, X.D.; Li, L.J. ATP-Coated Dual-Functionalized Titanium(IV) IMAC Material for Simultaneous Enrichment and Separation of Glycopeptides and Phosphopeptides. J. Proteome Res. 2023, 22, 2044–2054. [Google Scholar] [CrossRef]
- Xie, P.; Liu, J.; Liao, Z.; Zhou, Q.; Sun, J.; Liu, Z.; Xiong, H.; Wan, H. Profiling the differential phosphoproteome between breast milk and infant formula through a titanium (IV)-immobilized magnetic nanoplatform. Food Chem. 2024, 464, 141541. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, X.Y.; Yan, Y.H.; Xuan, R.R. Facile preparation of nitrogen/titanium-rich porous organic polymers for specific enrichment of N-glycopeptides and phosphopeptides. Anal. Methods 2024, 16, 695–703. [Google Scholar] [CrossRef]
- Wang, K.X.; Yu, A.J.; Gao, Y.; Chen, M.; Yuan, H.; Zhang, S.S.; Ouyang, G.F. A nitrogen-doped graphene tube composite based on immobilized metal affinity chromatography for the capture of phosphopeptides. Talanta 2023, 261, 124617. [Google Scholar] [CrossRef]
- He, Y.T.; Zhang, S.S.; Zhong, C.; Yang, Y.X.; Li, G.R.; Ji, Y.; Lin, Z. Facile synthesis of Ti4+-immobilized magnetic covalent organic frameworks for enhanced phosphopeptide enrichment. Talanta 2021, 235, 122789. [Google Scholar] [CrossRef]
- Wang, D.Q.; Huang, J.F.; Zhang, H.R.; Gu, T.J.; Li, L.J. Cotton Ti-IMAC: Developing Phosphorylated Cotton as a Novel Platform for Phosphopeptide Enrichment. ACS Appl. Mater. Interfaces 2023, 15, 47893–47901. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Wang, B.; Luo, Y.T.; Wang, W.M.; Ding, C.F.; Yan, Y.H. Facile preparation of porphyrin-based porous organic polymers for specific enrichment and isolation of phosphopeptides and phosphorylated exosomes. Talanta 2023, 264, 124771. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Tang, R.Z.; Jia, S.C.; Ma, S.J.; Gong, B.L.; Ou, J.J. Monodisperse Ti4+-immobilized macroporous adsorbent resins with polymer brush for improved multi-phosphopeptides enrichment in milk. Microchim. Acta 2022, 189, 405. [Google Scholar] [CrossRef]
- Jiang, Y.F.; Liang, W.D.; Wang, B.B.; Feng, Q.S.; Xia, C.L.; Wang, Q.Y.; Yan, Y.H.; Zhao, L.L.; Cui, W.; Liang, H.Z. Magnetic mesoporous silica nanoparticles modified by phosphonate functionalized ionic liquid for selective enrichment of phosphopeptides. RSC Adv. 2022, 12, 26859–26865. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, X.M.; Chen, X.; Zhu, G.T.; Wang, R.Q.; Feng, Y.Q. Pyridoxal 5′-phosphate mediated preparation of immobilized metal affinity material for highly selective and sensitive enrichment of phosphopeptides. J. Chromatogr. A 2017, 1499, 30–37. [Google Scholar] [CrossRef]
- Samarasimhareddy, M.; Mayer, G.; Hurevich, M.; Friedler, A. Multiphosphorylated peptides: Importance, synthetic strategies, and applications for studying biological mechanisms. Org. Biomol. Chem. 2020, 18, 3405–3422. [Google Scholar] [CrossRef]
- Russell, C.L.; Mitra, V.; Hansson, K.; Blennow, K.; Gobom, J.; Zetterberg, H.; Hiltunen, M.; Ward, M.; Pike, I. Comprehensive Quantitative Profiling of Tau and Phosphorylated Tau Peptides in Cerebrospinal Fluid by Mass Spectrometry Provides New Biomarker Candidates. J. Alzheimer’s Dis. 2017, 55, 303–313. [Google Scholar] [CrossRef]
- Simrén, J.; Leuzy, A.; Karikari, T.K.; Hye, A.; Benedet, A.L.; Lantero-Rodriguez, J.; Mattsson-Carlgren, N.; Schöll, M.; Mecocci, P.; Vellas, B.; et al. The diagnostic and prognostic capabilities of plasma biomarkers in Alzheimer’s disease. Alzheimer’s Dement. 2021, 17, 1145–1156. [Google Scholar] [CrossRef]
- Planel, E.; Krishnamurthy, P.; Miyasaka, T.; Liu, L.; Herman, M.; Kumar, A.; Bretteville, A.; Figueroa, H.Y.; Yu, W.H.; Whittington, R.A.; et al. Anesthesia-Induced Hyperphosphorylation Detaches 3-Repeat Tau from Microtubules without Affecting Their Stability In Vivo. J. Neurosci. 2008, 28, 12798–12807. [Google Scholar] [CrossRef]
- Sultanakhmetov, G.; Kato, I.; Asada, A.; Saito, T.; Ando, K. Microtubule-affinity regulating kinase family members distinctively affect tau phosphorylation and promote its toxicity in a Drosophila model. Genes Cells 2024, 29, 337–346. [Google Scholar] [CrossRef]
- Avila, J.; Simón, D.; Díaz-Hernández, M.; Pintor, J.; Hernández, F. Sources of Extracellular Tau and its Signaling. J. Alzheimer’s Dis. 2014, 40, S7–S15. [Google Scholar] [CrossRef] [PubMed]
- Cowan, C.M.; Bossing, T.; Page, A.; Shepherd, D.; Mudher, A. Soluble hyper-phosphorylated tau causes microtubule breakdown and functionally compromises normal tau in vivo. Acta Neuropathol. 2010, 120, 593–604. [Google Scholar] [CrossRef]
- Ruan, Z.; Pathak, D.; Kalavai, S.V.; Yoshii-Kitahara, A.; Muraoka, S.; Bhatt, N.; Takamatsu-Yukawa, K.; Hu, J.Q.; Wang, Y.Z.; Hersh, S.; et al. Alzheimer’s disease brain-derived extracellular vesicles spread tau pathology in interneurons. Brain 2021, 144, 288–309. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Horwitz, A.R. Myosin light chain mono- and di-phosphorylation differentially regulate adhesion and polarity in migrating cells. Biochem. Biophys. Res. Commun. 2010, 402, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.N.; Ma, W.; Shen, S.S.; Li, L.P.; Bai, Y.; Liu, H.W. Hydrazide functionalized monodispersed silica microspheres: A novel probe with tunable selectivity for a versatile enrichment of phosphopeptides with different numbers of phosphorylation sites in MS analysis. Chem. Commun. 2016, 52, 1162–1165. [Google Scholar] [CrossRef]
- Mamone, G.; Picariello, G.; Ferranti, P.; Addeo, F. Hydroxyapatite affinity chromatography for the highly selective enrichment of mono- and multi-phosphorylated peptides in phosphoproteome analysis. Proteomics 2010, 10, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.Y.; Xiao, X.; Zheng, S.; Zhang, W.Y.; Ding, M.J.; Jiang, H.Y.; Huang, L.L.; Kang, J. Mass spectrometric analysis of mono- and multi-phosphopeptides by selective binding with NiZnFeO magnetic nanoparticles. Nat. Commun. 2013, 4, 1656. [Google Scholar] [CrossRef]
- Qing, G.Y.; Lu, Q.; Li, X.L.; Liu, J.; Ye, M.L.; Liang, X.M.; Sun, T.L. Hydrogen bond based smart polymer for highly selective and tunable capture of multiply phosphorylated peptides. Nat. Commun. 2017, 8, 461. [Google Scholar] [CrossRef]
- Xiao, J.; Yang, S.S.; Wu, J.X.; Wang, H.; Yu, X.Z.; Shang, W.B.; Chen, G.Q.; Gu, Z.Y. Highly Selective Capture of Monophosphopeptides by Two-Dimensional Metal-Organic Framework Nanosheets. Anal. Chem. 2019, 91, 9093–9101. [Google Scholar] [CrossRef]
- Li, J.Y.; Cao, Z.M.; Hua, Y.; Wei, G.; Yu, X.Z.; Shang, W.B.; Lian, H.Z. Solvothermal Synthesis of Novel Magnetic Nickel Based Iron Oxide Nanocomposites for Selective Capture of Global- and Mono-Phosphopeptides. Anal. Chem. 2020, 92, 1058–1067. [Google Scholar] [CrossRef]
- Yu, L.Z.; Luo, B.; Li, Z.Y.; He, J.; Lan, F.; Wu, Y. PAMAM-PMAA brush-functionalized magnetic composite nanospheres: A smart nanoprobe with tunable selectivity for effective enrichment of mono-, multi-, or global phosphopeptides. J. Mater. Chem. B 2020, 8, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- La Barbera, G.; Capriotti, A.L.; Cavaliere, C.; Ferraris, F.; Montone, C.M.; Piovesana, S.; Chiozzi, R.Z.; Laganà, A. Saliva as a source of new phosphopeptide biomarkers: Development of a comprehensive analytical method based on shotgun peptidomics. Talanta 2018, 183, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Kaczor-Urbanowicz, K.E.; Carreras-Presas, C.M.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T.W. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef]
- Mantini, G.; Pham, T.; Piersma, S.R.; Jimenez, C.R. Computational Analysis of Phosphoproteomics Data in Multi-Omics Cancer Studies. Proteomics 2021, 21, 1900312. [Google Scholar] [CrossRef] [PubMed]
- Alduais, Y.; Zhang, H.J.; Fan, F.; Chen, J.; Chen, B.A. Non-small cell lung cancer (NSCLC): A review of risk factors, diagnosis, and treatment. Medicine 2023, 102, e32899. [Google Scholar] [CrossRef]
- Taekker, M.; Kristjánsdóttir, B.; Graumann, O.; Laursen, C.B.; Pietersen, P. Diagnostic accuracy of low-dose and ultra-low-dose CT in detection of chest pathology: A systematic review. Clin. Imag. 2021, 74, 139–148. [Google Scholar] [CrossRef]
- Wang, C.T.; Qian, L.Q.; Ji, L.Y.; Liu, S.; Wahid, A.; Jiang, X.T.; Sohail, A.; Ji, Y.; Zhang, Y.; Wang, P.; et al. Affinity chromatography assisted comprehensive phosphoproteomics analysis of human saliva for lung cancer. Anal. Chim. Acta 2020, 1111, 103–113. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Xu, W.H.; Zheng, H.J.; Jia, Q. Light, pH, and Temperature Triple-Responsive Magnetic Composites for Highly Efficient Phosphopeptide Enrichment. Anal. Chem. 2023, 95, 9043–9051. [Google Scholar] [CrossRef]
- Yang, C.H.; Chou, H.C.; Fu, Y.N.; Yeh, C.L.; Cheng, H.W.; Chang, I.C.; Liu, K.J.; Chang, G.C.; Tsai, T.F.; Tsai, S.F.; et al. EGFR over-expression in non-small cell lung cancers harboring EGFR mutations is associated with marked down-regulation of CD82. Biochim. Biophys. Acta-Mol. Basis Dis. 2015, 1852, 1540–1549. [Google Scholar] [CrossRef]
- Cavazzoni, A.; La Monica, S.; Alfieri, R.; Ravelli, A.; Van Der Steen, N.; Sciarrillo, R.; Madeddu, D.; Lagrasta, C.A.M.; Quaini, F.; Bonelli, M.; et al. Enhanced efficacy of AKT and FAK kinase combined inhibition in squamous cell lung carcinomas with stable reduction in PTEN. Oncotarget 2017, 8, 53068–53083. [Google Scholar] [CrossRef]
- Wahid, A.; Sohail, A.; Wang, H.Y.; Guo, M.; Zhang, L.; Ji, Y.; Wang, P.; Xiao, H. Titanium(IV) immobilized affinity chromatography facilitated phosphoproteomics analysis of salivary extracellular vesicles for lung cancer. Anal. Bioanal. Chem. 2022, 414, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Liu, Y.B.; Cao, S.; Tian, Z.W.; Zhou, H.H. RIF1 promotes tumor growth and cancer stem cell-like traits in NSCLC by protein phosphatase 1-mediated activation of Wnt/β-catenin signaling. Cell Death Dis. 2018, 9, 2–17. [Google Scholar] [CrossRef]
- Wang, M.Y.; Chen, B.H.; Zhang, W.R.; Zhang, F.C.; Qiu, Y.M.; Lin, Y.Y.; Yang, S.F. Dematin inhibits glioblastoma malignancy through RhoA-mediated CDKs downregulation and cytoskeleton remodeling. Exp. Cell Res. 2022, 417, 113196. [Google Scholar] [CrossRef]
- Saltiel, A.R. Insulin signaling in health and disease. J. Clin. Investig. 2021, 131, e142241. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Lero, M.W.; Mercado-Matos, J.; Zhu, S.; Jo, M.; Tocheny, C.E.; Morgan, J.S.; Shaw, L.M. The insulin and IGF signaling pathway sustains breast cancer stem cells by IRS2/PI3K-mediated regulation of MYC. Cell Rep. 2022, 41, 111759. [Google Scholar] [CrossRef]
- Fan, L.L.; Zheng, M.Y.; Zhou, X.Y.; Yu, Y.J.; Ning, Y.D.; Fu, W.Z.; Xu, J.; Zhang, S.W. Molecular mechanism of vimentin nuclear localization associated with the migration and invasion of daughter cells derived from polyploid giant cancer cells. J. Transl. Med. 2023, 21, 719. [Google Scholar] [CrossRef] [PubMed]
- Kuburich, N.A.; den Hollander, P.; Castaneda, M.; Pietila, M.; Tang, X.M.; Batra, H.; Martinez-Pena, F.; Visal, T.H.; Zhou, T.L.; Demestichas, B.R.; et al. Stabilizing vimentin phosphorylation inhibits stem-like cell properties and metastasis of hybrid epithelial/mesenchymal carcinomas. Cell Rep. 2023, 42, 113470. [Google Scholar] [CrossRef]
- Kumagai, S.; Koyama, S.; Nishikawa, H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 2021, 21, 181–197. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Chen, Z.; Wang, D.; Liang, W.; Wang, B.; Xia, C.; Yan, Y.; Ding, C.; Meng, X.; Liang, H. Dynamic 3D-Network Coating Composite Enables Global Isolation of Phosphopeptides, Stepwise Separation of Mono- and Multi-Phosphopeptides, and Phosphoproteomics of Human Lung Cells. Biomolecules 2025, 15, 894. https://doi.org/10.3390/biom15060894
Liu L, Chen Z, Wang D, Liang W, Wang B, Xia C, Yan Y, Ding C, Meng X, Liang H. Dynamic 3D-Network Coating Composite Enables Global Isolation of Phosphopeptides, Stepwise Separation of Mono- and Multi-Phosphopeptides, and Phosphoproteomics of Human Lung Cells. Biomolecules. 2025; 15(6):894. https://doi.org/10.3390/biom15060894
Chicago/Turabian StyleLiu, Linlin, Zhenhua Chen, Danni Wang, Weida Liang, Binbin Wang, Chenglong Xia, Yinghua Yan, Chuanfan Ding, Xiaodan Meng, and Hongze Liang. 2025. "Dynamic 3D-Network Coating Composite Enables Global Isolation of Phosphopeptides, Stepwise Separation of Mono- and Multi-Phosphopeptides, and Phosphoproteomics of Human Lung Cells" Biomolecules 15, no. 6: 894. https://doi.org/10.3390/biom15060894
APA StyleLiu, L., Chen, Z., Wang, D., Liang, W., Wang, B., Xia, C., Yan, Y., Ding, C., Meng, X., & Liang, H. (2025). Dynamic 3D-Network Coating Composite Enables Global Isolation of Phosphopeptides, Stepwise Separation of Mono- and Multi-Phosphopeptides, and Phosphoproteomics of Human Lung Cells. Biomolecules, 15(6), 894. https://doi.org/10.3390/biom15060894







