Muscle Stem Cell Microenvironment and Functions in Muscle Regeneration
Abstract
1. Introduction
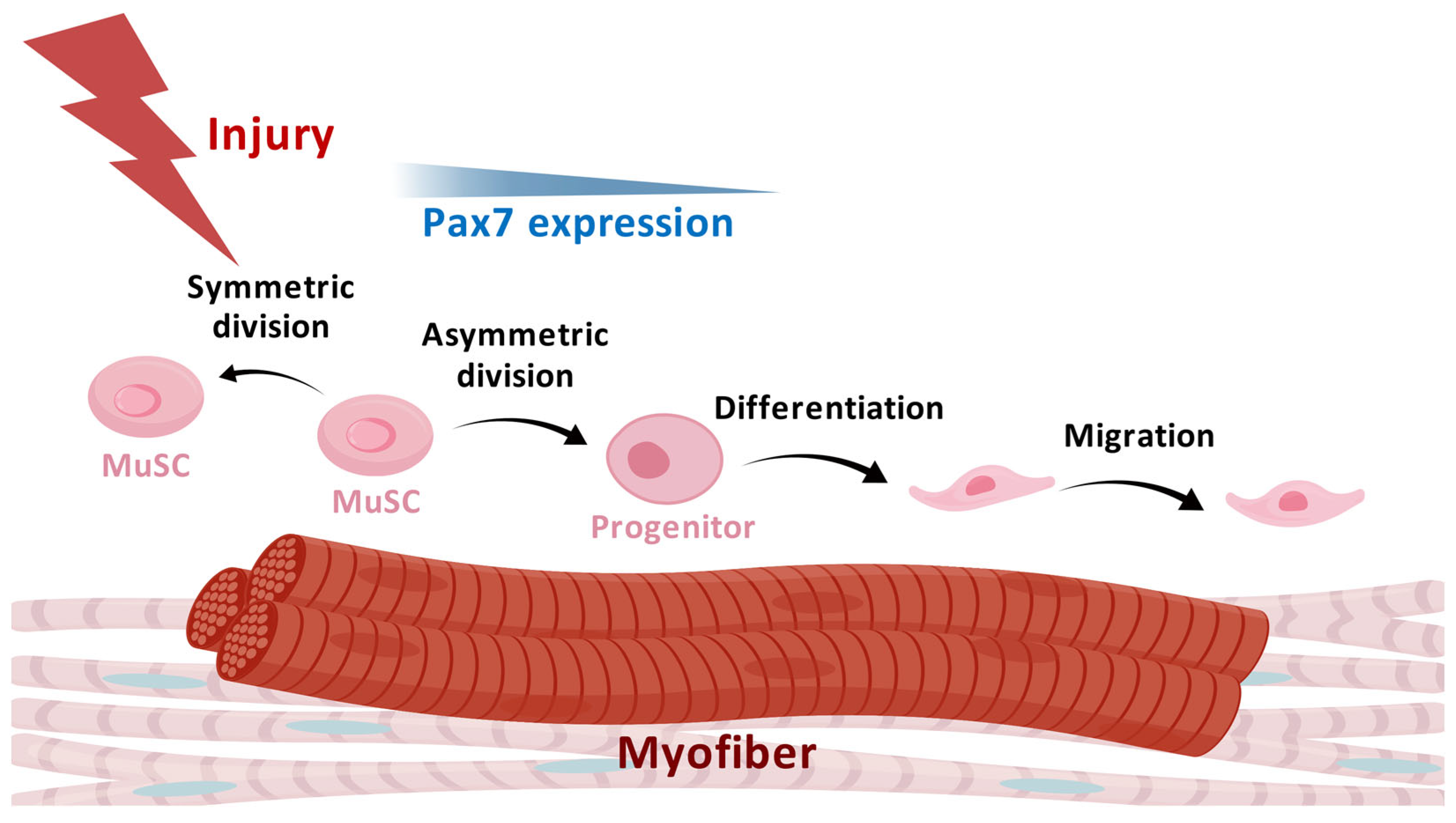
2. Characteristics and Heterogeneity of MuSCs
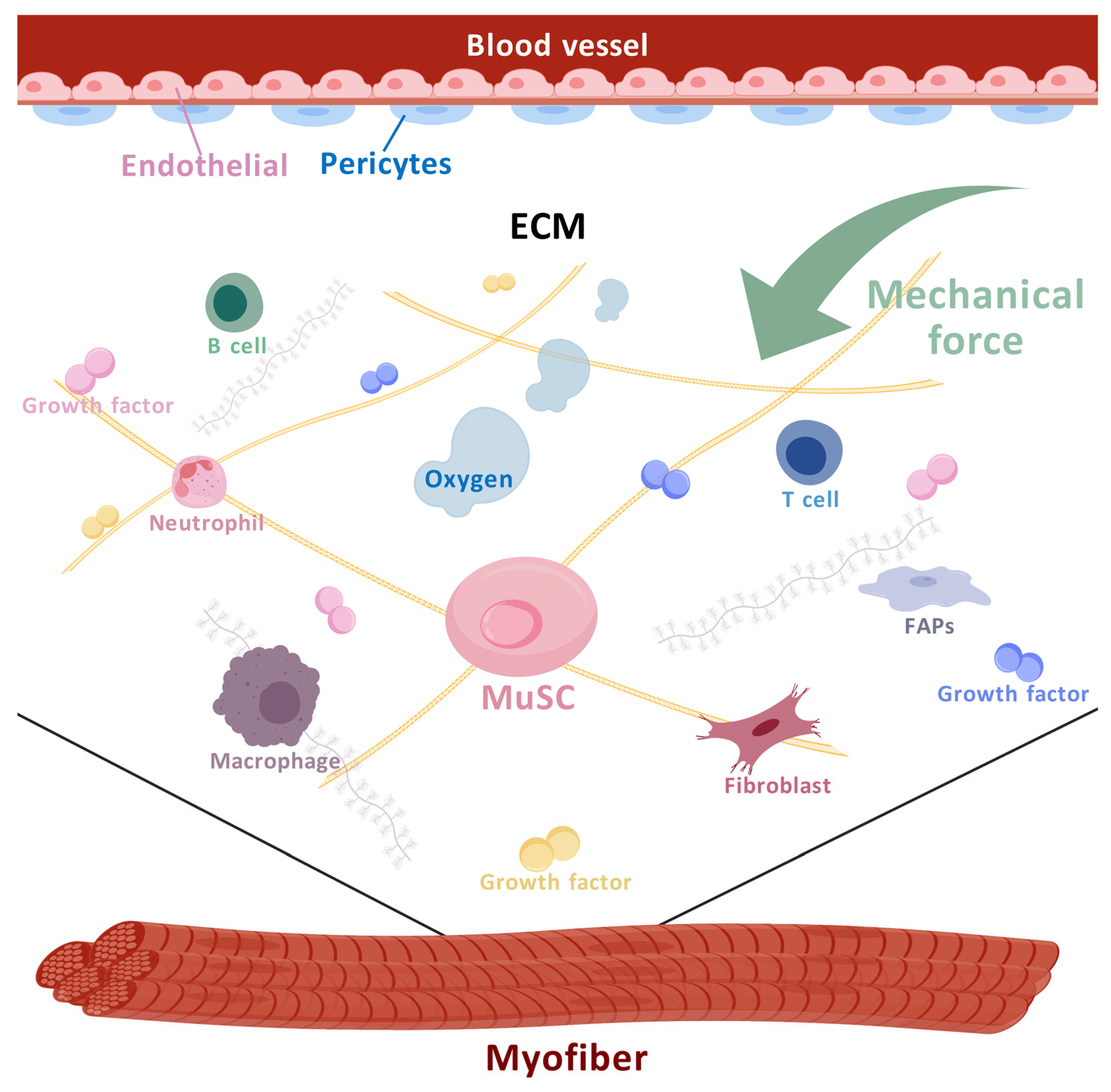
3. Microenvironment of MuSCs and Their Regulation in Muscle Regeneration
3.1. The Cell Components of the MuSC Niche
3.1.1. Myofibers
3.1.2. Other Cellular Components of the MuSC Niche
3.1.3. Immune Cells
3.2. Non-Cellular Niches
3.2.1. ECM and Factors
3.2.2. Mechanical Signals and MuSCs
3.2.3. Oxygen Tension and MuSCs
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Severinsen, M.C.K.; Pedersen, B.K. Muscle-Organ Crosstalk: The Emerging Roles of Myokines. Endocr. Rev. 2020, 41, 594–609. [Google Scholar] [CrossRef] [PubMed]
- Peault, B.; Rudnicki, M.; Torrente, Y.; Cossu, G.; Tremblay, J.P.; Partridge, T.; Gussoni, E.; Kunkel, L.M.; Huard, J. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol. Ther. 2007, 15, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Yue, L.; Lam, K.S.W.; Zhang, W.; So, W.K.; Tse, E.H.Y.; Cheung, T.H. CPEB1 directs muscle stem cell activation by reprogramming the translational land. Nat. Commun. 2022, 13, 947. [Google Scholar] [CrossRef]
- Zhu, H.; Lin, X.; Diao, Y. Function and regulation of muscle stem cells in skeletal muscle development and regeneration: A narrative review. J. Bio-X Res. 2021, 4, 89–96. [Google Scholar] [CrossRef]
- Loreti, M.; Sacco, A. The jam session between muscle stem cells and the extracellular matrix in the tissue microenvironment. NPJ Regen. Med. 2022, 7, 16. [Google Scholar] [CrossRef]
- Rodgers, J.T.; King, K.Y.; Brett, J.O.; Cromie, M.J.; Charville, G.W.; Maguire, K.K.; Brunson, C.; Mastey, N.; Liu, L.; Tsai, C.R.; et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 2014, 510, 393–396. [Google Scholar] [CrossRef] [PubMed]
- De Micheli, A.J.; Laurilliard, E.J.; Heinke, C.L.; Ravichandran, H.; Fraczek, P.; Soueid-Baumgarten, S.; De Vlaminck, I.; Elemento, O.; Cosgrove, B.D. Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Rep. 2020, 30, 3583–3595.e5. [Google Scholar] [CrossRef]
- Brondolin, M.; Herzog, D.; Sultan, S.; Warburton, F.; Vigilante, A.; Knight, R.D. Migration and differentiation of muscle stem cells are coupled by RhoA signalling during regeneration. Open Biol. 2023, 13, 230037. [Google Scholar] [CrossRef]
- Li, C.; Vargas-Franco, D.; Saha, M.; Davis, R.M.; Manko, K.A.; Draper, I.; Pacak, C.A.; Kang, P.B. Megf10 deficiency impairs skeletal muscle stem cell migration and muscle regeneration. FEBS Open Bio 2021, 11, 114–123. [Google Scholar] [CrossRef]
- Seale, P.; Asakura, A.; Rudnicki, M.A. The potential of muscle stem cells. Dev. Cell 2001, 1, 333–342. [Google Scholar] [CrossRef]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, J.R.; Morgan, J.E.; Pagel, C.N.; Partridge, T.A. Dynamics of myoblast transplantation reveal a discrete minority of precursors with stem cell-like properties as the myogenic source. J. Cell Biol. 1999, 144, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef]
- Zammit, P.S.; Relaix, F.; Nagata, Y.; Ruiz, A.P.; Collins, C.A.; Partridge, T.A.; Beauchamp, J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Oustanina, S.; Hause, G.; Braun, T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004, 23, 3430–3439. [Google Scholar] [CrossRef]
- Von Maltzahn, J.; Jones, A.E.; Parks, R.J.; Rudnicki, M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. USA 2013, 110, 16474–16479. [Google Scholar] [CrossRef] [PubMed]
- Sincennes, M.C.; Brun, C.E.; Lin, A.Y.T.; Rosembert, T.; Datzkiw, D.; Saber, J.; Ming, H.; Kawabe, Y.I.; Rudnicki, M.A. Acetylation of PAX7 controls muscle stem cell self-renewal and differentiation potential in mice. Nat. Commun. 2021, 12, 3253. [Google Scholar] [CrossRef]
- Ortuste Quiroga, H.P.; Fujimaki, S.; Ono, Y. Pax7 reporter mouse models: A pocket guide for satellite cell research. Eur. J. Transl. Myol. 2023, 33, 12174. [Google Scholar] [CrossRef]
- Liu, L.; Cheung, T.H.; Charville, G.W.; Rando, T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015, 10, 1612–1624. [Google Scholar] [CrossRef]
- Maesner, C.C.; Almada, A.E.; Wagers, A.J. Established cell surface markers efficiently isolate highly overlapping populations of skeletal muscle satellite cells by fluorescence-activated cell sorting. Skelet. Muscle 2016, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Tierney, M.T.; Stec, M.J.; Rulands, S.; Simons, B.D.; Sacco, A. Muscle Stem Cells Exhibit Distinct Clonal Dynamics in Response to Tissue Repair and Homeostatic Aging. Cell Stem Cell 2018, 22, 119–127.e3. [Google Scholar] [CrossRef]
- Scaramozza, A.; Park, D.; Kollu, S.; Beerman, I.; Sun, X.; Rossi, D.J.; Lin, C.P.; Scadden, D.T.; Crist, C.; Brack, A.S. Lineage Tracing Reveals a Subset of Reserve Muscle Stem Cells Capable of Clonal Expansion under Stress. Cell Stem Cell 2019, 24, 944–957.e5. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Prat, L.; Perdiguero, E.; Alonso-Martin, S.; Dell’Orso, S.; Ravichandran, S.; Brooks, S.R.; Juan, A.H.; Campanario, S.; Jiang, K.; Hong, X.; et al. FoxO maintains a genuine muscle stem-cell quiescent state until geriatric age. Nat. Cell Biol 2020, 22, 1307–1318. [Google Scholar] [CrossRef]
- Shi, Y.; He, G.; Lee, W.C.; McKenzie, J.A.; Silva, M.J.; Long, F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017, 8, 2043. [Google Scholar] [CrossRef]
- Peng, J.; Han, L.; Liu, B.; Song, J.; Wang, Y.; Wang, K.; Guo, Q.; Liu, X.; Li, Y.; Zhang, J.; et al. Gli1 marks a sentinel muscle stem cell population for muscle regeneration. Nat. Commun. 2023, 14, 6993. [Google Scholar] [CrossRef]
- Barruet, E.; Garcia, S.M.; Striedinger, K.; Wu, J.; Lee, S.; Byrnes, L.; Wong, A.; Xuefeng, S.; Tamaki, S.; Brack, A.S.; et al. Functionally heterogeneous human satellite cells identified by single cell RNA sequencing. eLife 2020, 9, e51576. [Google Scholar] [CrossRef] [PubMed]
- Byun, W.S.; Lee, J.; Baek, J.H. Beyond the bulk: Overview and novel insights into the dynamics of muscle satellite cells during muscle regeneration. Inflamm. Regen. 2024, 44, 39. [Google Scholar] [CrossRef]
- Cho, I.J.; Lui, P.P.; Obajdin, J.; Riccio, F.; Stroukov, W.; Willis, T.L.; Spagnoli, F.; Watt, F.M. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Rep. 2019, 12, 1190–1200. [Google Scholar] [CrossRef]
- De Morree, A.; Rando, T.A. Regulation of adult stem cell quiescence and its functions in the maintenance of tissue integrity. Nat. Rev. Mol. Cell Biol. 2023, 24, 334–354. [Google Scholar] [CrossRef]
- Sousa-Victor, P.; García-Prat, L.; Muñoz-Cánoves, P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2022, 23, 204–226. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.; Kuroda, K.; Le Grand, F.; Rudnicki, M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 2007, 129, 999–1010. [Google Scholar] [CrossRef]
- Feige, P.; Brun, C.E.; Ritso, M.; Rudnicki, M.A. Orienting Muscle Stem Cells for Regeneration in Homeostasis, Aging, and Disease. Cell Stem Cell 2018, 23, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhuang, C.L.; Hu, P. Regulation of muscle stem cell fate. Cell Regen. 2022, 11, 40. [Google Scholar] [CrossRef]
- Borycki, A.G.; Emerson, C.P. Muscle determination: Another key player in myogenesis? Curr. Biol. 1997, 7, R620–R623. [Google Scholar] [CrossRef]
- Han, W.M.; Mohiuddin, M.; Anderson, S.E.; Garcia, A.J.; Jang, Y.C. Co-delivery of Wnt7a and muscle stem cells using synthetic bioadhesive hydrogel enhances murine muscle regeneration and cell migration during engraftment. Acta Biomater. 2019, 94, 243–252. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, L.; Mu, W.; Zhang, Y.; Chen, T.; Wu, J.; Tang, H.; Zheng, S.; Liu, Y.; et al. Generic Diagramming Platform (GDP): A comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2025, 53, D1670–D1676. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Christ, B.; Ordahl, C.P. Early stages of chick somite development. Anat. Embryol. 1995, 191, 381–396. [Google Scholar] [CrossRef]
- Buckingham, M.; Bajard, L.; Chang, T.; Daubas, P.; Hadchouel, J.; Meilhac, S.; Montarras, D.; Rocancourt, D.; Relaix, F. The formation of skeletal muscle: From somite to limb. J. Anat. 2003, 202, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Grefte, S.; Kuijpers-Jagtman, A.M.; Torensma, R.; Von den Hoff, J.W. Skeletal muscle development and regeneration. Stem Cells Dev. 2007, 16, 857–868. [Google Scholar] [CrossRef]
- Kassar-Duchossoy, L.; Giacone, E.; Gayraud-Morel, B.; Jory, A.; Gomès, D.; Tajbakhsh, S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005, 19, 1426–1431. [Google Scholar] [CrossRef]
- Esteves de Lima, J.; Relaix, F. Master regulators of skeletal muscle lineage development and pluripotent stem cells differentiation. Cell Regen. 2021, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Wosczyna, M.N.; Rando, T.A. A Muscle Stem Cell Support Group: Coordinated Cellular Responses in Muscle Regeneration. Dev. Cell 2018, 46, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Baht, G.S.; Bareja, A.; Lee, D.E.; Rao, R.R.; Huang, R.; Huebner, J.L.; Bartlett, D.B.; Hart, C.R.; Gibson, J.R.; Lanza, I.R.; et al. Meteorin-like facilitates skeletal muscle repair through a Stat3/IGF-1 mechanism. Nat. Metab. 2020, 2, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Haroon, M.; Klein-Nulend, J.; Bakker, A.D.; Jin, J.; Seddiqi, H.; Offringa, C.; De Wit, G.M.J.; Le Grand, F.; Giordani, L.; Liu, K.J.; et al. Myofiber stretch induces tensile and shear deformation of muscle stem cells in their native niche. Biophys. J. 2021, 120, 2665–2678. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Kitajima, Y.; Masumoto, H.; Ono, Y. Damaged Myofiber-Derived Metabolic Enzymes Act as Activators of Muscle Satellite Cells. Stem Cell Rep. 2020, 15, 926–940. [Google Scholar] [CrossRef]
- Wang, C.; Rabadan Ros, R.; Martinez-Redondo, P.; Ma, Z.; Shi, L.; Xue, Y.; Guillen-Guillen, I.; Huang, L.; Hishida, T.; Liao, H.K.; et al. In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nat. Commun. 2021, 12, 3094. [Google Scholar] [CrossRef]
- Sciorati, C.; Rigamonti, E.; Manfredi, A.A.; Rovere-Querini, P. Cell death, clearance and immunity in the skeletal muscle. Cell Death Differ. 2016, 23, 927–937. [Google Scholar] [CrossRef]
- Lievens, E.; Klass, M.; Bex, T.; Derave, W. Muscle fiber typology substantially influences time to recover from high-intensity exercise. J. Appl. Physiol. 2020, 128, 648–659. [Google Scholar] [CrossRef]
- Leng, M.; Yang, F.; Zhao, J.; Xiong, Y.; Zhou, Y.; Zhao, M.; Jia, S.; Liu, L.; Zheng, Q.; Gan, L.; et al. Mitophagy-mediated S1P facilitates muscle adaptive responses to endurance exercise through SPHK1-S1PR1/S1PR2 in slow-twitch myofibers. Autophagy 2025, 1–19. [Google Scholar] [CrossRef]
- Liu, J.; Liang, X.; Zhou, D.; Lai, L.; Xiao, L.; Liu, L.; Fu, T.; Kong, Y.; Zhou, Q.; Vega, R.B.; et al. Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol. Med. 2016, 8, 1212–1228. [Google Scholar] [CrossRef]
- Widrick, J.J.; Trappe, S.W.; Costill, D.L.; Fitts, R.H. Force-velocity and force-power properties of single muscle fibers from elite master runners and sedentary men. Am. J. Physiol. 1996, 271, C676–C683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lin, S.; Luo, W.; Ren, T.; Huang, X.; Li, W.; Zhang, X. Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles. Int. J. Mol. Sci. 2022, 23, 11327. [Google Scholar] [CrossRef] [PubMed]
- Zimowska, M.; Kasprzycka, P.; Bocian, K.; Delaney, K.; Jung, P.; Kuchcinska, K.; Kaczmarska, K.; Gladysz, D.; Streminska, W.; Ciemerych, M.A. Inflammatory response during slow- and fast-twitch muscle regeneration. Muscle Nerve 2017, 55, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Cappellari, O.; Cossu, G. Pericytes in development and pathology of skeletal muscle. Circ. Res. 2013, 113, 341–347. [Google Scholar] [CrossRef]
- Kruger-Genge, A.; Blocki, A.; Franke, R.P.; Jung, F. Vascular Endothelial Cell Biology: An Update. Int. J. Mol. Sci. 2019, 20, 4411. [Google Scholar] [CrossRef] [PubMed]
- Uezumi, A.; Ito, T.; Morikawa, D.; Shimizu, N.; Yoneda, T.; Segawa, M.; Yamaguchi, M.; Ogawa, R.; Matev, M.M.; Miyagoe-Suzuki, Y.; et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J. Cell Sci. 2011, 124, 3654–3664. [Google Scholar] [CrossRef]
- Chapman, M.A.; Meza, R.; Lieber, R.L. Skeletal muscle fibroblasts in health and disease. Differentiation 2016, 92, 108–115. [Google Scholar] [CrossRef]
- Klingler, W.; Jurkat-Rott, K.; Lehmann-Horn, F.; Schleip, R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol. 2012, 31, 184–195. [Google Scholar]
- Zhou, S.; Zhang, W.; Cai, G.; Ding, Y.; Wei, C.; Li, S.; Yang, Y.; Qin, J.; Liu, D.; Zhang, H.; et al. Myofiber necroptosis promotes muscle stem cell proliferation via releasing Tenascin-C during regeneration. Cell Res. 2020, 30, 1063–1077. [Google Scholar] [CrossRef]
- Beltra, M.; Pin, F.; Costamagna, D.; Duelen, R.; Renzini, A.; Ballaro, R.; Garcia-Castillo, L.; Iannuzzi, A.; Moresi, V.; Coletti, D.; et al. PGC-1alpha in the myofibers regulates the balance between myogenic and adipogenic progenitors affecting muscle regeneration. iScience 2022, 25, 105480. [Google Scholar] [CrossRef]
- Webster, M.T.; Manor, U.; Lippincott-Schwartz, J.; Fan, C.M. Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration. Cell Stem Cell 2016, 18, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Asakura, Y.; Wang, X.; Zhou, K.; Unverdi, M.; Kann, A.P.; Krauss, R.S.; Asakura, A. Endothelial cell signature in muscle stem cells validated by VEGFA-FLT1-AKT1 axis promoting survival of muscle stem cell. eLife 2024, 13, e73592. [Google Scholar] [CrossRef]
- Zhang, J.; Muri, J.; Fitzgerald, G.; Gorski, T.; Gianni-Barrera, R.; Masschelein, E.; D’Hulst, G.; Gilardoni, P.; Turiel, G.; Fan, Z.; et al. Endothelial Lactate Controls Muscle Regeneration from Ischemia by Inducing M2-like Macrophage Polarization. Cell Metab. 2020, 31, 1136–1153.e7. [Google Scholar] [CrossRef]
- Kang, X.; Qian, J.; Shi, Y.X.; Bian, X.T.; Zhang, L.D.; Li, G.M.; Wang, L.T.; Zhao, J.; Dong, Z.Y.; Yang, M.M.; et al. Exercise-induced Musclin determines the fate of fibro-adipogenic progenitors to control muscle homeostasis. Cell Stem Cell 2024, 31, 212–226.e7. [Google Scholar] [CrossRef]
- Sastourne-Arrey, Q.; Mathieu, M.; Contreras, X.; Monferran, S.; Bourlier, V.; Gil-Ortega, M.; Murphy, E.; Laurens, C.; Varin, A.; Guissard, C.; et al. Adipose tissue is a source of regenerative cells that augment the repair of skeletal muscle after injury. Nat. Commun. 2023, 14, 80. [Google Scholar] [CrossRef]
- Yu, Y.; Su, Y.; Wang, G.; Lan, M.; Liu, J.; Garcia Martin, R.; Brandao, B.B.; Lino, M.; Li, L.; Liu, C.; et al. Reciprocal communication between FAPs and muscle cells via distinct extracellular vesicle miRNAs in muscle regeneration. Proc. Natl. Acad. Sci. USA 2024, 121, e2316544121. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yang, J.; An, Y.; Wang, J.; Tan, S.; Xu, H.; Dong, Y. MST1/2 regulates fibro/adipogenic progenitor fate decisions in skeletal muscle regeneration. Stem Cell Rep. 2024, 19, 501–514. [Google Scholar] [CrossRef] [PubMed]
- De Lima, J.E.; Blavet, C.; Bonnin, M.A.; Hirsinger, E.; Comai, G.; Yvernogeau, L.; Delfini, M.C.; Bellenger, L.; Mella, S.; Nassari, S.; et al. Unexpected contribution of fibroblasts to muscle lineage as a mechanism for limb muscle patterning. Nat. Commun. 2021, 12, 3851. [Google Scholar] [CrossRef]
- Sicherer, S.T.; Venkatarama, R.S.; Grasman, J.M. Recent Trends in Injury Models to Study Skeletal Muscle Regeneration and Repair. Bioengineering 2020, 7, 76. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Liu, Y. Skeletal Muscle Regeneration in Cardiotoxin-Induced Muscle Injury Models. Int. J. Mol. Sci. 2022, 23, 13380. [Google Scholar] [CrossRef]
- Kumar, A.; Hardy, D.; Besnard, A.; Latil, M.; Jouvion, G.; Briand, D.; Thépenier, C.; Pascal, Q.; Guguin, A.; Gayraud-Morel, B.; et al. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS ONE 2016, 11, e0147198. [Google Scholar] [CrossRef]
- Marshall, J.S.; Warrington, R.; Watson, W.; Kim, H.L. An introduction to immunology and immunopathology. Allergy Asthma Clin. Immunol. 2018, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.J.; Szymczak-Workman, A.L.; Collison, L.W.; Pillai, M.R.; Vignali, D.A. The development and function of regulatory T cells. Cell. Mol. Life Sci. 2009, 66, 2603–2622. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010, 125, S33–S40. [Google Scholar] [CrossRef] [PubMed]
- Kushioka, J.; Chow, S.K.; Toya, M.; Tsubosaka, M.; Shen, H.; Gao, Q.; Li, X.; Zhang, N.; Goodman, S.B. Bone regeneration in inflammation with aging and cell-based immunomodulatory therapy. Inflamm. Regen. 2023, 43, 29. [Google Scholar] [CrossRef]
- Prame Kumar, K.; Nicholls, A.J.; Wong, C.H.Y. Partners in crime: Neutrophils and monocytes/macrophages in inflammation and disease. Cell Tissue Res. 2018, 371, 551–565. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, L. The multifaceted role of macrophages in homeostatic and injured skeletal muscle. Front. Immunol. 2023, 14, 1274816. [Google Scholar] [CrossRef]
- Ziemkiewicz, N.; Hilliard, G.; Pullen, N.A.; Garg, K. The Role of Innate and Adaptive Immune Cells in Skeletal Muscle Regeneration. Int. J. Mol. Sci. 2021, 22, 3265. [Google Scholar] [CrossRef]
- Yahara, Y.; Ma, X.; Gracia, L.; Alman, B.A. Monocyte/Macrophage Lineage Cells From Fetal Erythromyeloid Progenitors Orchestrate Bone Remodeling and Repair. Front. Cell. Dev. Biol. 2021, 9, 622035. [Google Scholar] [CrossRef]
- Larouche, J.A.; Fraczek, P.M.; Kurpiers, S.J.; Yang, B.A.; Davis, C.; Castor-Macias, J.A.; Sabin, K.; Anderson, S.; Harrer, J.; Hall, M.; et al. Neutrophil and natural killer cell imbalances prevent muscle stem cell-mediated regeneration following murine volumetric muscle loss. Proc. Natl. Acad. Sci. USA 2022, 119, e2111445119. [Google Scholar] [CrossRef]
- Graca, F.A.; Stephan, A.; Minden-Birkenmaier, B.A.; Shirinifard, A.; Wang, Y.D.; Demontis, F.; Labelle, M. Platelet-derived chemokines promote skeletal muscle regeneration by guiding neutrophil recruitment to injured muscles. Nat. Commun. 2023, 14, 2900. [Google Scholar] [CrossRef]
- Hoeffel, G.; Debroas, G.; Roger, A.; Rossignol, R.; Gouilly, J.; Laprie, C.; Chasson, L.; Barbon, P.V.; Balsamo, A.; Reynders, A.; et al. Sensory neuron-derived TAFA4 promotes macrophage tissue repair functions. Nature 2021, 594, 94–99. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Nayer, B.; Singh, S.K.; Alshoubaki, Y.K.; Yuan, E.; Park, A.J.; Maruyama, K.; Akira, S.; Martino, M.M. CGRP sensory neurons promote tissue healing via neutrophils and macrophages. Nature 2024, 628, 604–611. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Kikuta, J.; Matsui, T.; Hasegawa, T.; Fujii, K.; Okuzaki, D.; Liu, Y.C.; Yoshioka, T.; Seno, S.; Motooka, D.; et al. Periportal macrophages protect against commensal-driven liver inflammation. Nature 2024, 629, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Cappellesso, F.; Amorim, R.; Serneels, J.; Virga, F.; Eelen, G.; Carobbio, S.; Rincon, M.Y.; Maechler, P.; De Bock, K.; et al. Macrophage-derived glutamine boosts satellite cells and muscle regeneration. Nature 2020, 587, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, D.; Nguyen, P.D.; Rossello, F.J.; Wimmer, V.C.; Tan, J.L.; Galvis, L.A.; Julier, Z.; Wood, A.J.; Boudier, T.; Isiaku, A.I.; et al. Macrophages provide a transient muscle stem cell niche via NAMPT secretion. Nature 2021, 591, 281–287. [Google Scholar] [CrossRef]
- He, Y.; Heng, Y.; Qin, Z.; Wei, X.; Wu, Z.; Qu, J. Intravital microscopy of satellite cell dynamics and their interaction with myeloid cells during skeletal muscle regeneration. Sci. Adv. 2023, 9, eadi1891. [Google Scholar] [CrossRef] [PubMed]
- Babaeijandaghi, F.; Cheng, R.; Kajabadi, N.; Soliman, H.; Chang, C.K.; Smandych, J.; Tung, L.W.; Long, R.; Ghassemi, A.; Rossi, F.M.V. Metabolic reprogramming of skeletal muscle by resident macrophages points to CSF1R inhibitors as muscular dystrophy therapeutics. Sci. Transl. Med. 2022, 14, eabg7504. [Google Scholar] [CrossRef]
- Krasniewski, L.K.; Chakraborty, P.; Cui, C.Y.; Mazan-Mamczarz, K.; Dunn, C.; Piao, Y.; Fan, J.; Shi, C.; Wallace, T.; Nguyen, C.; et al. Single-cell analysis of skeletal muscle macrophages reveals age-associated functional subpopulations. eLife 2022, 11, e77974. [Google Scholar] [CrossRef]
- Sousa, N.S.; Bica, M.; Bras, M.F.; Sousa, A.C.; Antunes, I.B.; Encarnacao, I.A.; Costa, T.M.; Martins, I.B.; Barbosa-Morais, N.L.; Sousa-Victor, P.; et al. The immune landscape of murine skeletal muscle regeneration and aging. Cell Rep. 2024, 43, 114975. [Google Scholar] [CrossRef]
- Burzyn, D.; Kuswanto, W.; Kolodin, D.; Shadrach, J.L.; Cerletti, M.; Jang, Y.; Sefik, E.; Tan, T.G.; Wagers, A.J.; Benoist, C.; et al. A special population of regulatory T cells potentiates muscle repair. Cell 2013, 155, 1282–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, Z.; Qu, C.; Cui, W.; Wang, X.; Du, J. CD8 T cells are involved in skeletal muscle regeneration through facilitating MCP-1 secretion and Gr1(high) macrophage infiltration. J. Immunol. 2014, 193, 5149–5160. [Google Scholar] [CrossRef] [PubMed]
- Kuswanto, W.; Burzyn, D.; Panduro, M.; Wang, K.K.; Jang, Y.C.; Wagers, A.J.; Benoist, C.; Mathis, D. Poor Repair of Skeletal Muscle in Aging Mice Reflects a Defect in Local, Interleukin-33-Dependent Accumulation of Regulatory T Cells. Immunity 2016, 44, 355–367. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Mann, D.M.; Ruoslahti, E. Negative regulation of transforming growth factor-beta by the proteoglycan decorin. Nature 1990, 346, 281–284. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, D.; Xu, L.; Xu, Y.; Gao, Z.; Pan, Y.; Jiang, M.; Wei, Y.; Wang, L.; Liao, Y.; et al. Harnessing matrix stiffness to engineer a bone marrow niche for hematopoietic stem cell rejuvenation. Cell Stem Cell 2023, 30, 378–395.e378. [Google Scholar] [CrossRef]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef]
- Mavropalias, G.; Boppart, M.; Usher, K.M.; Grounds, M.D.; Nosaka, K.; Blazevich, A.J. Exercise builds the scaffold of life: Muscle extracellular matrix biomarker responses to physical activity, inactivity, and aging. Biol. Rev. Camb. Philos. Soc. 2023, 98, 481–519. [Google Scholar] [CrossRef] [PubMed]
- Lukjanenko, L.; Jung, M.J.; Hegde, N.; Perruisseau-Carrier, C.; Migliavacca, E.; Rozo, M.; Karaz, S.; Jacot, G.; Schmidt, M.; Li, L.; et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 2016, 22, 897–905. [Google Scholar] [CrossRef]
- Martins, S.G.; Ribeiro, V.; Melo, C.; Paulino-Cavaco, C.; Antonini, D.; Dayalan Naidu, S.; Murtinheira, F.; Fonseca, I.; Saget, B.; Pita, M.; et al. Laminin-α2 chain deficiency in skeletal muscle causes dysregulation of multiple cellular mechanisms. Life Sci. Alliance 2024, 7, e202402829. [Google Scholar] [CrossRef]
- Mousavizadeh, R.; Hojabrpour, P.; Eltit, F.; McDonald, P.C.; Dedhar, S.; McCormack, R.G.; Duronio, V.; Jafarnejad, S.M.; Scott, A. β1 integrin, ILK and mTOR regulate collagen synthesis in mechanically loaded tendon cells. Sci. Rep. 2020, 10, 12644. [Google Scholar] [CrossRef] [PubMed]
- Adair-Kirk, T.L.; Atkinson, J.J.; Broekelmann, T.J.; Doi, M.; Tryggvason, K.; Miner, J.H.; Mecham, R.P.; Senior, R.M. A site on laminin alpha 5, AQARSAASKVKVSMKF, induces inflammatory cell production of matrix metalloproteinase-9 and chemotaxis. J. Immunol. 2003, 171, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Sarate, R.M.; Hochstetter, J.; Valet, M.; Hallou, A.; Song, Y.; Bansaccal, N.; Ligare, M.; Aragona, M.; Engelman, D.; Bauduin, A.; et al. Dynamic regulation of tissue fluidity controls skin repair during wound healing. Cell 2024, 187, 5298–5315.e19. [Google Scholar] [CrossRef] [PubMed]
- Gillies, A.R.; Lieber, R.L. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef]
- Meyer, G.A.; Lieber, R.L. Elucidation of extracellular matrix mechanics from muscle fibers and fiber bundles. J. Biomech. 2011, 44, 771–773. [Google Scholar] [CrossRef]
- Urciuolo, A.; Quarta, M.; Morbidoni, V.; Gattazzo, F.; Molon, S.; Grumati, P.; Montemurro, F.; Tedesco, F.S.; Blaauw, B.; Cossu, G.; et al. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 2013, 4, 1964. [Google Scholar] [CrossRef]
- Metti, S.; Da Ros, F.; Toniato, G.; Cescon, M.; Bonaldo, P. Native collagen VI delays early muscle stem cell differentiation. J. Cell Sci. 2024, 137, jcs261419. [Google Scholar] [CrossRef]
- Van Ry, P.M.; Minogue, P.; Hodges, B.L.; Burkin, D.J. Laminin-111 improves muscle repair in a mouse model of merosin-deficient congenital muscular dystrophy. Hum. Mol. Genet. 2014, 23, 383–396. [Google Scholar] [CrossRef]
- Yao, Y.; Norris, E.H.; Mason, C.E.; Strickland, S. Laminin regulates PDGFRbeta(+) cell stemness and muscle development. Nat. Commun. 2016, 7, 11415. [Google Scholar] [CrossRef]
- Rayagiri, S.S.; Ranaldi, D.; Raven, A.; Mohamad Azhar, N.I.F.; Lefebvre, O.; Zammit, P.S.; Borycki, A.G. Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat. Commun. 2018, 9, 1075. [Google Scholar] [CrossRef]
- Stearns-Reider, K.M.; Hicks, M.R.; Hammond, K.G.; Reynolds, J.C.; Maity, A.; Kurmangaliyev, Y.Z.; Chin, J.; Stieg, A.Z.; Geisse, N.A.; Hohlbauch, S.; et al. Myoscaffolds reveal laminin scarring is detrimental for stem cell function while sarcospan induces compensatory fibrosis. NPJ Regen. Med. 2023, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Hayward, M.K.; Muncie, J.M.; Weaver, V.M. Tissue mechanics in stem cell fate, development, and cancer. Dev. Cell 2021, 56, 1833–1847. [Google Scholar] [CrossRef] [PubMed]
- Ladoux, B.; Mege, R.M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017, 18, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Wickstrom, S.A. Mechanical state transitions in the regulation of tissue form and function. Nat. Rev. Mol. Cell Biol. 2024, 25, 654–670. [Google Scholar] [CrossRef]
- Lemke, S.B.; Schnorrer, F. Mechanical forces during muscle development. Mech. Dev. 2017, 144, 92–101. [Google Scholar] [CrossRef]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef]
- Tao, J.; Choudhury, M.I.; Maity, D.; Kim, T.; Sun, S.X.; Fan, C.M. Mechanical compression creates a quiescent muscle stem cell niche. Commun. Biol. 2023, 6, 43. [Google Scholar] [CrossRef]
- Ma, N.; Chen, D.; Lee, J.H.; Kuri, P.; Hernandez, E.B.; Kocan, J.; Mahmood, H.; Tichy, E.D.; Rompolas, P.; Mourkioti, F. Piezo1 regulates the regenerative capacity of skeletal muscles via orchestration of stem cell morphological states. Sci. Adv. 2022, 8, eabn0485. [Google Scholar] [CrossRef]
- Hirano, K.; Tsuchiya, M.; Shiomi, A.; Takabayashi, S.; Suzuki, M.; Ishikawa, Y.; Kawano, Y.; Takabayashi, Y.; Nishikawa, K.; Nagao, K.; et al. The mechanosensitive ion channel PIEZO1 promotes satellite cell function in muscle regeneration. Life Sci. Alliance 2023, 6, e202201783. [Google Scholar] [CrossRef]
- Mohyeldin, A.; Garzon-Muvdi, T.; Quinones-Hinojosa, A. Oxygen in stem cell biology: A critical component of the stem cell niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Pouysségur, J. Oxygen, a source of life and stress. FEBS Lett. 2007, 581, 3582–3591. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wen, Y.; Bi, P.; Lai, X.; Liu, X.S.; Liu, X.; Kuang, S. Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development 2012, 139, 2857–2865. [Google Scholar] [CrossRef] [PubMed]
- Redshaw, Z.; Loughna, P.T. Oxygen concentration modulates the differentiation of muscle stem cells toward myogenic and adipogenic fates. Differentiation 2012, 84, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Couture, V.; Lauzon, M.A.; Balg, F.; Faucheux, N.; Grenier, G. Muscle injury-induced hypoxia alters the proliferation and differentiation potentials of muscle resident stromal cells. Skelet. Muscle 2019, 9, 18. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, J.; Zhou, Q.; He, X.; Zheng, Z.; Wei, Y.; Zhou, K.; Lin, Y.; Yu, H.; Zhang, H.; et al. Hypoxia induces mitochondrial protein lactylation to limit oxidative phosphorylation. Cell Res. 2024, 34, 13–30. [Google Scholar] [CrossRef]
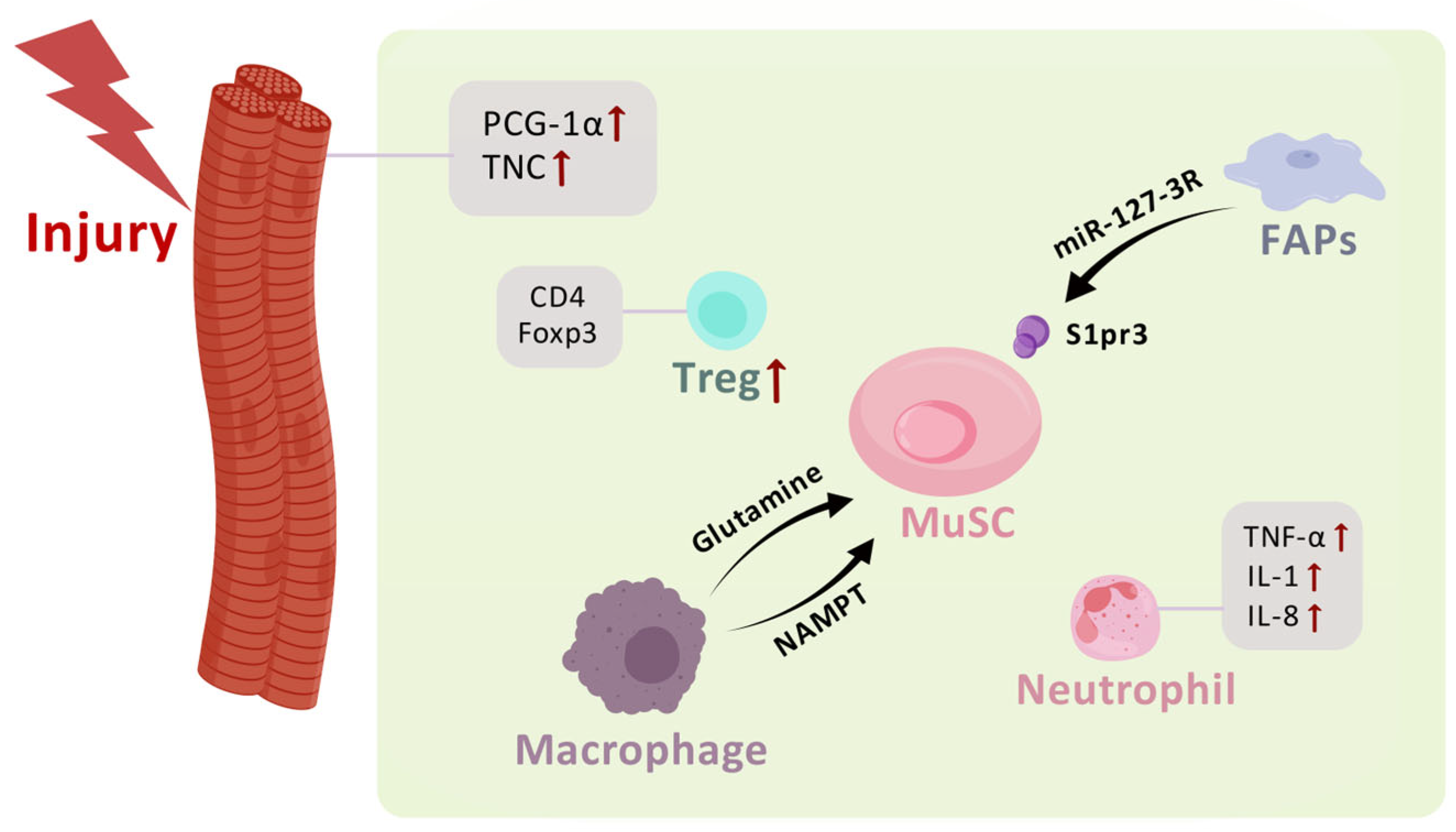
| Factor | Effects |
|---|---|
| PCG-1α | Increased myogenic progenitors |
| TNC | Promoted MuSC proliferation |
| Glutamine | Promoted MuSC proliferation and differentiation |
| NAMPT | Promoted MuSC proliferation |
| mir-127-3R | Activated myogenesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Chen, M.; Zhang, L. Muscle Stem Cell Microenvironment and Functions in Muscle Regeneration. Biomolecules 2025, 15, 765. https://doi.org/10.3390/biom15060765
Li W, Chen M, Zhang L. Muscle Stem Cell Microenvironment and Functions in Muscle Regeneration. Biomolecules. 2025; 15(6):765. https://doi.org/10.3390/biom15060765
Chicago/Turabian StyleLi, Wenjing, Minyou Chen, and Lingli Zhang. 2025. "Muscle Stem Cell Microenvironment and Functions in Muscle Regeneration" Biomolecules 15, no. 6: 765. https://doi.org/10.3390/biom15060765
APA StyleLi, W., Chen, M., & Zhang, L. (2025). Muscle Stem Cell Microenvironment and Functions in Muscle Regeneration. Biomolecules, 15(6), 765. https://doi.org/10.3390/biom15060765







