Proteomic Analysis Reveals the Phenotypic Heterogeneity and Tolerance Mechanisms of Halophilic Vibrio parahaemolyticus Under Dual Stress of Low Salinity and Bile Salts in the Human Intestine
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Cell Lines, and Culture Conditions
2.2. Preparation of Media with Different Salinities
2.3. Bacterial Growth Curve Determination
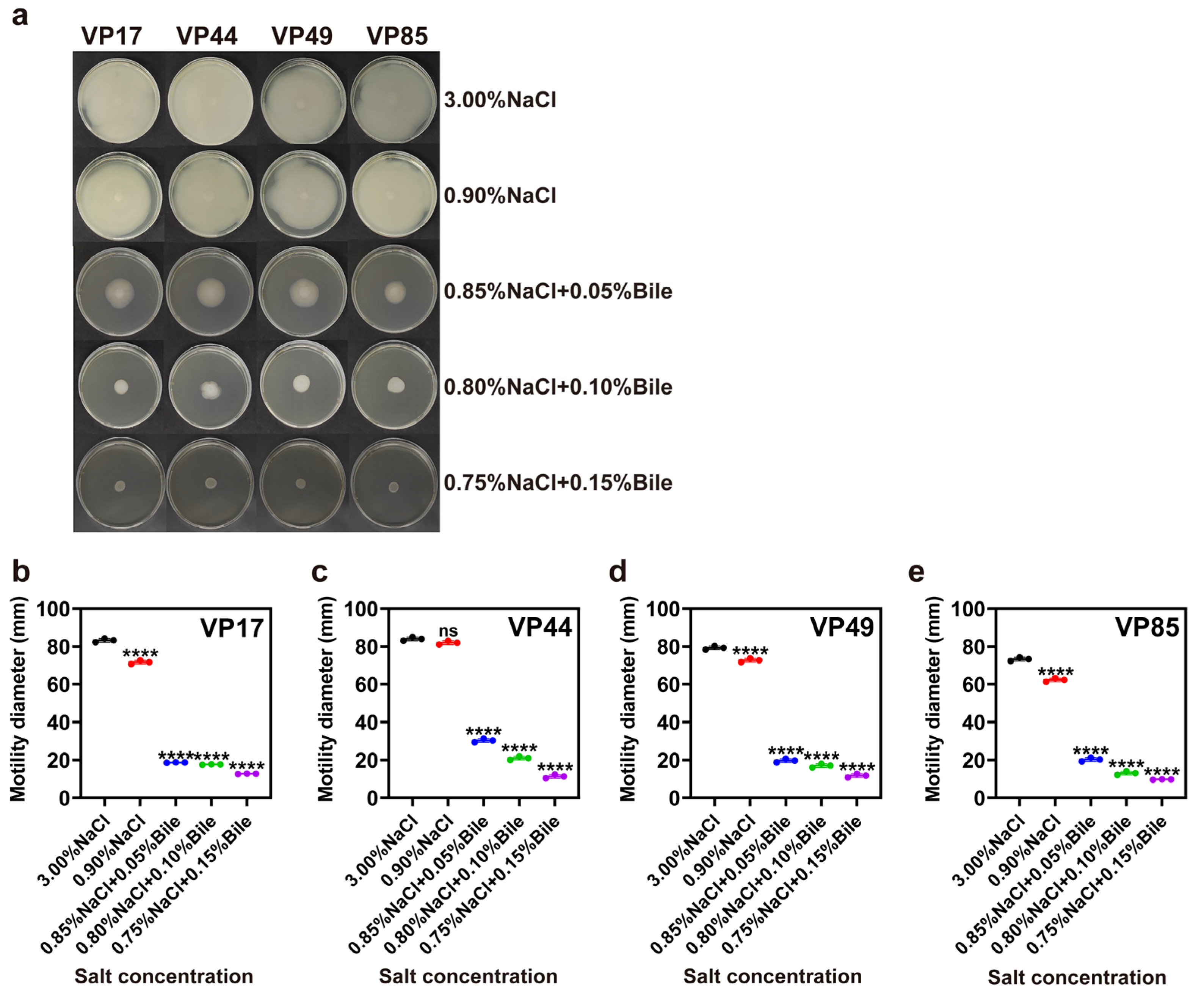
2.4. Bacterial Motility Assay
2.5. Measurement of Biofilm Production
2.6. Confocal Laser Scanning Microscope (CLSM) Observation of Biofilm Structure
2.7. CLSM for Cell Membrane Damage Study
2.8. Scanning Electron Microscopy (SEM) for Cell Membrane Damage Study
2.9. Proteomic Analysis of VP17 Under Different Salinity Stresses
2.10. Intracellular ROS of V. parahaemolyticus Under Different Salinity Stresses
2.11. CCK-8 Assay for Cytostatic Rate
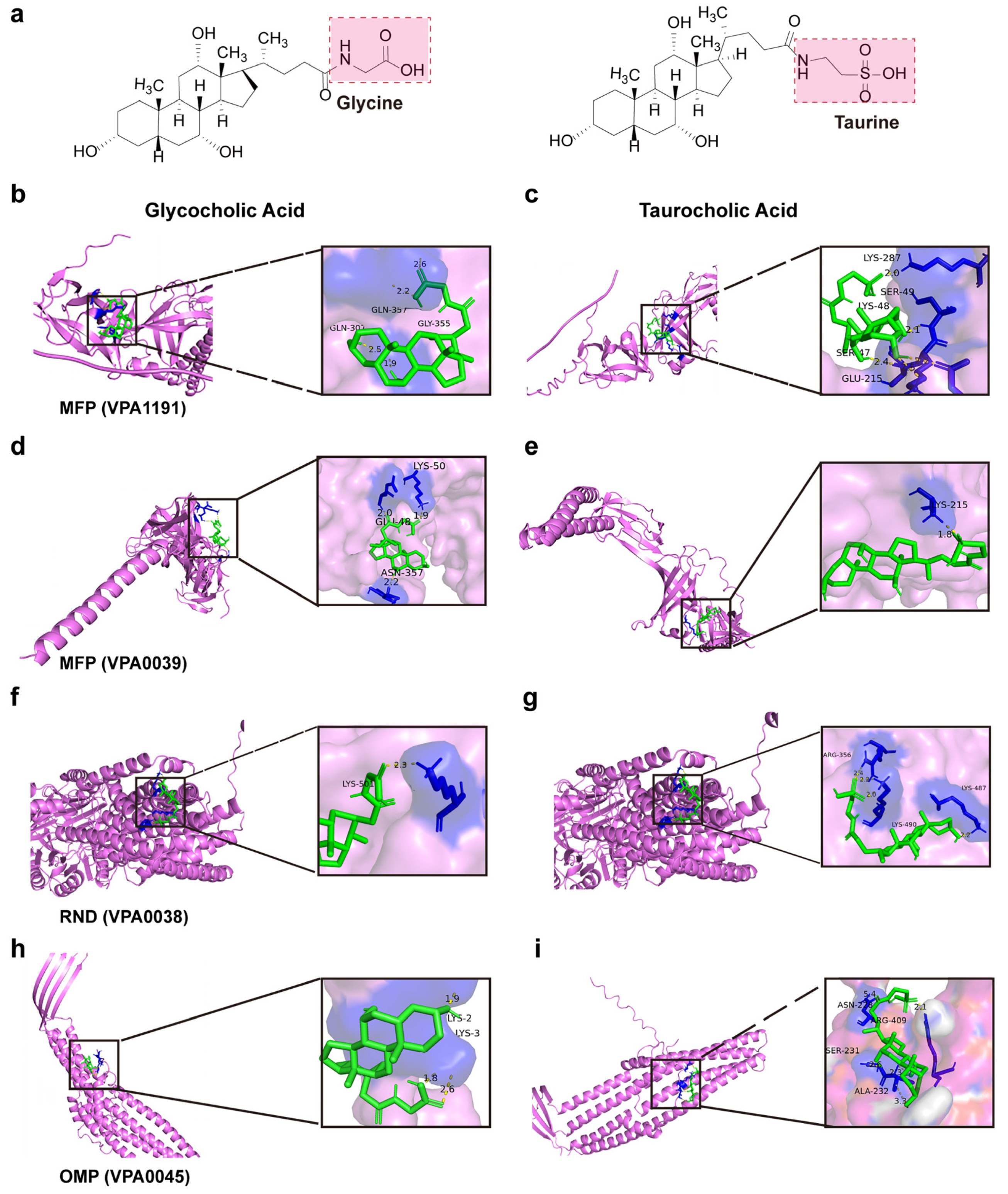
2.12. Visualization of the Molecular Docking Between Efflux Pump Proteins and Bile Salts Molecules
2.13. Data Processing and Statistical Analysis
3. Results
3.1. Growth
3.2. Motility
3.3. Biofilm Membrane Production
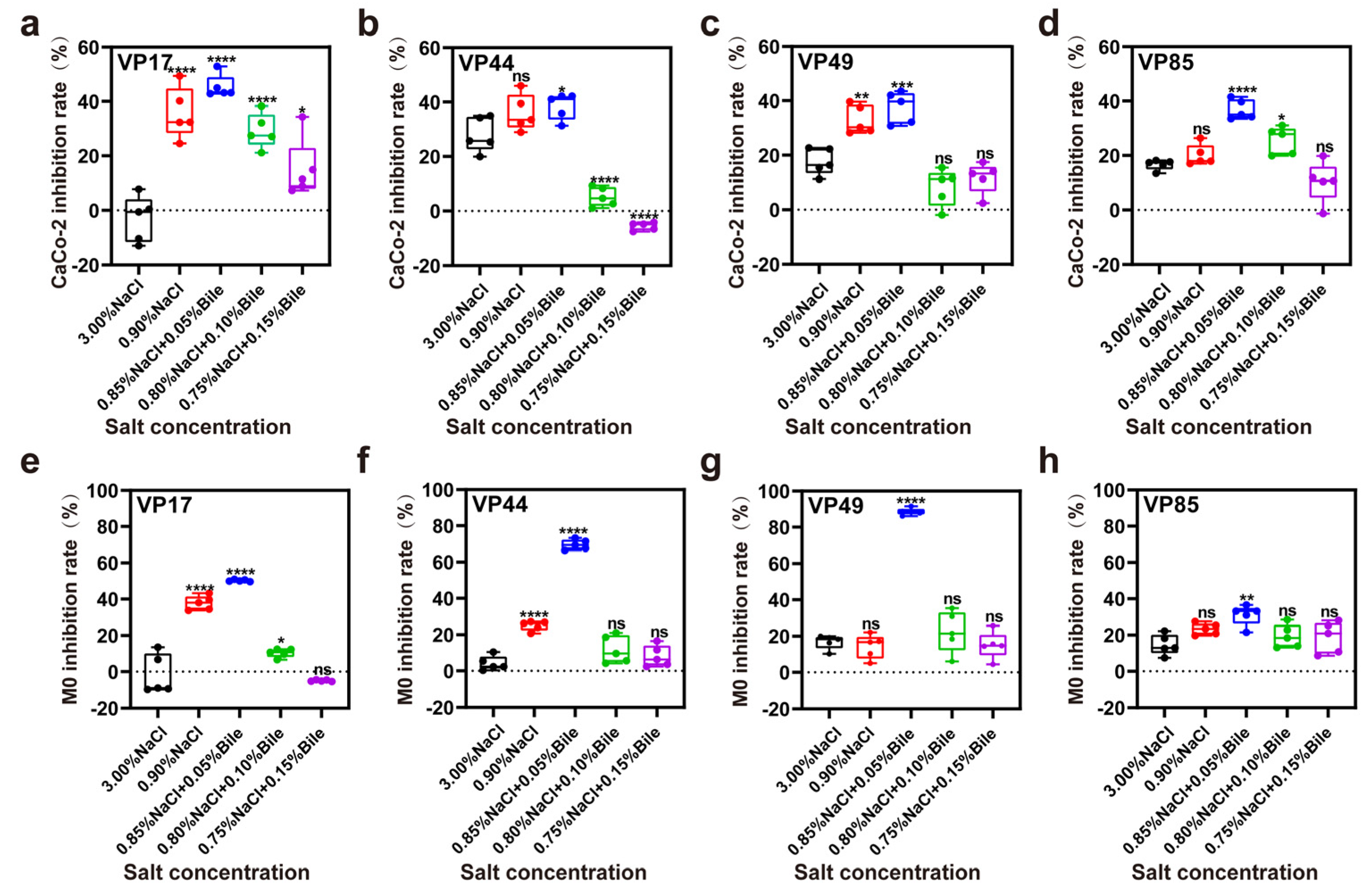
3.4. ROS Levels
3.5. Inhibition Rate of Cell
3.6. Proteomic Analysis of V. parahaemolyticus Under Dual Stress of Low Salinity and Bile Salts
3.6.1. PCA Analysis of V. parahaemolyticus Protein Data Under Different Salinity Stresses
3.6.2. Differential Expression Protein Analysis
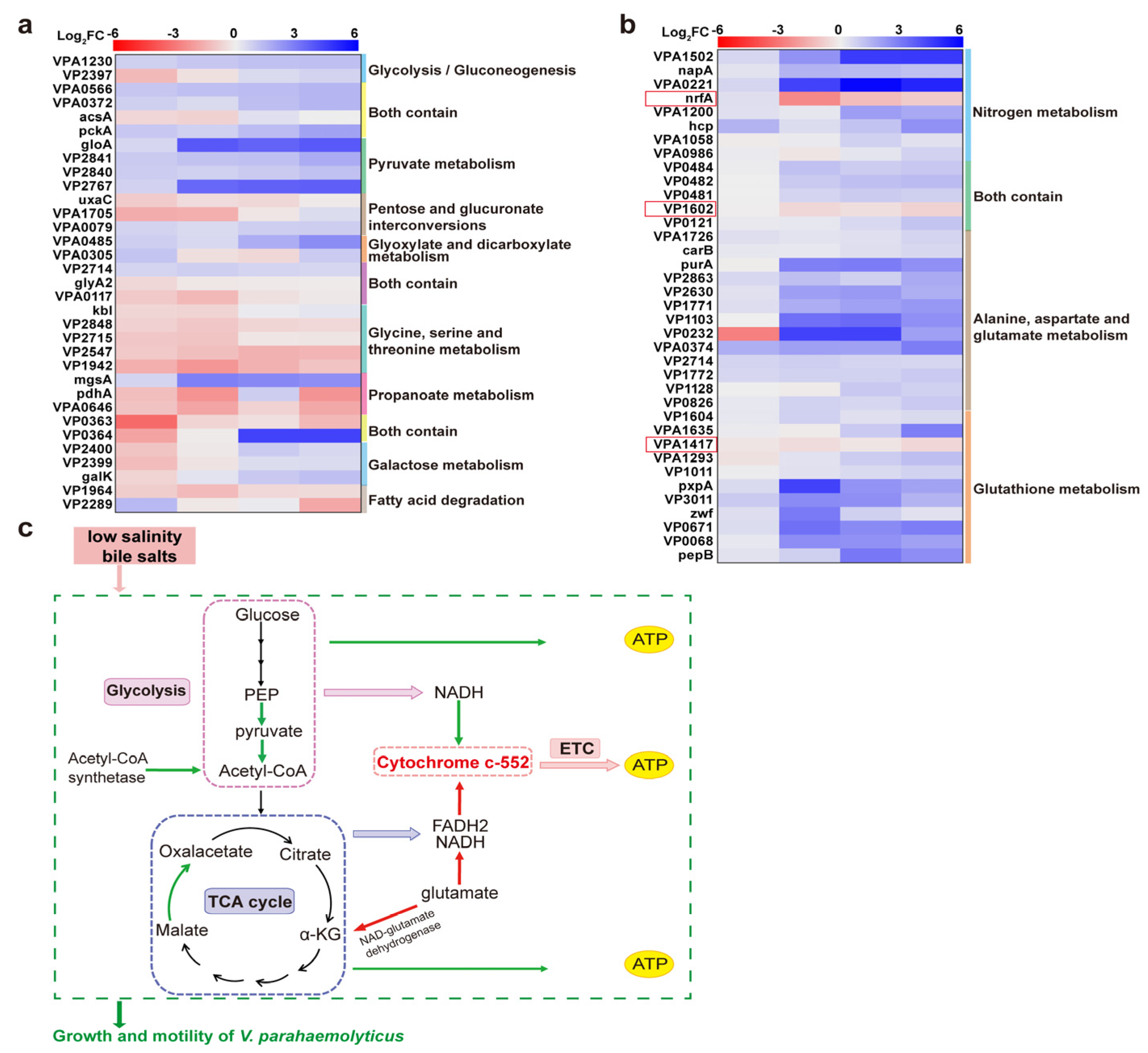
3.6.3. GO Annotation and KEGG Enrichment Results of DEPs
4. Discussion
4.1. Downregulation of Energy Metabolism-Related Proteins Inhibited the Growth of Strains
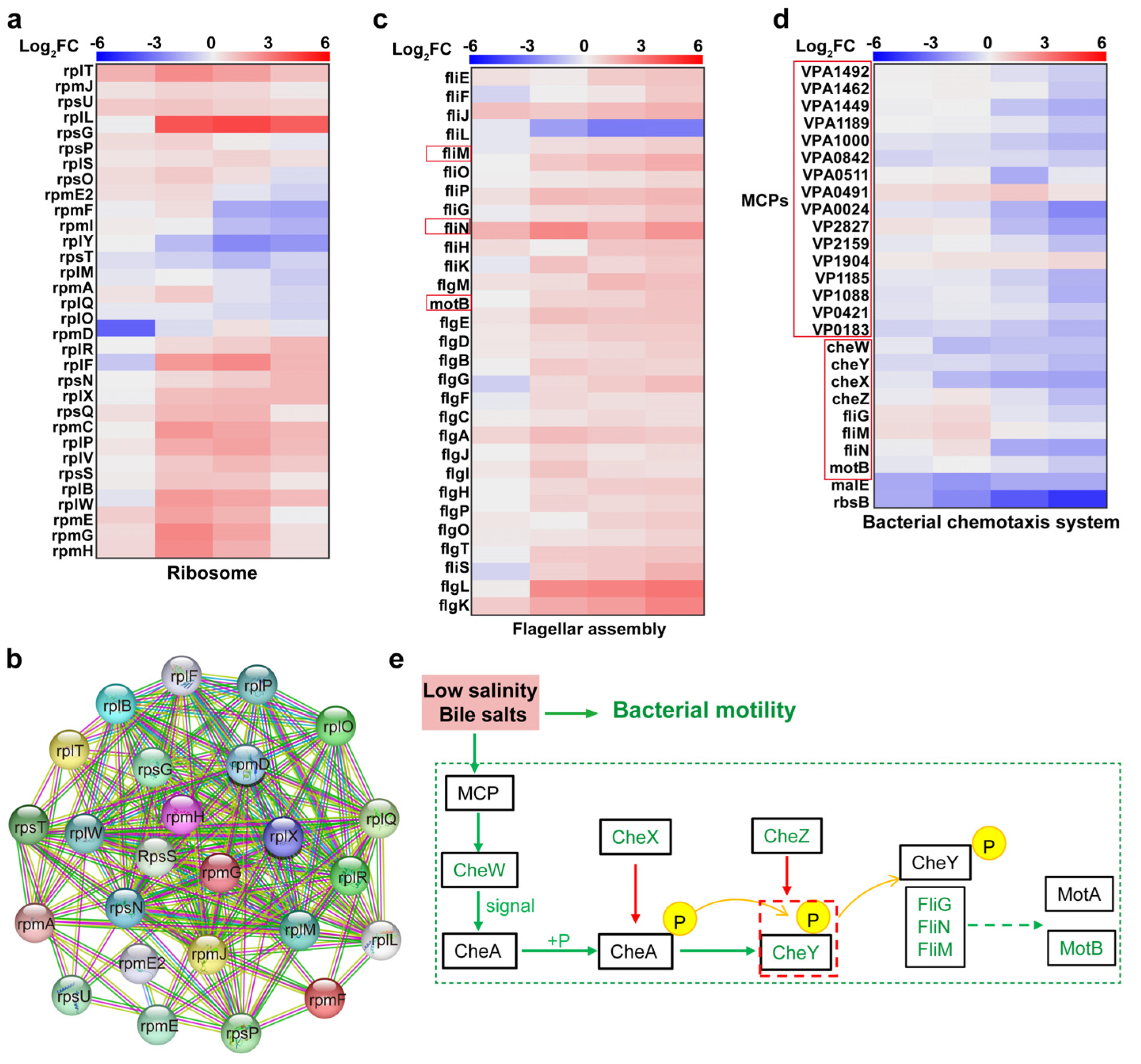
4.2. Upregulation of Ribosome-Associated Proteins to Synthesise Cell Membrane Repair-Associated Proteins
4.3. Downregulation of Chemotactic System and Flagellar Motor Proteins Inhibited V. parahaemolyticus Motility
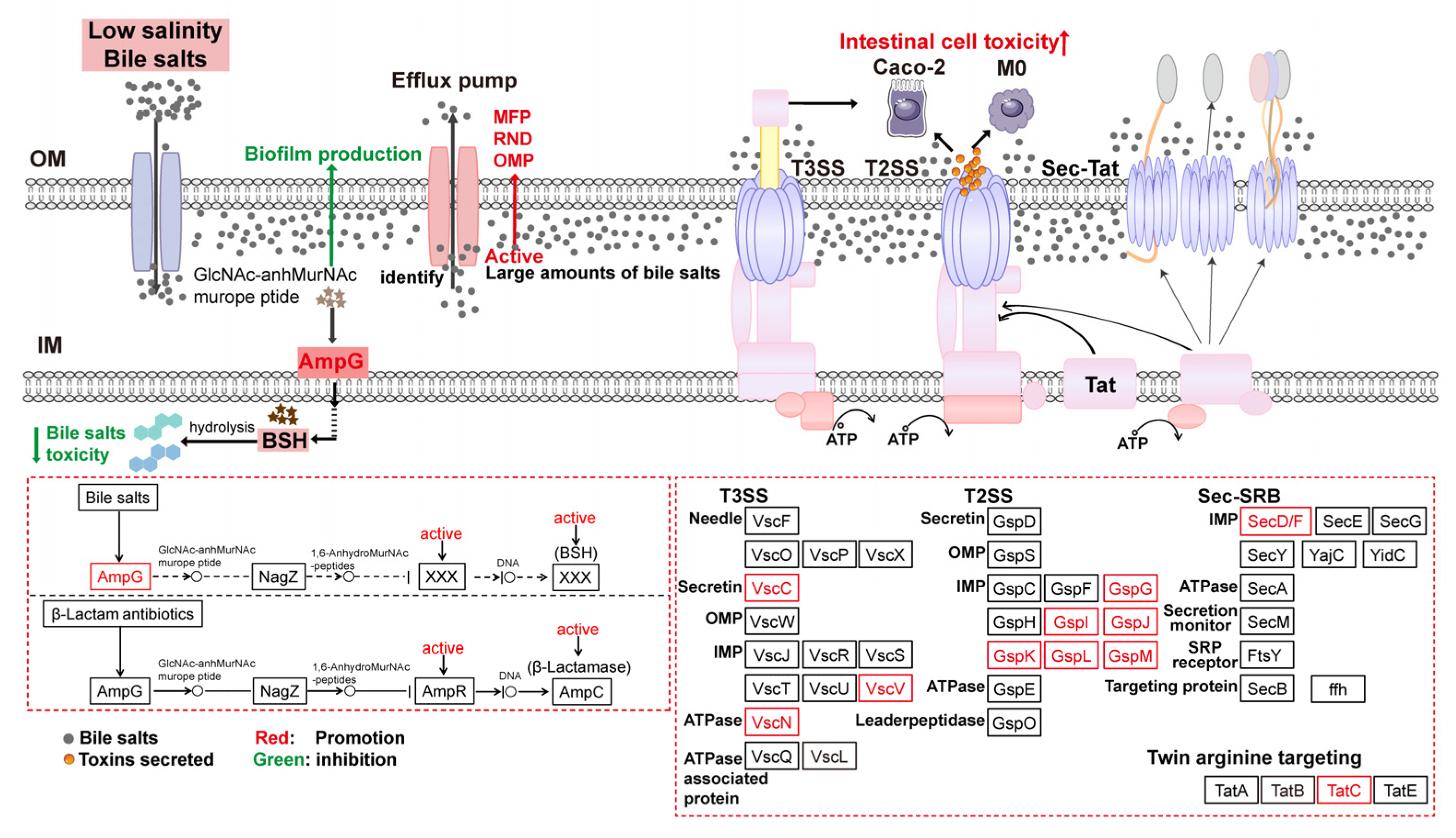
4.4. Regulation of Secretion System-Related Proteins Affected V. parahaemolyticus Virulence
4.5. Activation of the Beta-Lactam Resistance Pathway Promoted Bile Salts Efflux to Reduce Cytotoxicity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fujino, T.; Okuno, Y.; Nakada, D.; Aoyama, A.; Mukai, T.; Ueho, T. On the bacteriological examination of shirasu food poisoning. Med. J. Osaka Univ. 1953, 4, 299–304. [Google Scholar]
- Zou, A.J.; Kinch, L.; Chimalapati, S.; Garcia, N.; Tomchick, D.R.; Orth, K. Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus. J. Biol. Chem. 2023, 299, 104591. [Google Scholar]
- Zakaria, N.H.; Abd Rahim, N.D.E.; Rosilan, N.F.; Sung, Y.Y.; Waiho, K.; Harun, S.; Zainal Abidin, R.A.; Afiqah-Aleng, N. Global landscape of Vibrio parahaemolyticus research: A bibliometric analysis. World J. Microbiol. Biotechnol. 2025, 41, 45. [Google Scholar]
- Takkella, D.; Sharma, S.; Vishwakarma, J.; Gavvala, K. Bile salt induced aggregation and nanostructure formation of β-lactoglobulin in gastrointestinal environments. Food Hydrocolloids 2025, 162, 110944. [Google Scholar]
- Pratama, A.; Ishii, E.; Kodama, T.; Iida, T.; Matsuda, S. The xenogeneic silencer histone-like nucleoid-structuring protein mediates the temperature and salinity-dependent regulation of the type III secretion system 2 in Vibrio parahaemolyticus. J. Bacteriol. 2023, 205, e00266-22. [Google Scholar]
- Kumar, R.; Ng, T.H.; Chang, C.C.; Tung, T.C.; Lin, S.S.; Lo, C.F.; Wang, H.C. Bile acid and bile acid transporters are involved in the pathogenesis of acute hepatopancreatic necrosis disease in white shrimp Litopenaeus vannamei. Cell. Microbiol. 2020, 22, e13127. [Google Scholar]
- Gubensäk, N.; Sagmeister, T.; Buhlheller, C.; Di Geronimo, B.; Wagner, G.E.; Petrowitsch, L.; Gräwert, M.A.; Rotzinger, M.; Berger, T.M.I.; Schäfer, J.; et al. Vibrio cholerae’s ToxRS bile sensing system. eLife 2023, 12, e88721. [Google Scholar]
- Kaval, K.G.; Chimalapati, S.; Siegel, S.D.; Garcia, N.; Jaishankar, J.; Dalia, A.B.; Orth, K. Membrane-localized expression, production and assembly of Vibrio parahaemolyticus T3SS2 provides evidence for transertion. Nat. Commun. 2023, 14, 1178. [Google Scholar]
- Zheng, J.Y.; Shi, B.; Sun, J.Y.; Pan, Y.; Ding, Y.K.; Shi, X.N.; Zhang, J.; Zhang, H.L.; He, J.T.; Zhang, K.L.; et al. Global phylogeography and genomic characterization of Vibrio parahaemolyticus infections in Jilin province, China (2016–2022). Int. J. Food Microbiol. 2025, 428, 110993. [Google Scholar]
- Song, Z.Y.; Chen, L.; Tang, S.Y.; Pan, Y.J.; Xie, Q.C.; Zhao, Y.; Liu, H.Q. Effects of low-salt stress on biological characteristics and transcriptomic profiles of Vibrio parahaemolyticus. Int. J. Food Microbiol. 2025, 430, 111047. [Google Scholar]
- Zhang, Y.K.; Tan, X.T.; Li, M.Z.; Liu, P.; Jiao, X.N.; Gu, D. Transcriptome analysis reveals the effect of low NaCl concentration on osmotic stress and type III secretion system in Vibrio parahaemolyticus. Int. J. Mol. Sci. 2023, 24, 2621. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, J.F.; Zhang, M.M.; Xue, X.F.; Wu, Q.M.; Yang, W.H.; Yin, Z.; Zhou, D.S.; Lu, R.F.; Zhang, Y.Q. The effect of salinity on biofilm formation and c-di-GMP production in Vibrio parahaemolyticus. Curr. Microbiol. 2022, 79, 25. [Google Scholar]
- Saito, S.; Iwade, Y.; Tokuoka, E.; Nishio, T.; Otomo, Y.; Araki, E.; Konuma, H.; Nakagawa, H.; Tanaka, H.; Sugiyama, K.; et al. Epidemiological evidence of lesser role of Thermostable Direct Hemolysin (TDH)-Related Hemolysin (TRH) than TDH on Vibrio parahaemolyticus pathogenicity. Foodborne Pathog. Dis. 2015, 12, 131–138. [Google Scholar] [PubMed]
- Gotoh, K.; Kodama, T.; Hiyoshi, H.; Izutsu, K.; Park, K.S.; Dryselius, R.; Akeda, Y.; Honda, T.; Iida, T. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS ONE 2010, 5, e13365. [Google Scholar]
- Zhang, C.X.; Liu, D.K. Characterization of bacterial swarming motility: A review. Chin. J. Biotechnol. 2023, 39, 3188–3203. [Google Scholar]
- Shi, J.; Zhao, W.; Xie, J.; Zhu, Y.H.; Pan, Y.J.; Ou, J.; Zhao, Y.; Liu, H.Q. Comparison on the growth heterogeneity of Vibrio parahaemolyticus coupled with strain source and genotype analyses in different oligotrophic conditions. J. Food Prot. 2021, 84, 1904–1910. [Google Scholar]
- Chen, B.W.; Huang, J.M.; Li, H.H.; Zeng, Q.H.; Wang, J.J.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. Eradication of planktonic Vibrio parahaemolyticus and its sessile biofilm by curcumin-mediated photodynamic inactivation. Food Control 2020, 113, 107181. [Google Scholar]
- Tan, L.; Zhao, F.; Han, Q.; Zhao, A.J.; Malakar, P.K.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. High correlation between structure development and chemical variation during biofilm formation by Vibrio parahaemolyticus. Front. Microbiol. 2018, 9, 1181. [Google Scholar]
- Liu, H.; Wang, Y.; Cao, J.J.; Jiang, H.Y.; Yao, J.J.; Gong, G.L.; Chen, X.F.; Xu, W.S.; He, X.X. Antimicrobial activity and virulence attenuation of citral against the fish pathogen Vibrio alginolyticus. Aquaculture 2020, 515, 734578. [Google Scholar]
- Bai, Y.; Yang, Q.P.; Sun, Y.N.; Li, F.Q.; Sun, J.L.; Yang, S.R.; Yang, D.J.; Peng, Z.X.; Yang, B.W.; Xu, J.; et al. Antimicrobial susceptibility and genomic characterization of Vibrio parahaemolyticus isolated from aquatic foods in 15 provinces, China, 2020. Int. J. Food Microbiol. 2024, 418, 110737. [Google Scholar]
- Liu, T.Z.; Huang, T.; Li, J.J.; Li, A.Q.; Li, C.; Huang, X.X.; Li, D.X.; Wang, S.W.; Liang, M.F. Optimization of differentiation and transcriptomic profile of THP-1 cells into macrophage by PMA. PLoS ONE 2023, 18, e0286056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Yang, Z.; Dai, K.; Hu, B.D.; Xu, S.Y.; Wang, Y.; Lei, L.; Du, S.Y.; Zhao, Q.; Huang, X.B.; et al. Rab4b promotes cytolethal distending toxin from glaesserella parasuis-induced cytotoxicity in PK-15 cells. Toxins 2024, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.L.; Dai, X.F. Shaping of microbial phenotypes by trade-offs. Nat. Commun. 2024, 15, 4238. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.T.; Liu, L.; He, J. Biofilms: The microbial “Protective Clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef]
- Urdaneta, V.; Casadesus, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Pan, Q.Q.; Shen, X.D.; Yu, L.L.; Tian, F.W.; Zhao, J.X.; Zhang, H.; Chen, W.; Zhai, Q.X. Comparative genomic analysis determines the functional genes related to bile salt resistance in Lactobacillus salivarius. Microorganisms 2021, 9, 2038. [Google Scholar] [CrossRef]
- Lin, J.Z.; Zhou, D.J.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-targeting antibiotics: Modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef]
- Aseev, L.V.; Koledinskaya, L.S.; Boni, I.V. Regulation of ribosomal protein operons rplM-rpsI, rpmB-rpmG, and rplU-rpmA at the transcriptional and translational levels. J. Bacteriol. 2016, 198, 2494–2502. [Google Scholar] [CrossRef]
- Tan, J.X.; Zhang, L.; Zhou, X.T.; Han, S.Y.; Zhou, Y.; Zhu, Y.Q. Structural basis of the bacterial flagellar motor rotational switching. Cell Res. 2024, 34, 788–801. [Google Scholar] [CrossRef]
- Han, Q.; Wang, S.F.; Qian, X.X.; Guo, L.; Shi, Y.F.; He, R.; Yuan, J.H.; Hou, Y.J.; Li, D.F. Flagellar brake protein YcgR interacts with motor proteins MotA and FliG to regulate the flagellar rotation speed and direction. Front. Microbiol. 2023, 14, 1159974. [Google Scholar]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Microbiol. Spectr. 2016, 4, 26999395. [Google Scholar]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G.J. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [PubMed]
- Wu, C.; Zhao, Z.; Liu, Y.P.; Zhu, X.Y.; Liu, M.; Luo, P.; Shi, Y. Type III secretion 1 effector gene diversity among Vibrio isolates from coastal areas in China. Front. Cell. Infect. Microbiol. 2020, 10, 301. [Google Scholar] [CrossRef]
- Li, P.Z.; Ying, J.; Yang, G.J.; Li, A.F.; Wang, J.; Lu, J.W.; Wang, J.R.; Xu, T.; Yi, H.G.; Li, K.W.; et al. Structure-function analysis of the transmembrane protein AmpG from Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0168060. [Google Scholar] [CrossRef]
- Sun, J.J.; Deng, Z.Q.; Yan, A.X. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar]




| Strains/Cell Lines | trh | tdh | tlh | Source |
|---|---|---|---|---|
| VP17 | − | + | + | Fecal specimens with acute diarrhea |
| VP44 | − | − | + | Fecal specimens with acute diarrhea |
| VP49 | + | + | + | Fecal specimens with acute diarrhea |
| VP85 | + | − | + | Fecal specimens with acute diarrhea |
| THP-1 | Primary cells purchased from Beyotime Biotechnology | |||
| Caco-2 | Primary cells purchased from Beyotime Biotechnology |
| Efflux Pump Proteins | Bile Salts Molecule | Mole Dock Score (Kcal/mol) | Interaction Residues |
|---|---|---|---|
| MFP (VPA1191) | Glycocholic acid | −4.99 | Gln357, Gln307, and Gly355 |
| MFP (VPA1191) | Taurocholic acid | −2.92 | Lys48, Lys287, Ser47 |
| MFP (VP0039) | Glycocholic acid | −4.55 | Glu48, Lys50, and Asn357 |
| MFP (VP0039) | Taurocholic acid | −3.78 | Lys215 |
| RND (VP0038) | Glycocholic acid | −5.15 | Lys501 |
| RND (VP0038) | Taurocholic acid | −3.69 | Lys487, Lys490, Arg356 |
| OMP (VP0425) | Glycocholic acid | −4.37 | Lys2, Lys3 |
| OMP (VP0425) | Taurocholic acid | −4.33 | Asn228, Arg409, Ser2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Yang, B.; Zhou, X.; Gong, Z.; Wang, E.; Pan, Y.; Zhao, Y.; Liu, H. Proteomic Analysis Reveals the Phenotypic Heterogeneity and Tolerance Mechanisms of Halophilic Vibrio parahaemolyticus Under Dual Stress of Low Salinity and Bile Salts in the Human Intestine. Biomolecules 2025, 15, 518. https://doi.org/10.3390/biom15040518
Guo Y, Yang B, Zhou X, Gong Z, Wang E, Pan Y, Zhao Y, Liu H. Proteomic Analysis Reveals the Phenotypic Heterogeneity and Tolerance Mechanisms of Halophilic Vibrio parahaemolyticus Under Dual Stress of Low Salinity and Bile Salts in the Human Intestine. Biomolecules. 2025; 15(4):518. https://doi.org/10.3390/biom15040518
Chicago/Turabian StyleGuo, Yingying, Bing Yang, Xiaoyan Zhou, Zhangxi Gong, Enxiao Wang, Yingjie Pan, Yong Zhao, and Haiquan Liu. 2025. "Proteomic Analysis Reveals the Phenotypic Heterogeneity and Tolerance Mechanisms of Halophilic Vibrio parahaemolyticus Under Dual Stress of Low Salinity and Bile Salts in the Human Intestine" Biomolecules 15, no. 4: 518. https://doi.org/10.3390/biom15040518
APA StyleGuo, Y., Yang, B., Zhou, X., Gong, Z., Wang, E., Pan, Y., Zhao, Y., & Liu, H. (2025). Proteomic Analysis Reveals the Phenotypic Heterogeneity and Tolerance Mechanisms of Halophilic Vibrio parahaemolyticus Under Dual Stress of Low Salinity and Bile Salts in the Human Intestine. Biomolecules, 15(4), 518. https://doi.org/10.3390/biom15040518





