The Effects of P5CR Gene Function of Endophytic Fungus Alternaria oxytropis OW7.8 on Swainsonine Biosynthesis
Abstract
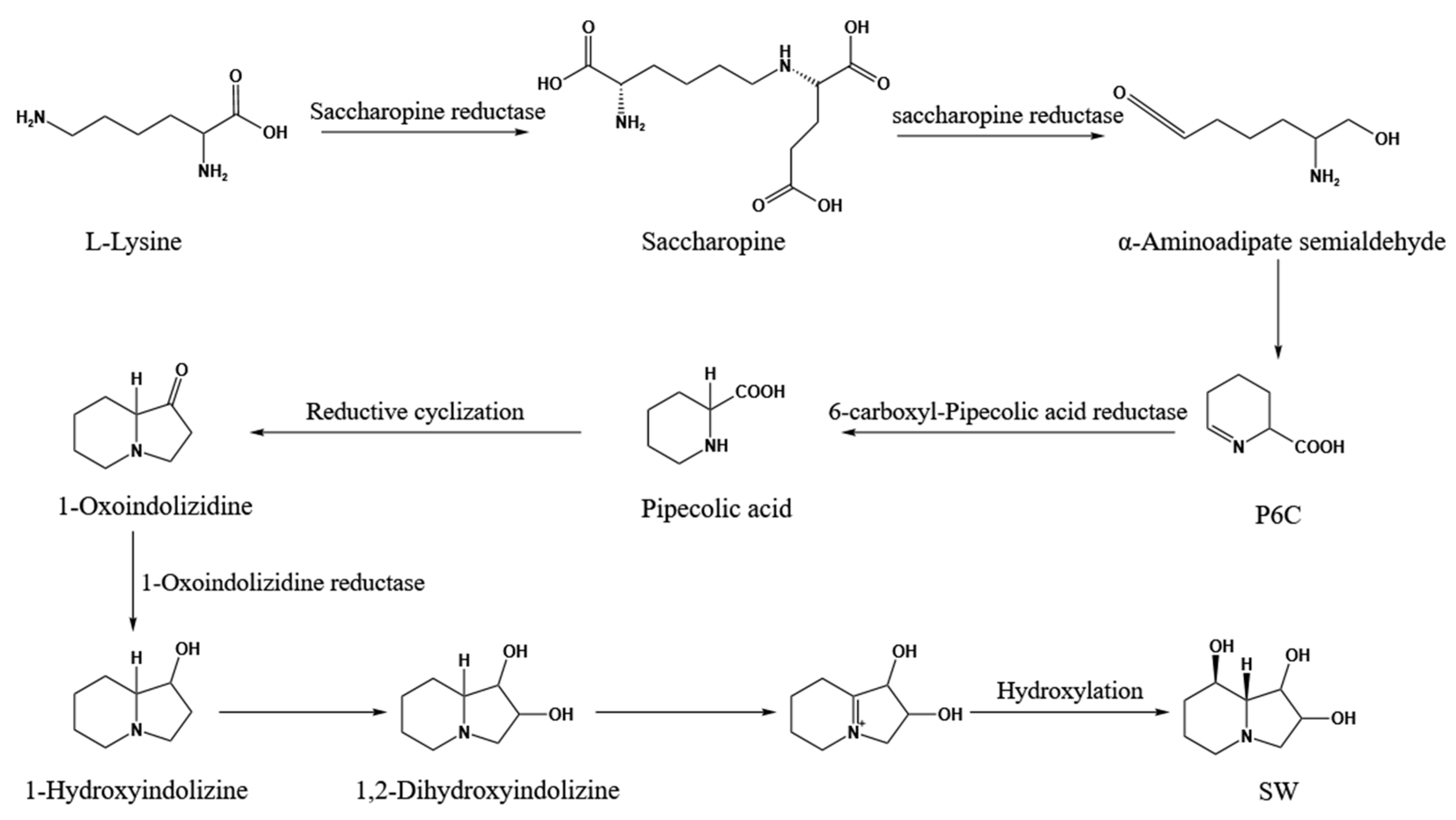
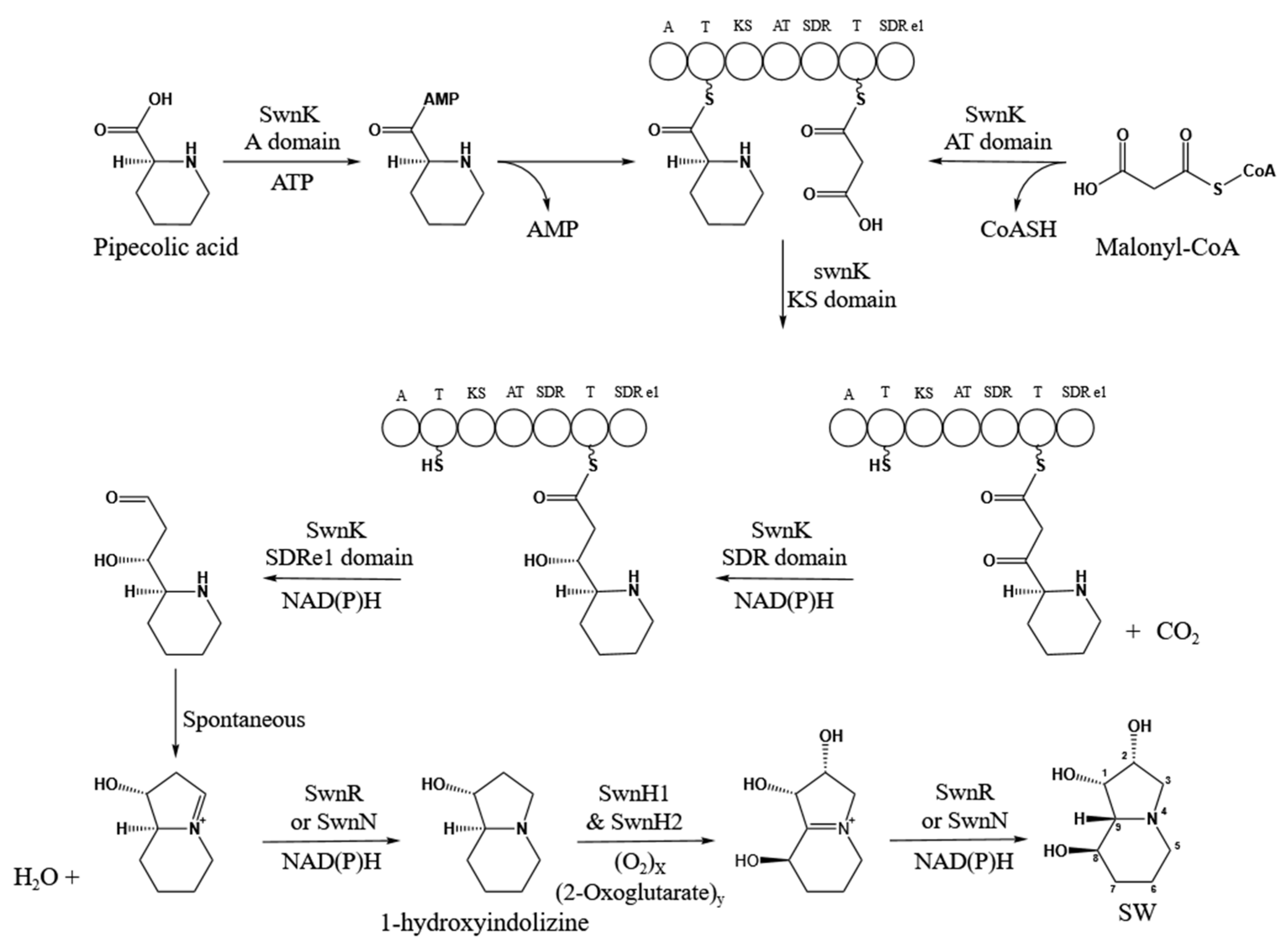
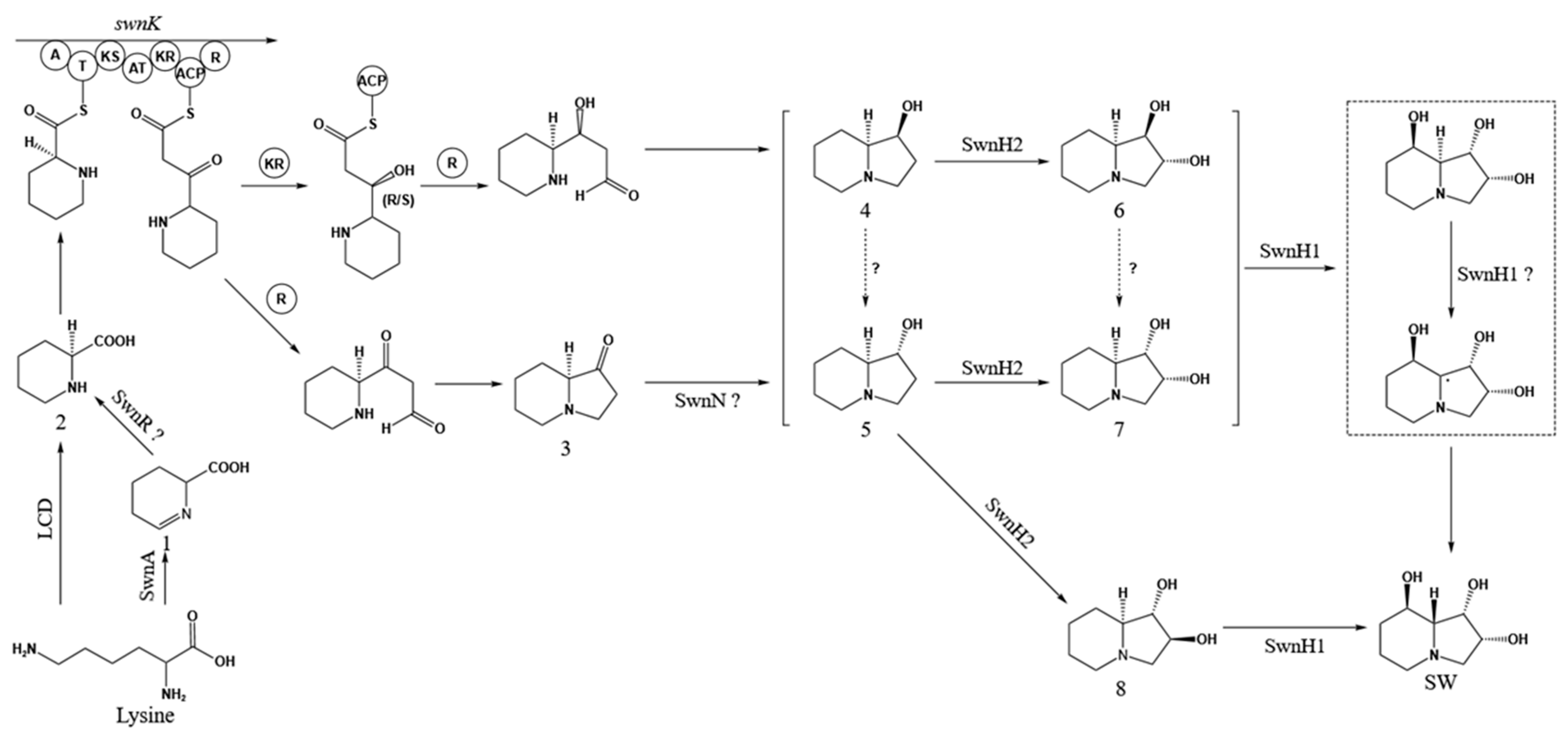
1. Introduction
2. Materials and Methods
2.1. Fungal Strain
2.2. Genomic DNA Extraction from A. oxytropis OW7.8
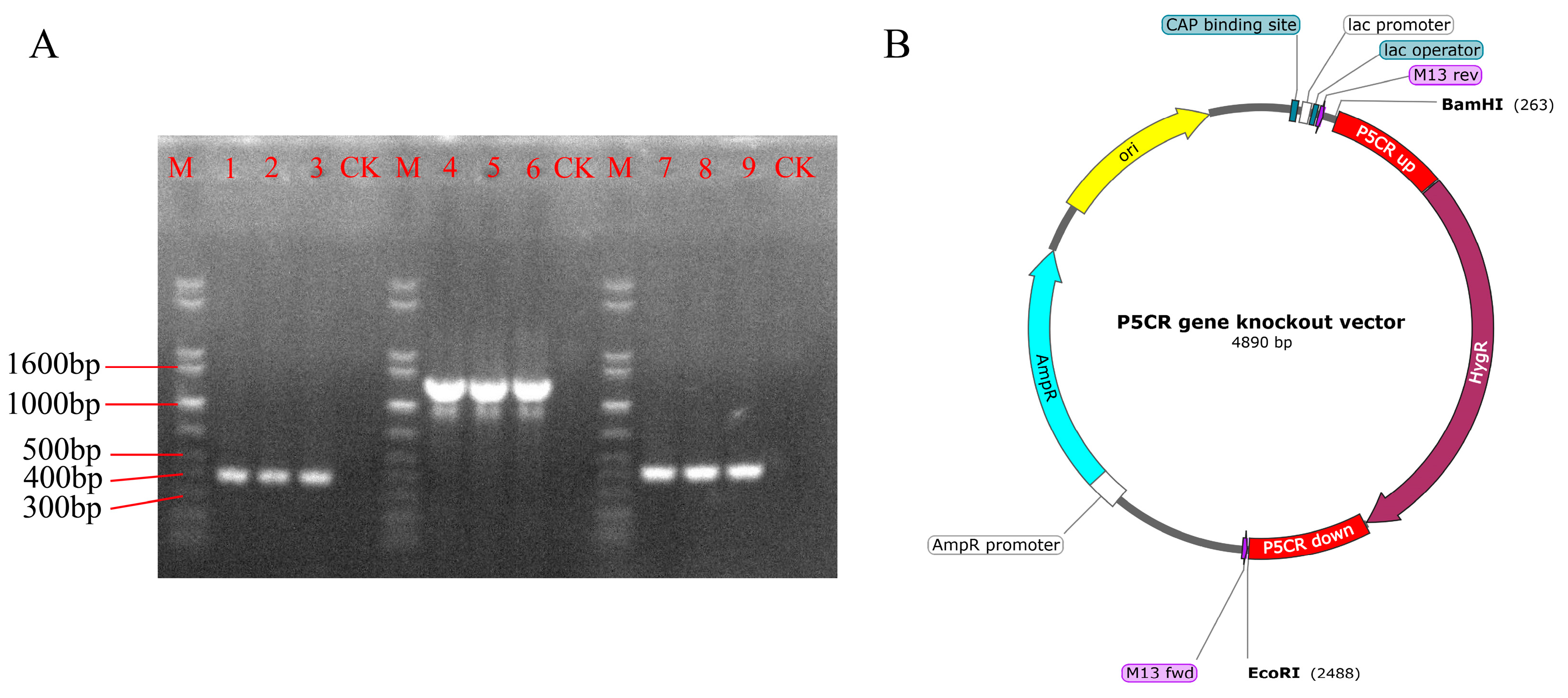
2.3. Construction of the P5CR Gene Knockout Vector
2.4. Protoplast Preparation and Transformation of A. oxytropis OW7.8
2.5. Screening and Identification of Transformants
2.6. Scanning Electron Microscopy of Mycelia
2.7. Extraction and Detection of SW in A. oxytropis OW7.8 and ΔP5CR Mycelia
2.8. Transcriptome Sequencing and Analysis of A. oxytropis OW7.8 and ΔP5CR
2.9. Metabolomic Profiling and Analysis of A. oxytropis OW7.8 and ΔP5CR
2.10. Extraction of SW Biosynthesis-Related Gene Expression Levels in A. oxytropis OW7.8 and ΔP5CR
3. Results
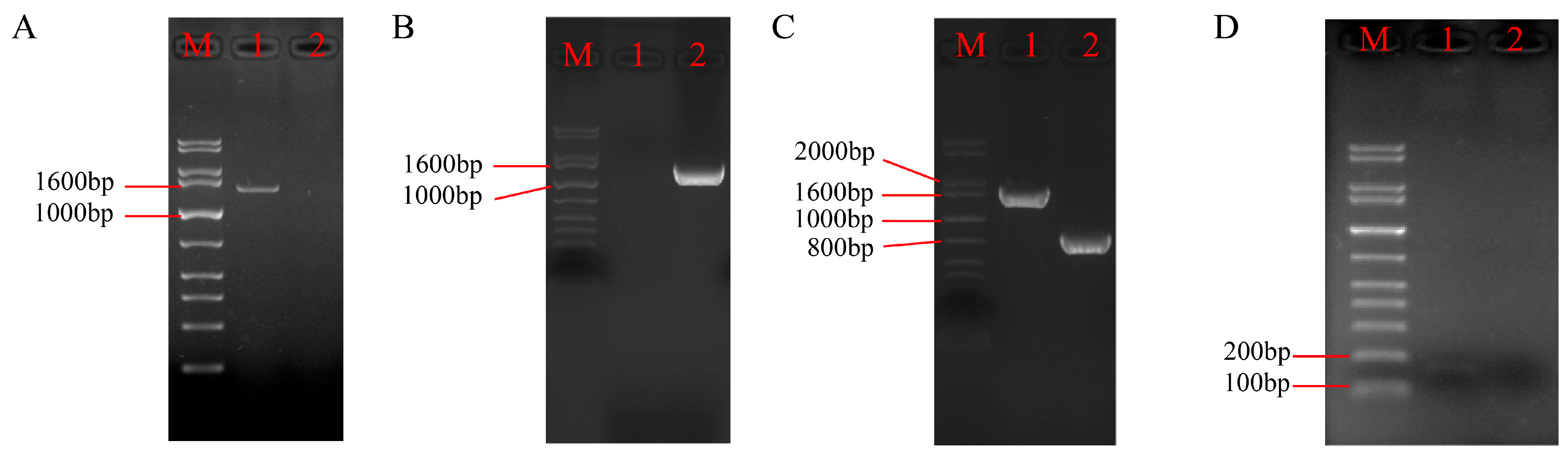
3.1. Construction of the P5CR Gene Knockout Vector in A. oxytropis
3.2. Screening and Identification of Knockout Transformants
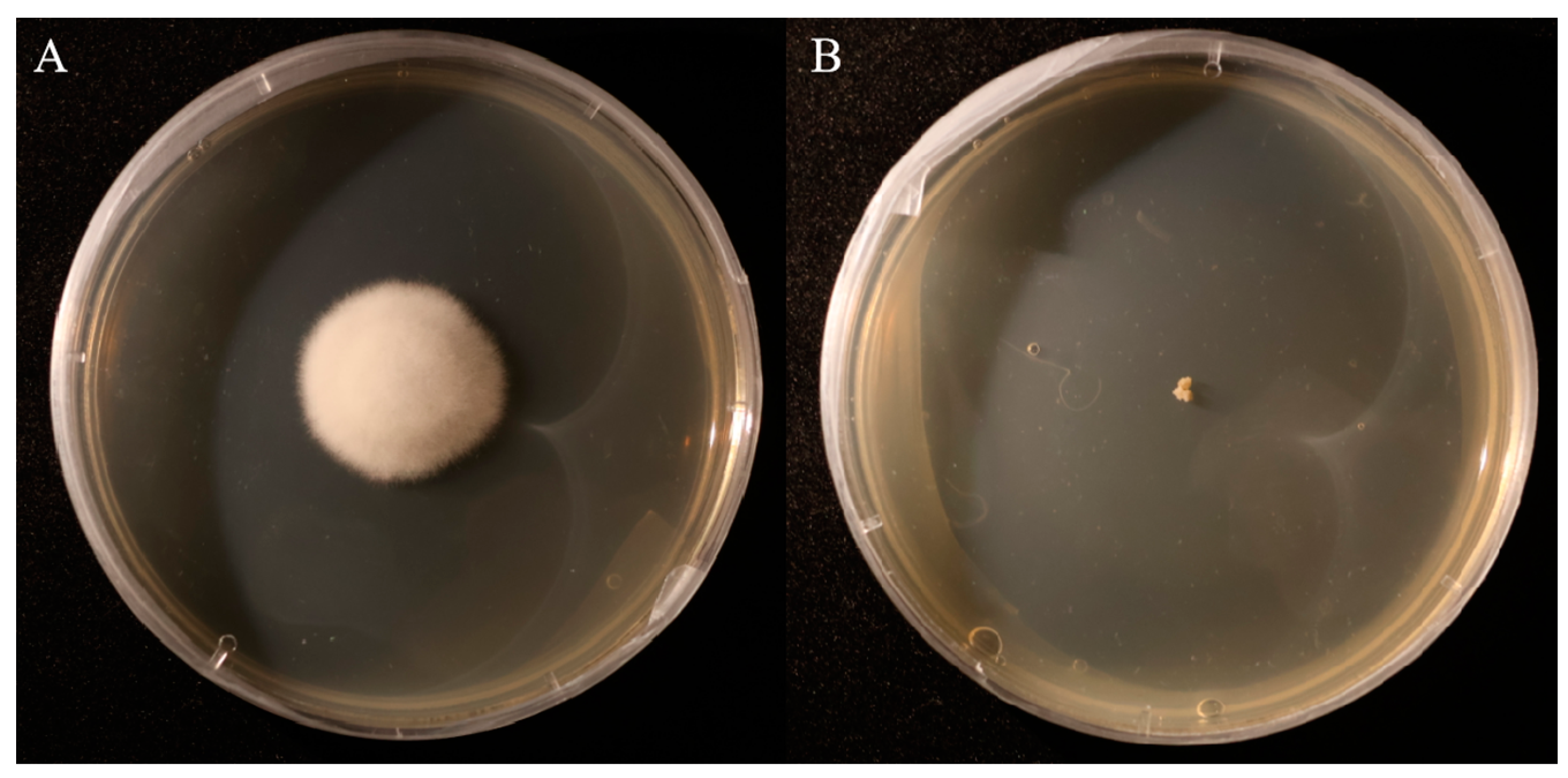
3.3. Comparison of Colony Morphology and Scanning Electron Microscopy (SEM) Structures Between A. oxytropis OW7.8 and ΔP5CR
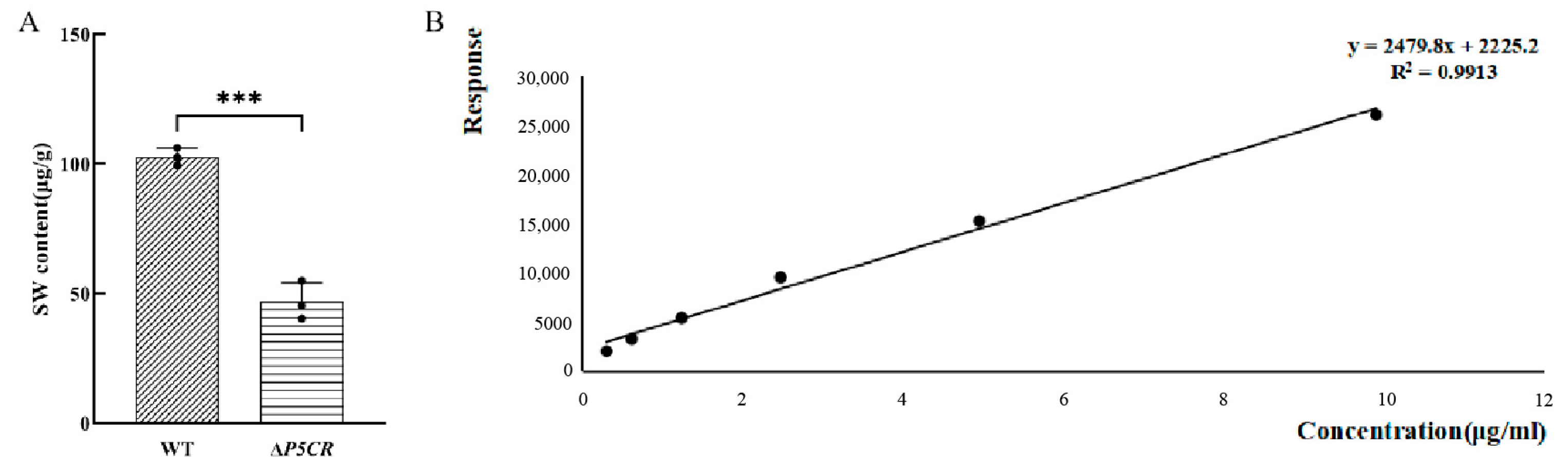
3.4. SW Levels in A. oxytropis OW7.8 and ΔP5CR Mycelia
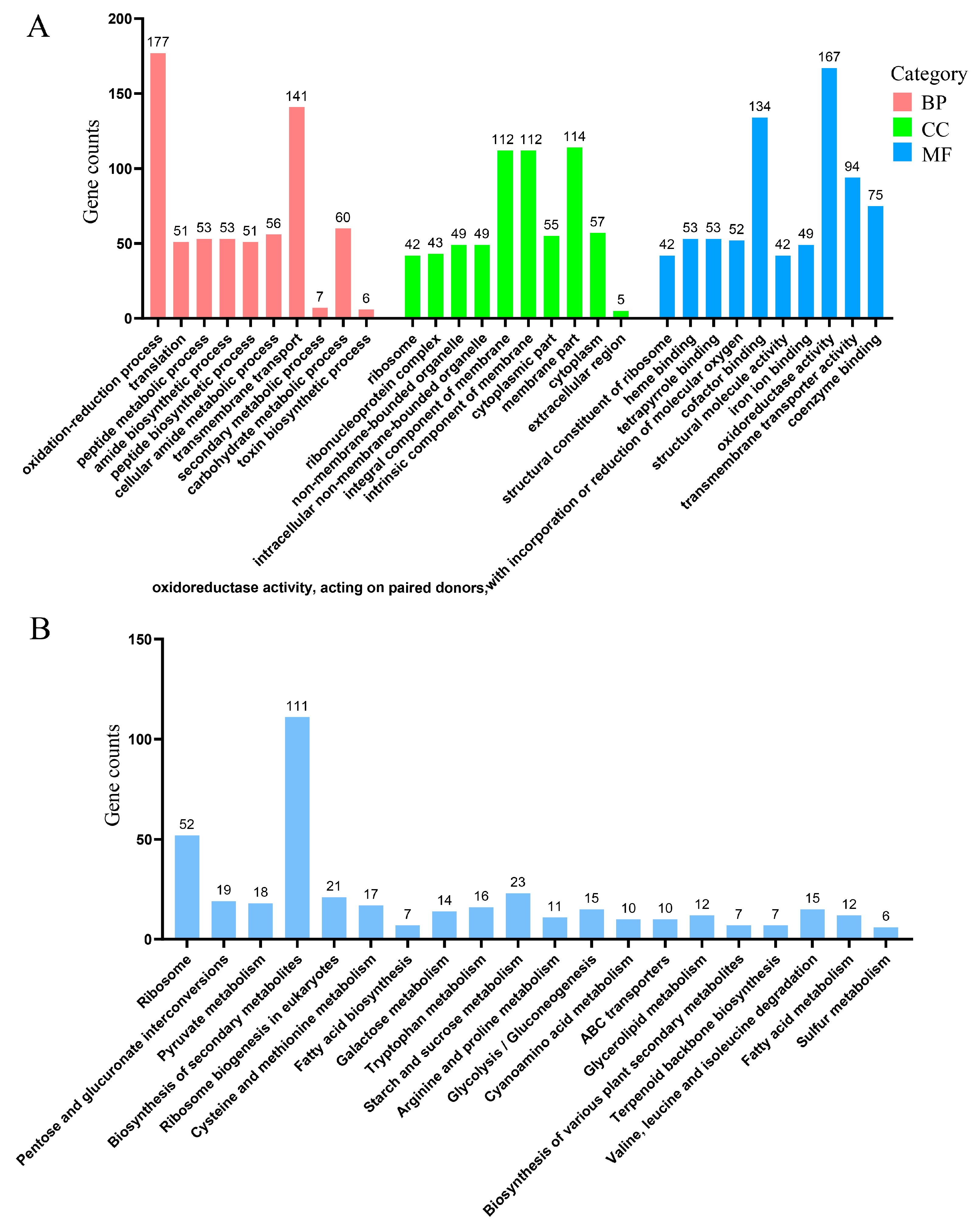
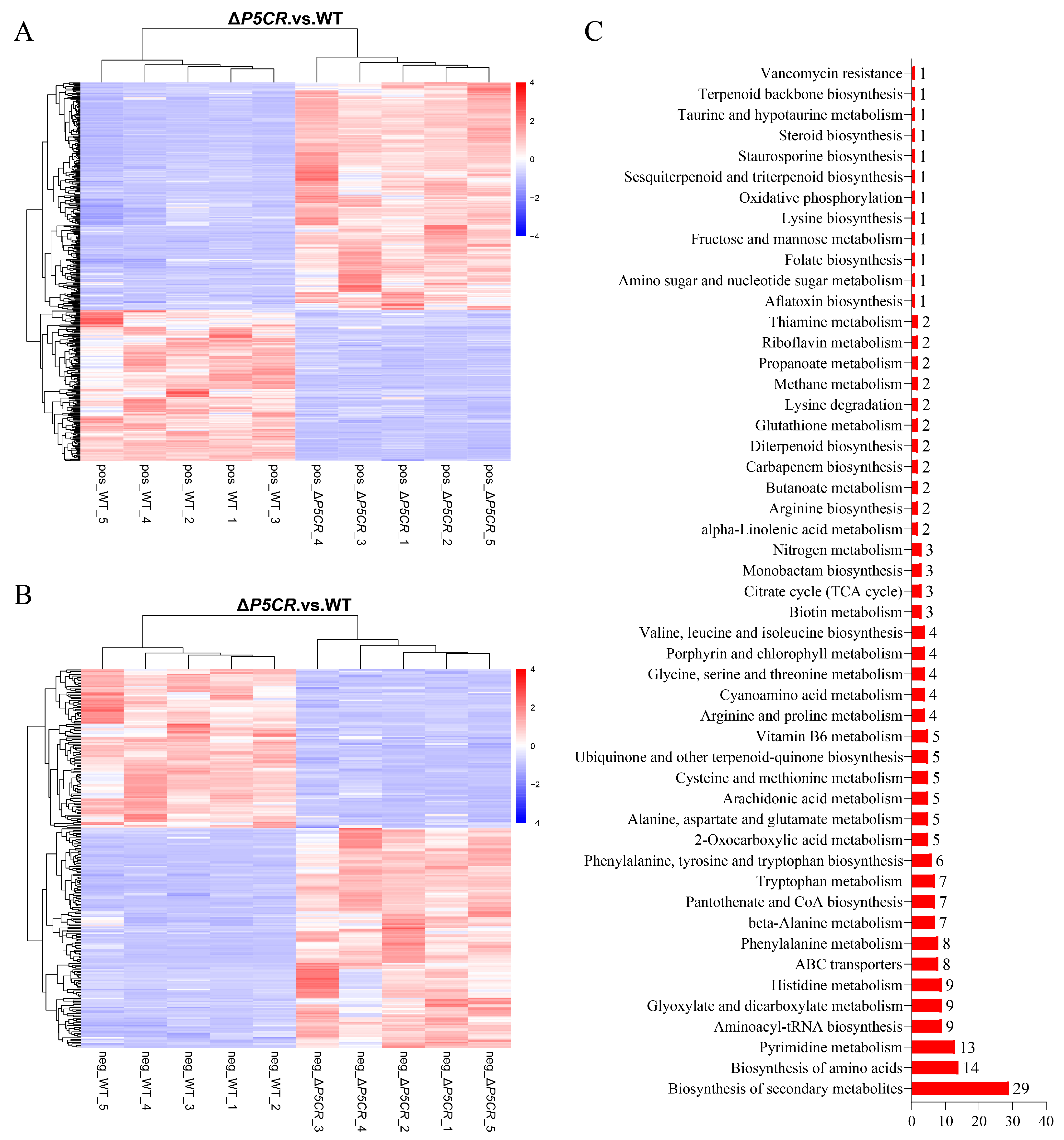
3.5. Transcriptomic Analysis of A. oxytropis OW7.8 and ΔP5CR
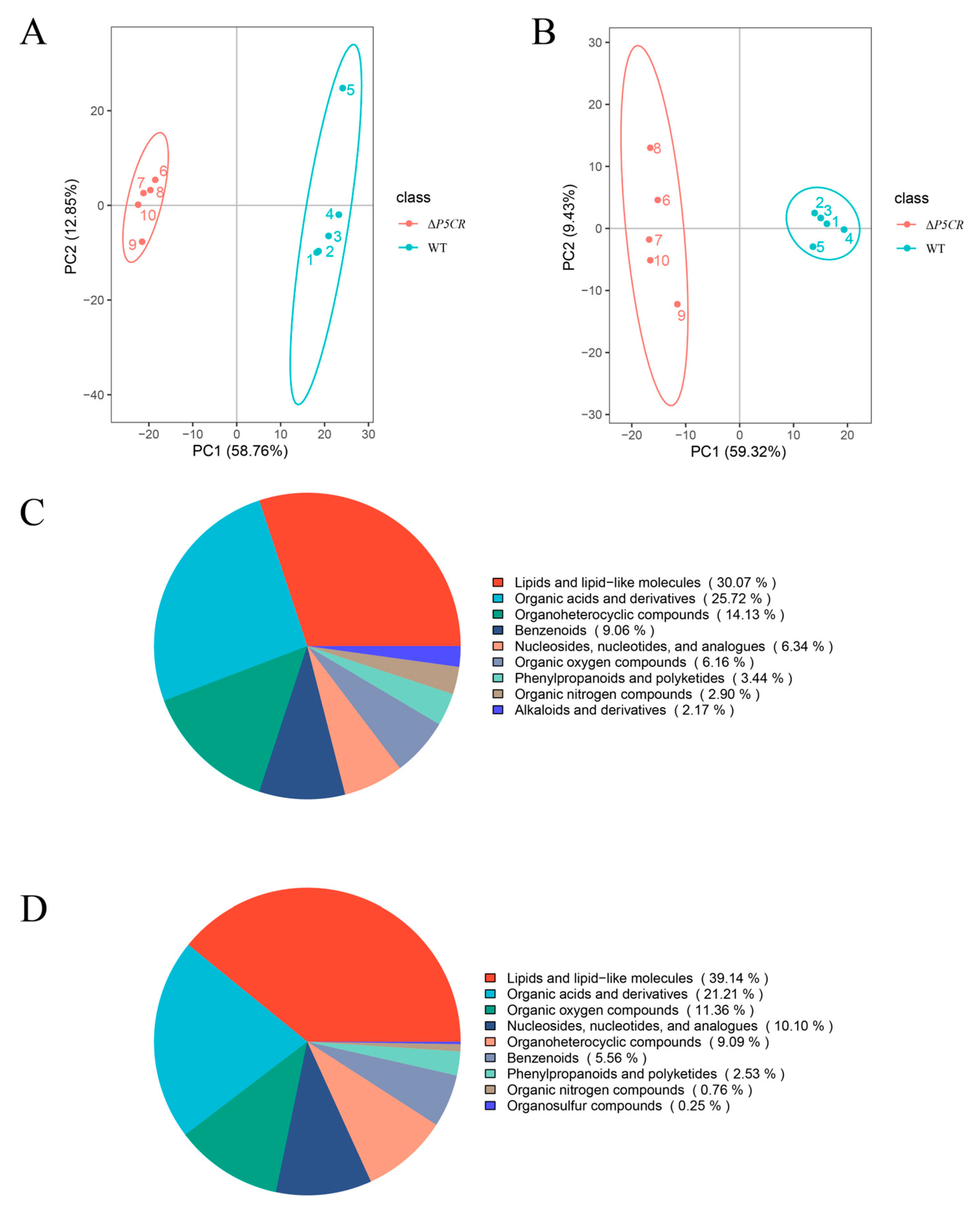
3.6. Metabolomic Profiling of A. oxytropis OW7.8 and ΔP5CR
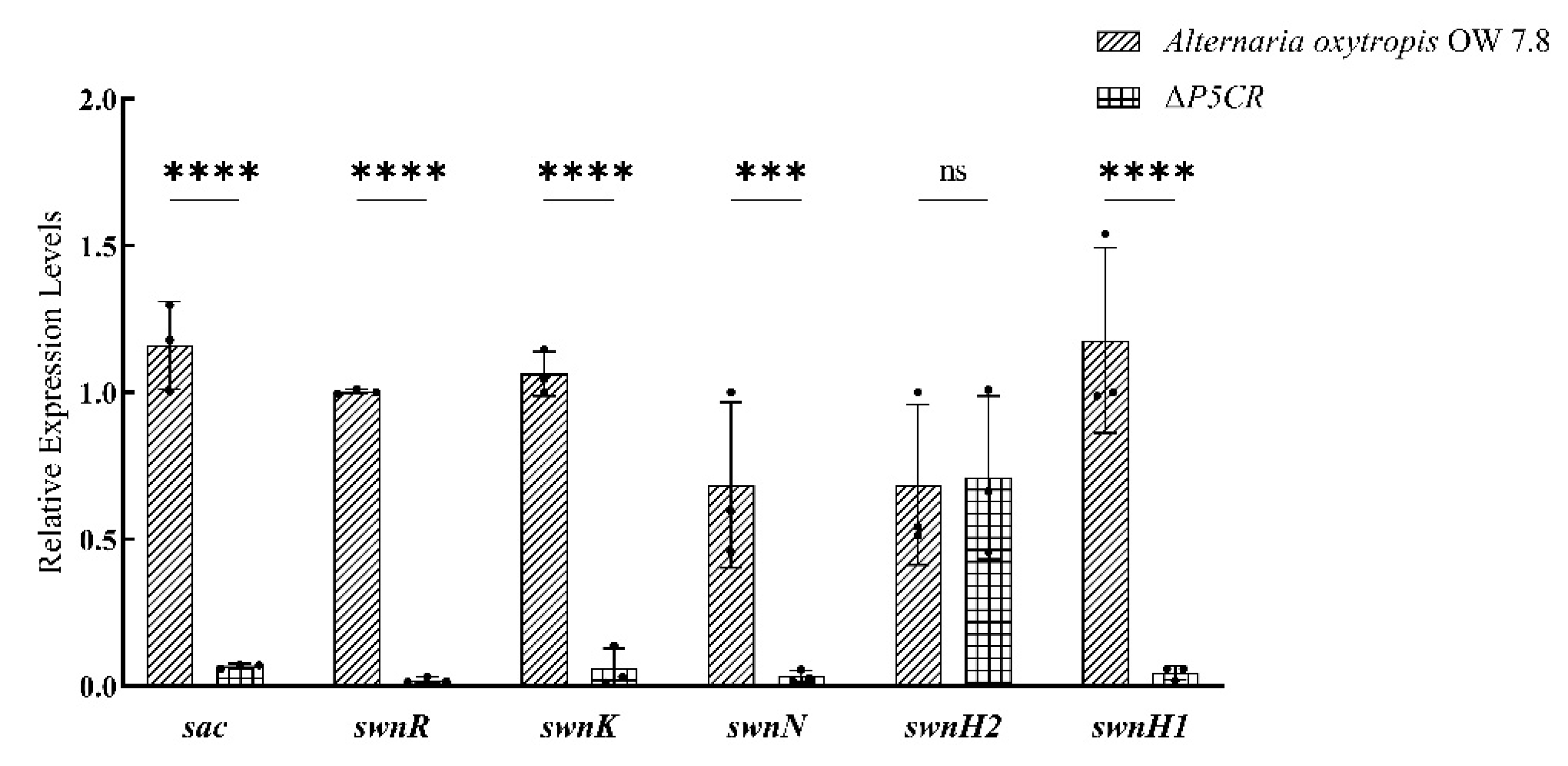
3.7. Expression of SW Biosynthesis-Related Genes in A. oxytropis OW7.8 and ΔP5CR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, Y.; Zhao, Q.; Wang, J.; Wang, J.; Wang, Y.; Song, Y.; Geng, G.; Li, Q. Swainsonine-Producing Fungal Endophytes from Major Locoweed Species in China. Toxicon 2010, 56, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhang, L.; Zhao, Q.; Beckmann, M.; Phillips, H.; Meng, H.; Mo, C.; Mur, L.A.J.; He, W. Host-Species Variation and Environment Influence Endophyte Symbiosis and Mycotoxin Levels in Chinese Oxytropis Species. Toxins 2022, 14, 181. [Google Scholar] [CrossRef]
- Fu, J.J.; Guo, Y.Z.; Huang, W.Y.; Guo, R.; Lu, H.; Wu, C.C.; Zhao, B.Y. The Distribution of Locoweed in Natural Grassland in the United States and the Current Status and Prospects of Research on Animal Poisoning. Acta Agrestia Sin. 2019, 27, 519–530. [Google Scholar]
- Cook, D.; Gardner, D.R.; Lee, S.T.; Pfister, J.A.; Stonecipher, C.A.; Welsh, S.L. A Swainsonine Survey of North American Astragalus and Oxytropis Taxa Implicated as Locoweeds. Toxicon 2016, 118, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, R.J.; McKenzie, R.A.; O’Sullivan, B.M.; Elbein, A.D. Identification of the Glycosidase Inhibitors Swainsonine and Calystegine B2 in Weir Vine (Ipomoea SP. Q6 {AFF. CALOBRA}) and Correlation with Toxicity. J. Nat. Prod. 1995, 58, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tan, P.; Wang, H.; Wang, J.; Zhang, C.; Lu, H.; Zhao, B. Swainsonine inhibits autophagic degradation and causes cytotoxicity by reducing CTSD O-GlcNAcylation. Chem.-Biol. Interact. 2023, 382, 110629. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.-S.; Peng, Y.; Chen, W.; Dong, N.; Wu, Q.; Pan, B.; Wang, B.; Guo, W. Golgi α-mannosidases regulate cell surface N-glycan type and ectodomain shedding of the transmembrane protease corin. J. Biol. Chem. 2023, 299, 105211. [Google Scholar] [CrossRef]
- Fu, K.; Chen, X.; Shou, N.; Wang, Z.; Yuan, X.; Wu, D.; Wang, Q.; Cheng, Y.; Ling, N.; Shi, Z. Swainsonine Induces Liver Inflammation in Mice via Disturbance of Gut Microbiota and Bile Acid Metabolism. J. Agric. Food Chem. 2023, 71, 1758–1767. [Google Scholar] [CrossRef]
- Xin, L.; Ping, L. Advances in research on swainsonine biosynthesis pathway in fungi. Chem. Life 2018, 38, 815–820. [Google Scholar]
- Cook, D.; Gardner, D.R.; Pfister, J.A. Swainsonine-Containing Plants and Their Relationship to Endophytic Fungi. J. Agric. Food Chem. 2014, 62, 7326–7334. [Google Scholar] [CrossRef]
- Quach, Q.N.; Clay, K.; Lee, S.T.; Gardner, D.R.; Cook, D. Phylogenetic Patterns of Bioactive Secondary Metabolites Produced by Fungal Endosymbionts in Morning Glories (Ipomoeeae, Convolvulaceae). New Phytol. 2023, 238, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Donzelli, B.G.G.; Creamer, R.; Baucom, D.L.; Gardner, D.R.; Pan, J.; Moore, N.; Krasnoff, S.B.; Jaromczyk, J.W.; Schardl, C.L. Swainsonine Biosynthesis Genes in Diverse Symbiotic and Pathogenic Fungi. G3 Genes Genomes Genet. 2017, 7, 1791–1797. [Google Scholar]
- Wang, C.; Liu, R.; Lim, G.-H.; De Lorenzo, L.; Yu, K.; Zhang, K.; Hunt, A.G.; Kachroo, A.; Kachroo, P. Pipecolic Acid Confers Systemic Immunity by Regulating Free Radicals. Sci. Adv. 2018, 4, eaar4509. [Google Scholar] [PubMed]
- Gao, X.; Cook, D.; Ralphs, M.H.; Yan, L.; Gardner, D.R.; Lee, S.T.; Panter, K.E.; Han, B.; Zhao, M.-L. Detection of Swainsonine and Isolation of the Endophyte Undifilum from the Major Locoweeds in Inner Mongolia. Biochem. Syst. Ecol. 2012, 45, 79–85. [Google Scholar] [CrossRef]
- Pryor, B.M.; Creamer, R.; Shoemaker, R.A.; McLain-Romero, J.; Hambleton, S. Undifilum, a New Genus for Endophytic Embellisia Oxytropis and Parasitic Helminthosporium Bornmuelleri on Legumes. Botany 2009, 87, 178–194. [Google Scholar] [CrossRef]
- Baucom, D.L.; Romero, M.; Belfon, R.; Creamer, R. Two New Species of Undifilum, Fungal Endophytes of Astragalus (Locoweeds) in the United States. Botany 2012, 90, 866–875. [Google Scholar]
- Braun, K.; Romero, J.; Liddell, C.; Creamer, R. Production of Swainsonine by Fungal Endophytes of Locoweed. Mycol. Res. 2003, 107, 980–988. [Google Scholar] [CrossRef]
- Guan, H.; Liu, X.; Mur, L.A.J.; Fu, Y.; Wei, Y.; Wang, J.; He, W. Rethinking of the Roles of Endophyte Symbiosis and Mycotoxin in Oxytropis Plants. J. Fungi 2021, 7, 400. [Google Scholar] [CrossRef]
- Yu, Y.T.; Wang, J.H.; Zhao, Q.M.; Li, H.L.; Song, Y.M.; Wang, Y.; Zhang, Z.; Cui, Z.; Geng, G.; Li, Q. Isolation and identification of swainsonine-producing fungal endophyte from Oxytropis kansuensis. J. Northwest AF Univ. 2009, 37, 40–46+51. [Google Scholar]
- Wu, K.X.; Tang, S.Y.; Zhu, Y.R.; Guo, Z.Y.; Mo, C.H.; Zhao, B.Y.; Lu, H. Isolation and Identification of Endophytic Fungi from Wild-Type Astragalus adsurgens and Analysis of Swainsonine. Acta Agrestia Sin. 2022, 30, 1692–1700. [Google Scholar]
- Zhang, L.; Wu, R. Assembly of High-quality Genomes of the Locoweed Oxytropis Ochrocephala and Its Endophyte Alternaria Oxytropis Provides New Evidence for Their Symbiotic Relationship and Swainsonine Biosynthesis. Mol. Ecol. Resour. 2023, 23, 253–272. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Li, Y.-Z. Alternaria Gansuense, a Plant Systematic Fungal Pathogen Producing Swainsonine In Vivo and In Vitro. Curr. Microbiol. 2023, 80, 232. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Ping, L. Transcriptome Profiles of Alternaria Oxytropis Provides Insights into Swainsonine Biosynthesis. Sci. Rep. 2019, 9, 6021. [Google Scholar]
- Lu, P.; Li, X.; Wang, S.; Gao, F.; Bo, Y.; Xi, L.; Du, L.; Hu, J.; Zhao, L. Saccharopine Reductase Influences Production of Swainsonine in Alternaria oxytropis. Sydowia 2021, 73, 69–74. [Google Scholar]
- Xi, L.J.; Lu, P.; Gao, F.; Du, L. The Influence of Different Additives on the Swainsonine Biosynthesis in Saccharopine Reductase Gene Disruption Mutant M1 of an Endophytic Fungus from Oxytropis Glabra. Life Sci. Res. 2018, 22, 298–304. [Google Scholar]
- Muhammad, F.; Rafiq, A.; Muhammad, S.; Saad Ur, R.; Yasar, S.; Amjad, H.; Mohammad Maroof, S.; Amber, A.; Sabaz Ali, K. Real-time expression and in silico characterization of pea genes involved in salt and water-deficit stress. Mol. Biol. Rep. 2023, 51, 18. [Google Scholar]
- Min, Y.; Yu, D.; Yang, J.; Zhao, W.; Zhang, L.; Bai, Y.; Guo, C. Bioinformatics and expression analysis of proline metabolism-related gene families in alfalfa under saline-alkali stress. Plant Physiol. Biochem. 2023, 205, 108182. [Google Scholar]
- Shah, T.; Khan, Z.; Alahmadi Tahani, A.; Imran, A.; Asad, M.; Khan Shah, R.; Ansari Mohammad, J. Mycorrhizosphere bacteria inhibit chromium uptake and phytotoxicity by regulating proline metabolism, antioxidant defense system, and aquaporin gene expression in tomato. Environ. Sci. Pollut. Res. Int. 2024, 31, 24836–24850. [Google Scholar]
- Zhou, B.; Zhang, T.; Wang, F. Unravelling the molecular and biochemical responses in cotton plants to biochar and biofertilizer amendments for Pb toxicity mitigation. Environ. Sci. Pollut. Res. Int. 2023, 30, 100799–100813. [Google Scholar]
- Cook, D.; Beaulieu, W.T.; Mott, I.W.; Riet-Correa, F.; Gardner, D.R.; Grum, D.; Pfister, J.A.; Clay, K.; Marcolongo-Pereira, C. Production of the Alkaloid Swainsonine by a Fungal Endosymbiont of the Ascomycete Order Chaetothyriales in the Host Ipomoea Carnea. J. Agric. Food Chem. 2013, 61, 3797–3803. [Google Scholar]
- Liu, C.; Ding, N.; Lu, P.; Yuan, B.; Li, Y.; Jiang, K. The Effects of swnN Gene Function of Endophytic Fungus Alternaria Oxytropis OW 7.8 on Its Swainsonine Biosynthesis. Int. J. Mol. Sci. 2024, 25, 10310. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhao, X.; Lu, P.; Min, Y. The Effects of swnH1 Gene Function of Endophytic Fungus Alternaria Oxytropis OW 7.8 on Its Swainsonine Biosynthesis. Microorganisms 2024, 12, 2081. [Google Scholar] [CrossRef]
- Luo, F.; Hong, S.; Chen, B.; Yin, Y.; Tang, G.; Hu, F.; Zhang, H.; Wang, C. Unveiling of Swainsonine Biosynthesis via a Multibranched Pathway in Fungi. ACS Chem. Biol. 2020, 15, 2476–2484. [Google Scholar] [PubMed]
- Wang, W.F.; Qian, Y.G.; Lu, P.; He, S.; Du, L.; Li, Y.L.; Gao, F. The Relationship between Swainsonine and Endophytic Fungi in Different Populations of Oxytropis Glabra from Inner Mongolia Chin. J. Anim. Vet. Sci. 2022, 53, 304–314. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar]
- Cooper, B.; Yang, R. An Assessment of AcquireX and Compound Discoverer Software 3.3 for Non-Targeted Metabolomics. Sci. Rep. 2024, 14, 4841. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic Profiles Delineate Potential Role for Sarcosine in Prostate Cancer Progression. Nature 2009, 457, 910–914. [Google Scholar]
- Haspel, J.A.; Chettimada, S.; Shaik, R.S.; Chu, J.-H.; Raby, B.A.; Cernadas, M.; Carey, V.; Process, V.; Hunninghake, G.M.; Ifedigbo, E.; et al. Circadian Rhythm Reprogramming during Lung Inflammation. Nat. Commun. 2014, 5, 4753. [Google Scholar]


| Gene | Encoding Product | Function Prediction |
|---|---|---|
| swnA | Aminotransferase | Catalyzing the synthesis of pyrroline-6-carboxylate (P6C) from L-lysine |
| swnR | Dehydrogenase or reductase | Catalyzing the synthesis of L-PA from P6C |
| swnK | Multifunctional protein | Catalyzing the synthesis of 1-oxoindolizidine (or 1-hydroxyindolizine) from L-PA |
| swnN | Dehydrogenase or reductase | Catalyzing the synthesis of 1-hydroxyindolizine from 1-oxoindolizidine |
| swnH1 | Fe(II)/α-Ketoglutarate-dependent dioxygenase | Catalyzing the synthesis of SW from 1,2-dihydroxyindolizine |
| swnH2 | Fe(II)/α-Ketoglutarate-dependent dioxygenase | Catalyzing the synthesis of 1,2-dihydroxyindolizine form 1-hydroxyindolizine |
| swnT | Transmembrane transporter | Transport of SW |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Li, Y.; Lu, P.; Wang, Y.; Gao, F.; Yuan, B.; Du, L.; Li, Y.; Jiang, K. The Effects of P5CR Gene Function of Endophytic Fungus Alternaria oxytropis OW7.8 on Swainsonine Biosynthesis. Biomolecules 2025, 15, 460. https://doi.org/10.3390/biom15040460
Yang F, Li Y, Lu P, Wang Y, Gao F, Yuan B, Du L, Li Y, Jiang K. The Effects of P5CR Gene Function of Endophytic Fungus Alternaria oxytropis OW7.8 on Swainsonine Biosynthesis. Biomolecules. 2025; 15(4):460. https://doi.org/10.3390/biom15040460
Chicago/Turabian StyleYang, Fan, Yinzhe Li, Ping Lu, Yu Wang, Feng Gao, Bo Yuan, Ling Du, Yuling Li, and Kai Jiang. 2025. "The Effects of P5CR Gene Function of Endophytic Fungus Alternaria oxytropis OW7.8 on Swainsonine Biosynthesis" Biomolecules 15, no. 4: 460. https://doi.org/10.3390/biom15040460
APA StyleYang, F., Li, Y., Lu, P., Wang, Y., Gao, F., Yuan, B., Du, L., Li, Y., & Jiang, K. (2025). The Effects of P5CR Gene Function of Endophytic Fungus Alternaria oxytropis OW7.8 on Swainsonine Biosynthesis. Biomolecules, 15(4), 460. https://doi.org/10.3390/biom15040460






