Prediction of circRNA–Disease Associations via Graph Isomorphism Transformer and Dual-Stream Neural Predictor
Abstract
1. Introduction
- We propose an efficient knowledge representation learning method using a Graph Isomorphism Transformer, which fully captures local and global circRNA–disease associations within a multi-source heterogeneous knowledge graph, enabling higher-order associative knowledge representation and addressing the data sparsity problem;
- We introduce a Dual-Stream Neural Predictor specifically designed for circRNA–disease association prediction, which captures complex non-linear associations and significantly improves prediction accuracy and computational efficiency;
- Extensive experiments have shown that the GIT-DSP model is superior in CDA prediction and has great potential in the fields of non-coding RNA (ncRNA) and protein association prediction.
2. Materials and Methods
2.1. Datasets
2.2. Methods
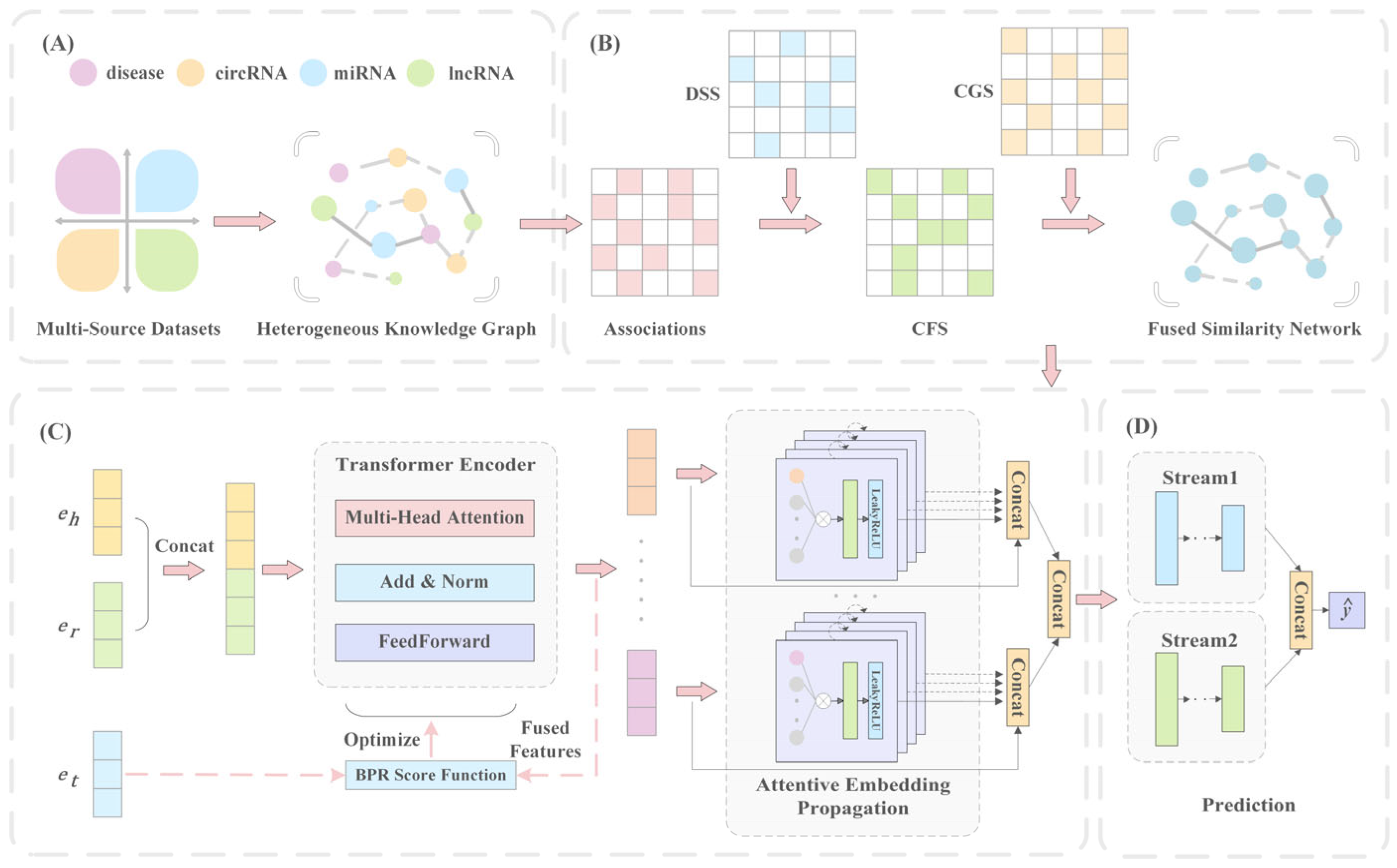
2.2.1. Heterogeneous Knowledge Graph
2.2.2. Transformer-Based Knowledge Representation
2.2.3. Graph Isomorphism Layers
- Information propagation
- Information aggregation
2.2.4. Dual-Stream Neural Predictor
3. Results and Discussion
3.1. Evaluation Metrics
3.2. Parameter Setting
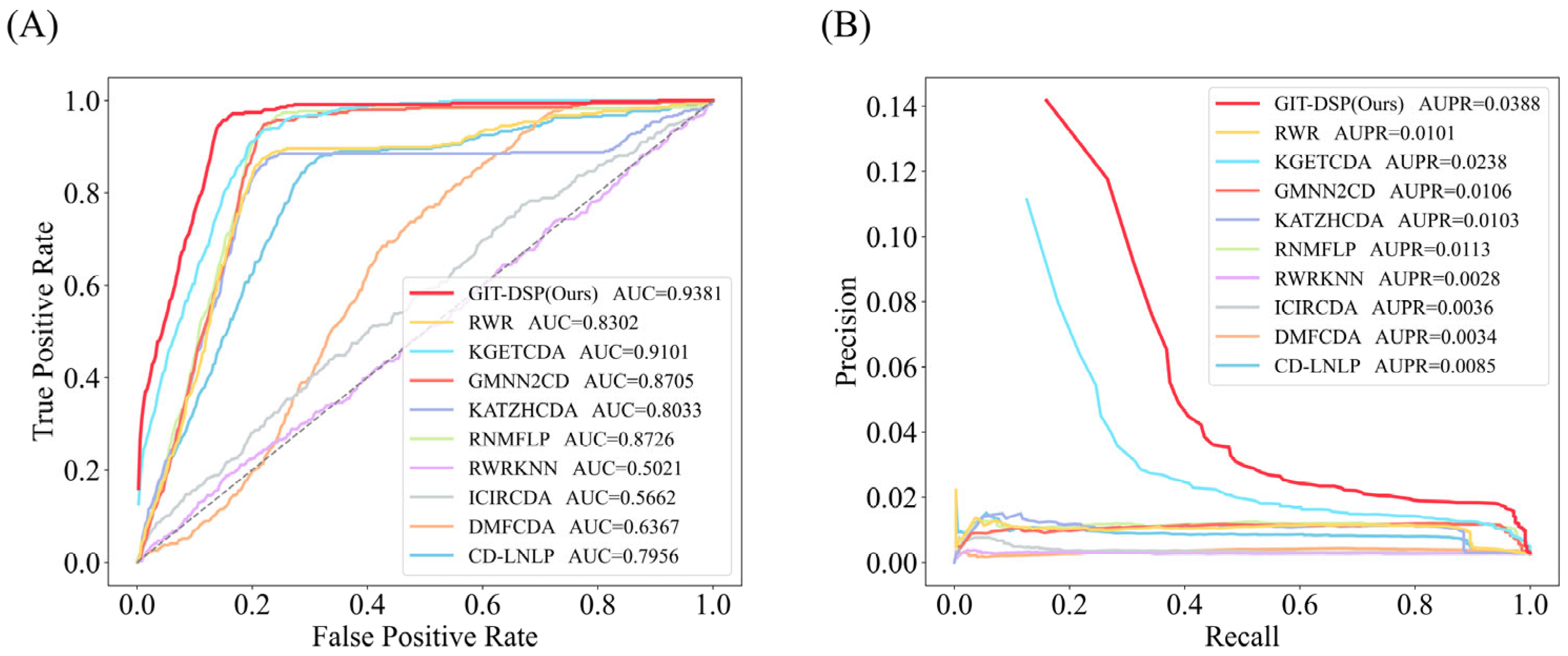
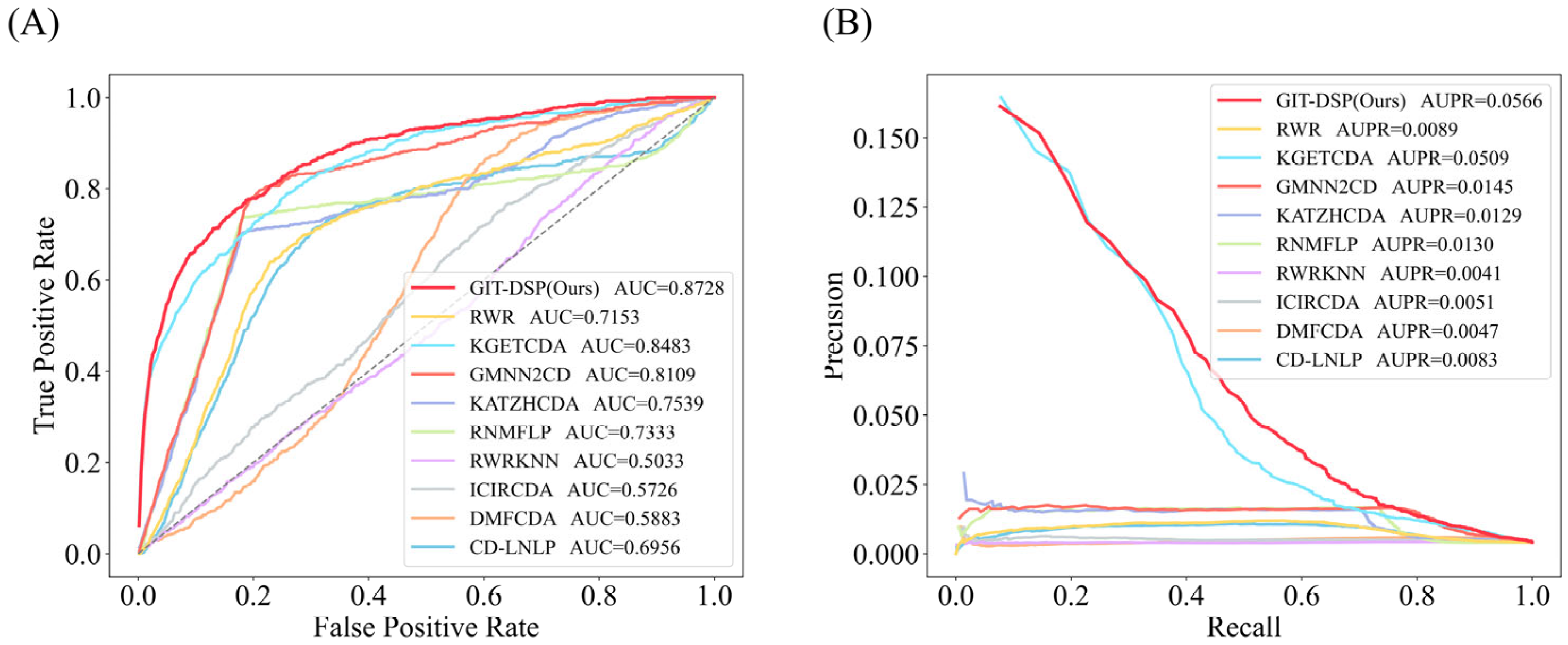
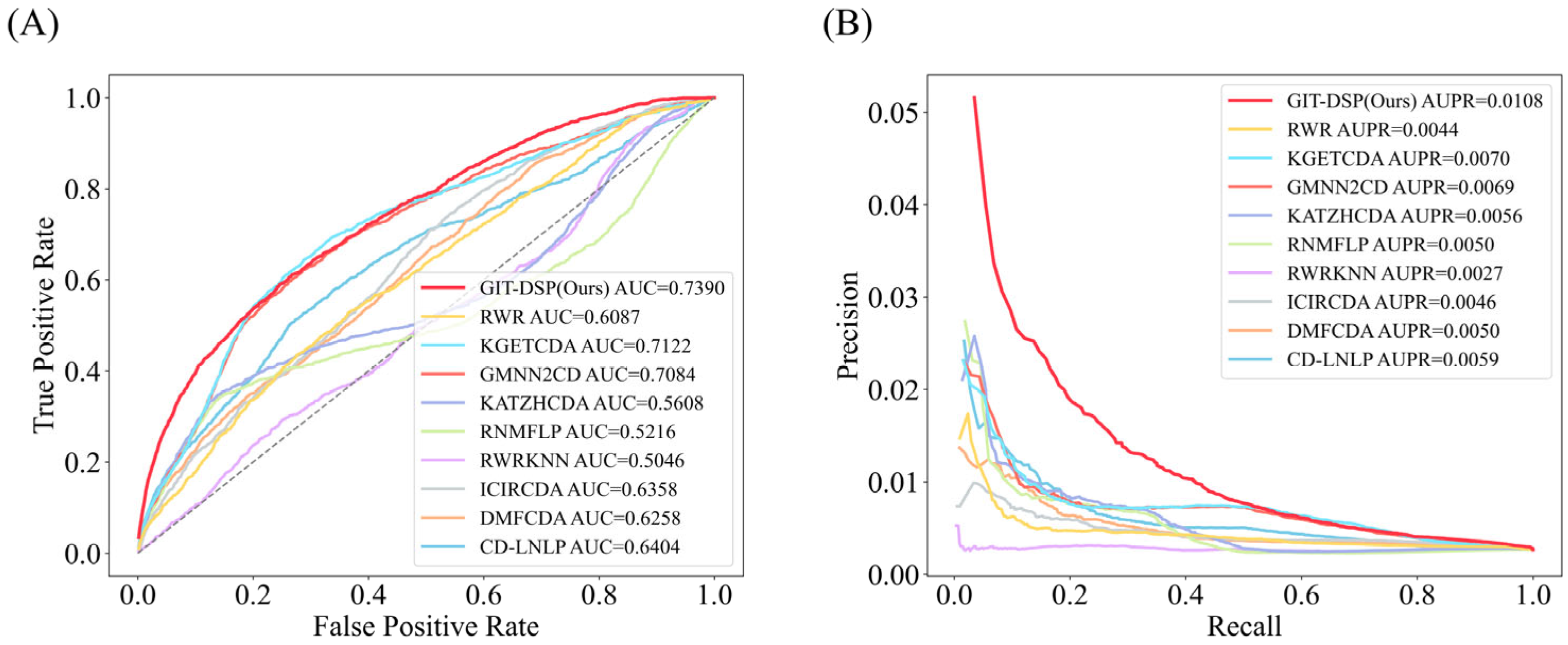
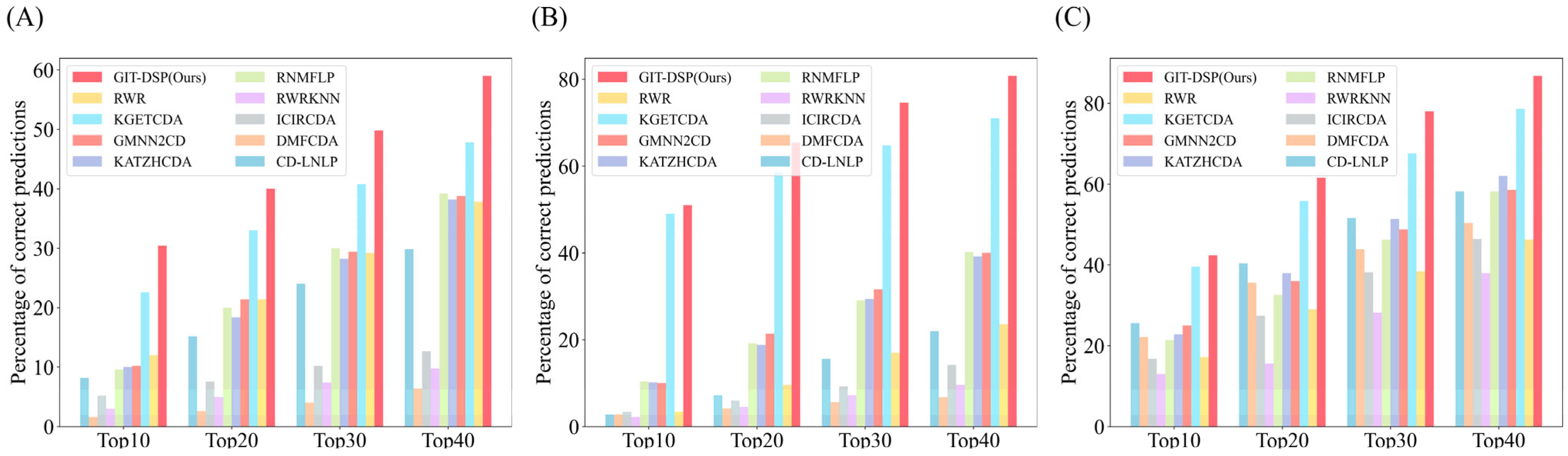
3.3. Performance Comparison
- KATZHCDA: Uses information from heterogeneous graphs to predict associations between circRNA and diseases through the KATZ algorithm.
- RWR: A restart random wander method is used to simulate the process of random wandering on the network, and the restart probability is introduced to regulate the direction of wandering, which achieves the prediction of potential CDAs.
- CD-LNLP: A linear neighborhood label propagation approach that uses known association graphs to propagate labels and thus predict CDAs.
- RWR-KNN: A method that combines the RWR algorithm and the K-nearest neighbor algorithm, with the former evaluating node similarity and the latter enhancing node classification accuracy to improve CDA prediction.
- ICIRCDA: Calculates initial circRNA–disease associations based on diverse biological information, corrects false-negative associations through local association profiles, and uses matrix factorization to compute the final CDA scores.
- RNMFLP: Uses robust non-negative matrix factorization to capture potential CDA pairs and employs the LP algorithm to enhance CDA prediction accuracy from the candidate association pairs.
- DMFCDA: Models non-linear associations through deep matrix factorization and multi-layer neural networks, automatically learning the potential representations of circRNA–disease associations.
- GMNN2CD: Uses graph Markov neural networks to obtain deep features from low-dimensional representations and propagates labels with a graph autoencoder to predict CDAs.
- KGETCDA: Uses a Transformer for knowledge representation and a multi-layer perceptron to calculate CDA affinity scores from embeddings for prediction.
3.4. Ablation Study
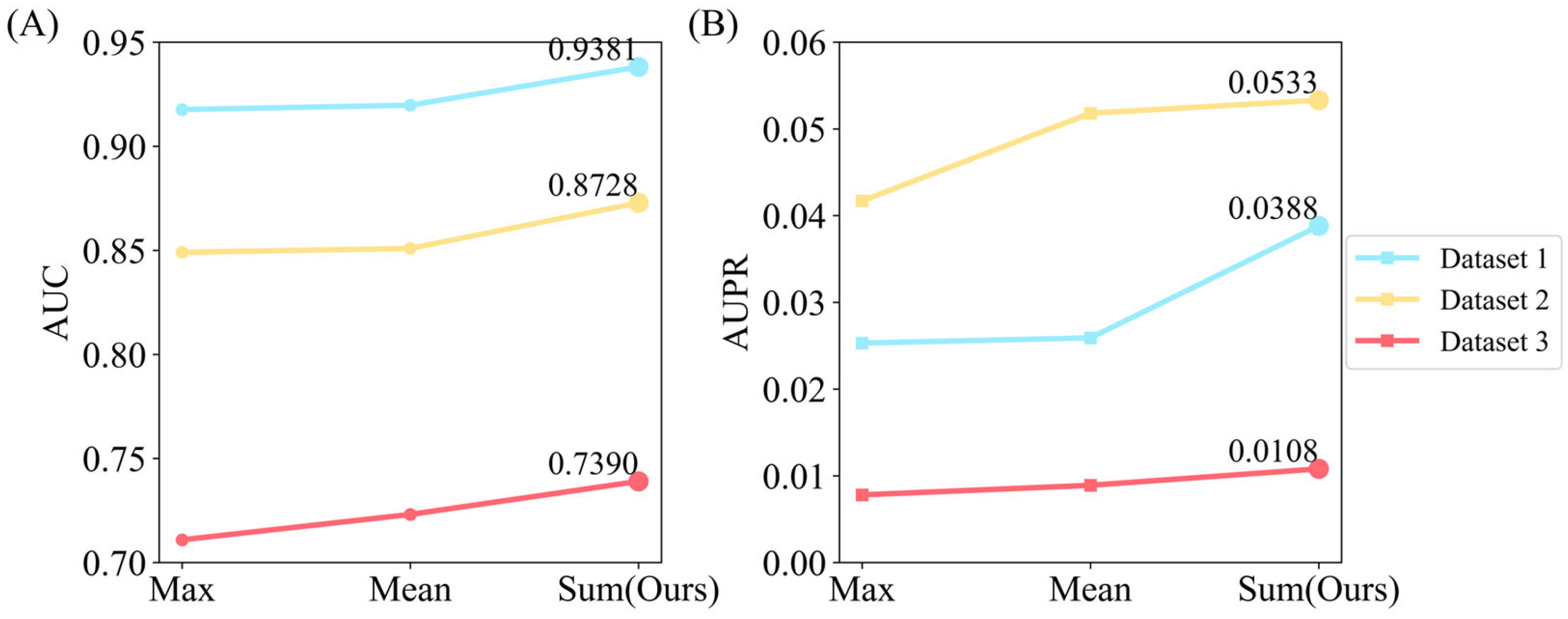
3.4.1. Effect of Different Aggregation and Prediction Modules
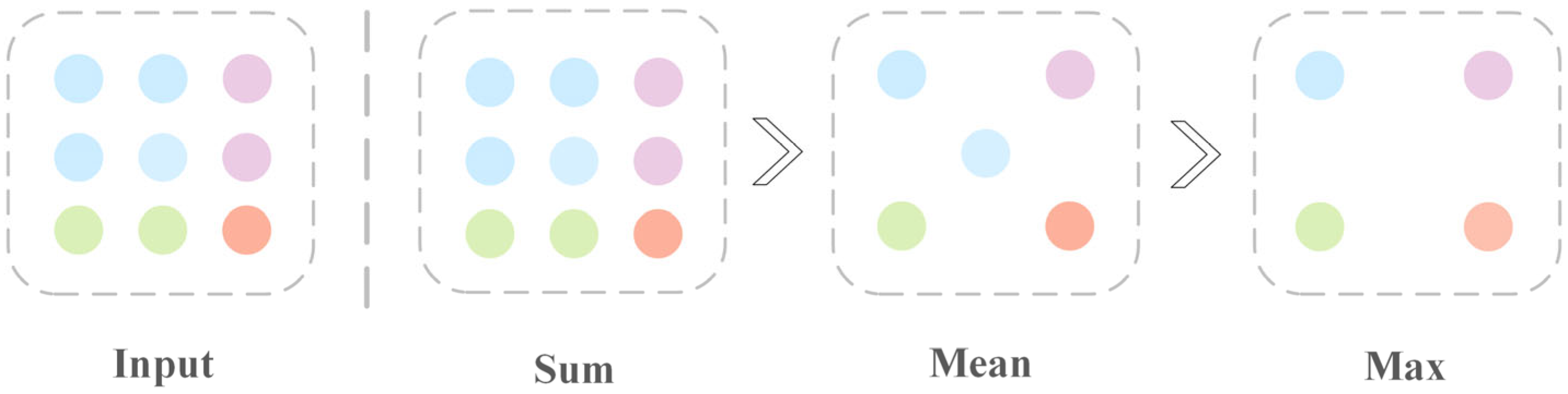
3.4.2. Effect of Different Neighborhood Feature Calculations
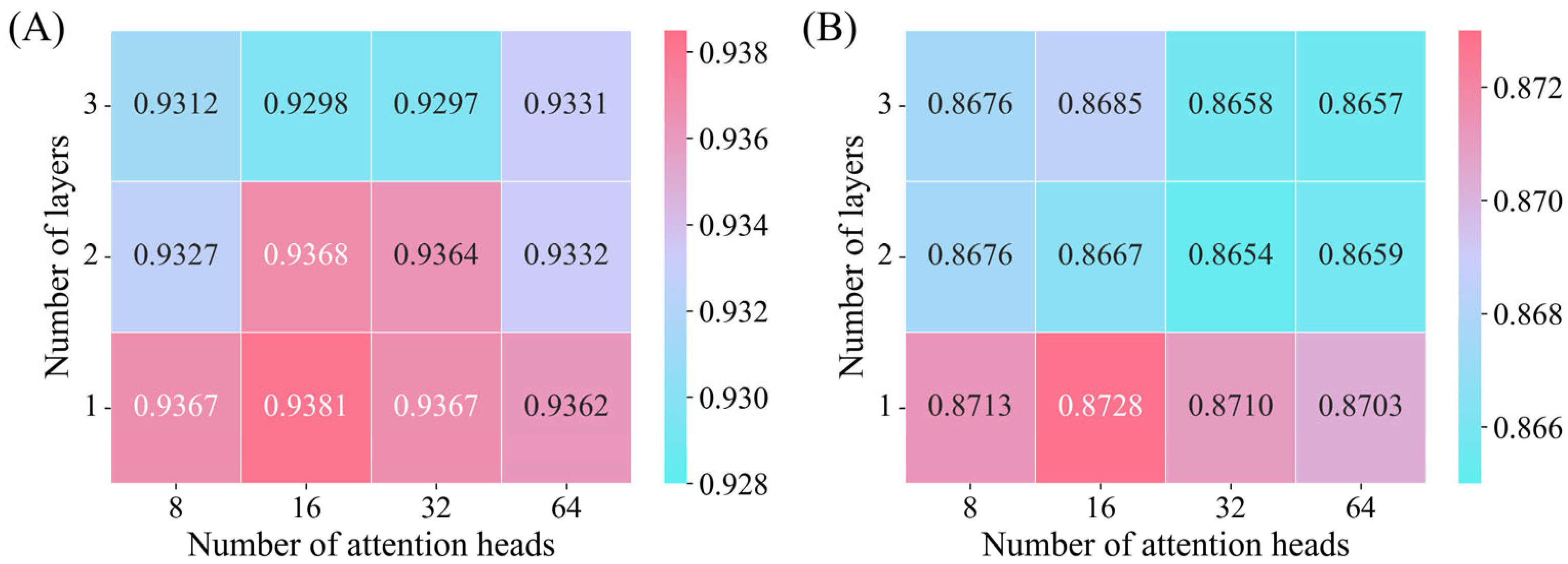
3.4.3. Effect of Different Attention Heads and Layers
3.5. Case Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gu, A.; Jaijyan, D.K.; Yang, S.A.-O.; Zeng, M.; Pei, S.; Zhu, H.A.-O. Functions of Circular RNA in Human Diseases and Illnesses. Non-Coding RNA 2023, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Lei, B.; Yin, Z.; Xu, F.; Liu, X. CircMTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3. Int. J. Mol. Sci. 2024, 25, 2817. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-m.; Kim, W.R.; Park, E.G.; Lee, D.H.; Lee, Y.J.; Shin, H.J.; Jeong, H.-s.; Roh, H.-Y.; Kim, H.-S. Exploring the Regulatory Landscape of Dementia: Insights from Non-Coding RNAs. Int. J. Mol. Sci. 2024, 25, 6190. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, K.; Tan, S.; Xin, J.; Yuan, Q.; Xu, H.; Xu, X.; Liang, Q.; Christiani, D.C.; Wang, M.; et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol. Cancer 2021, 20, 13. [Google Scholar] [CrossRef]
- Wang, C.C.; Han, C.D.; Zhao, Q.; Chen, X. Circular RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2021, 22, bbab286. [Google Scholar] [CrossRef]
- Lan, W.A.-O.; Dong, Y.; Zhang, H.; Li, C.; Chen, Q.A.-O.; Liu, J.; Wang, J.A.-O.; Chen, Y.A.-O. Benchmarking of computational methods for predicting circRNA-disease associations. Brief. Bioinform. 2023, 24, bbac613. [Google Scholar] [CrossRef]
- Fan, C.; Lei, X.; Wu, F.X. Prediction of CircRNA-Disease Associations Using KATZ Model Based on Heterogeneous Networks. Int. J. Biol. Sci. 2018, 14, 1950. [Google Scholar] [CrossRef]
- Deng, L.; Zhang, W.; Shi, Y.; Tang, Y. Fusion of multiple heterogeneous networks for predicting circRNA-disease associations. Sci. Rep. 2019, 9, 960. [Google Scholar] [CrossRef]
- Vural, H.; Kaya, M.; Alhajj, R. A Model Based on Random Walk with Restart to Predict CircRNA—Disease Associations on Heterogeneous Network. In Proceedings of the 2019 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM), Vancouver, BC, Canada, 27–30 August 2019; pp. 929–932. [Google Scholar]
- Wang, Y.; Nie, C.; Zang, T.; Wang, Y. Predicting circRNA-Disease Associations Based on circRNA Expression Similarity and Functional Similarity. Front. Genet. 2019, 10, 832. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, C.; Wang, X.; Liu, F. Predicting CircRNA-Disease Associations Through Linear Neighborhood Label Propagation Method. IEEE Access 2019, 7, 83474–83483. [Google Scholar] [CrossRef]
- Ge, E.; Yang, Y.; Gang, M.; Fan, C.; Zhao, Q. Predicting human disease-associated circRNAs based on locality-constrained linear coding. Genomics 2020, 112, 1335–1342. [Google Scholar] [CrossRef]
- Lei, X.; Fang, Z. GBDTCDA: Predicting circRNA-disease Associations Based on Gradient Boosting Decision Tree with Multiple Biological Data Fusion. Int. J. Biol. Sci. 2019, 15, 2911. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Bian, C. Integrating random walk with restart and k-Nearest Neighbor to identify novel circRNA-disease association. Sci. Rep. 2020, 10, 1943. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Liu, B. iCircDA-MF: Identification of circRNA-disease associations based on matrix factorization. Brief. Bioinform. 2020, 21, 1356–1367. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Yang, C.; Huang, L.; Chen, X.; Fu, X.; Liu, W. RNMFLP: Predicting circRNA-disease associations based on robust nonnegative matrix factorization and label propagation. Brief. Bioinform. 2022, 23, bbac155. [Google Scholar] [CrossRef]
- Lu, C.; Zeng, M.; Zhang, F.; Wu, F.X.; Li, M.; Wang, J. Deep Matrix Factorization Improves Prediction of Human CircRNA-Disease Associations. IEEE J. Biomed. Health Inform. 2020, 25, 891–899. [Google Scholar] [CrossRef]
- Lan, W.; Dong, Y.; Chen, Q.; Liu, J.; Wang, J.; Chen, Y.P.P.; Pan, S. IGNSCDA: Predicting CircRNA-Disease Associations Based on Improved Graph Convolutional Network and Negative Sampling. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 19, 3530–3538. [Google Scholar] [CrossRef]
- Lan, W.A.-O.; Dong, Y.; Chen, Q.A.-O.; Zheng, R.; Liu, J.; Pan, Y.; Chen, Y.A.-O. KGANCDA: Predicting circRNA-disease associations based on knowledge graph attention network. Brief. Bioinform. 2022, 23, bbab494. [Google Scholar] [CrossRef]
- Niu, M.A.-O.; Zou, Q.A.-O.; Wang, C.A.-O. GMNN2CD: Identification of circRNA-disease associations based on variational inference and graph Markov neural networks. Bioinformatics 2022, 38, 2246–2253. [Google Scholar] [CrossRef]
- Wu, J.; Ning, Z.; Ding, Y.; Wang, Y.; Peng, Q.; Fu, L.A.-O. KGETCDA: An efficient representation learning framework based on knowledge graph encoder from transformer for predicting circRNA-disease associations. Brief. Bioinform. 2023, 24, bbad292. [Google Scholar] [CrossRef]
- Rophina, M.; Sharma, D.; Poojary, M.; Scaria, V. Circad: A comprehensive manually curated resource of circular RNA associated with diseases. Database 2020, 2020, baaa019. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, K.; Wu, F.; Wang, W.; Zhang, K.; Hu, H.; Liu, Y.; Jiang, T. circRNA disease: A manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.-R.; Liu, W.; Zhang, Q.; Guo, A.-Y. lncRNASNP2: An updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2017, 46, D276–D280. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012, 41, D983–D986. [Google Scholar] [CrossRef]
- Lan, W.; Zhu, M.; Chen, Q.; Chen, B.; Liu, J.; Li, M.; Chen, Y.-P.P. CircR2Cancer: A manually curated database of associations between circRNAs and cancers. Database 2020, 2020, baaa085. [Google Scholar] [CrossRef]
- Chen, J.; Lin, J.; Hu, Y.; Ye, M.; Yao, L.; Wu, L.; Zhang, W.; Wang, M.; Deng, T.; Guo, F.; et al. RNADisease v4.0: An updated resource of RNA-associated diseases, providing RNA-disease analysis, enrichment and prediction. Nucleic Acids Res. 2022, 51, D1397–D1404. [Google Scholar] [CrossRef]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2020, 49, D1251–D1258. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2018, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Yao, D.; Zhang, L.; Zheng, M.; Sun, X.; Lu, Y.; Liu, P. Circ2Disease: A manually curated database of experimentally validated circRNAs in human disease. Sci. Rep. 2018, 8, 11018. [Google Scholar] [CrossRef]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA–disease associations. Nucleic Acids Res. 2018, 47, D1013–D1017. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lin, P.; Li, D.; Wang, W.; Qi, X.; Li, J.; Xiong, J. MGACL: Prediction Drug–Protein Interaction Based on Meta-Graph Association-Aware Contrastive Learning. Biomolecules 2024, 14, 1267. [Google Scholar] [CrossRef]
- Park, C.; Lee, H.; Jeong, O.-r. Leveraging Medical Knowledge Graphs and Large Language Models for Enhanced Mental Disorder Information Extraction. Future Internet 2024, 16, 260. [Google Scholar] [CrossRef]
- Yu, L.; Xu, Z.; Cheng, M.; Lin, W.; Qiu, W.; Xiao, X. MSEDDI: Multi-Scale Embedding for Predicting Drug—Drug Interaction Events. Int. J. Mol. Sci. 2023, 24, 4500. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, X.; Cao, Y.; Liu, M.; Chua, T.-S. KGAT: Knowledge Graph Attention Network for Recommendation. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 950–958. [Google Scholar]
- Jiang, P.; Zhang, Z.; Yu, Q.; Wang, Z.; Diao, L.; Li, D. ToxDAR: A Workflow Software for Analyzing Toxicologically Relevant Proteomic and Transcriptomic Data, from Data Preparation to Toxicological Mechanism Elucidation. Int. J. Mol. Sci. 2024, 25, 9544. [Google Scholar] [CrossRef]
- Hopp, M.-T.; Domingo-Fernández, D.; Gadiya, Y.; Detzel, M.S.; Graf, R.; Schmalohr, B.F.; Kodamullil, A.T.; Imhof, D.; Hofmann-Apitius, M. Linking COVID-19 and Heme-Driven Pathophysiologies: A Combined Computational–Experimental Approach. Biomolecules 2021, 11, 644. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; He, C.; Wei, P.; Su, Y.; Xia, J.; Zheng, C. Prediction of circRNA-Disease Associations Based on the Combination of Multi-Head Graph Attention Network and Graph Convolutional Network. Biomolecules 2022, 12, 932. [Google Scholar] [CrossRef]
- Gao, Z.; Fu, G.; Ouyang, C.; Tsutsui, S.; Liu, X.; Yang, J.; Gessner, C.; Foote, B.; Wild, D.; Ding, Y.; et al. edge2vec: Representation learning using edge semantics for biomedical knowledge discovery. BMC Bioinform. 2019, 20, 306. [Google Scholar] [CrossRef]
- Kabir, A.; Shehu, A. GOProFormer: A Multi-Modal Transformer Method for Gene Ontology Protein Function Prediction. Biomolecules 2022, 12, 1709. [Google Scholar] [CrossRef]
- Wang, X.; Yang, L.; Wang, R. mRCat: A Novel CatBoost Predictor for the Binary Classification of mRNA Subcellular Localization by Fusing Large Language Model Representation and Sequence Features. Biomolecules 2024, 14, 767. [Google Scholar] [CrossRef]
- Xie, W.; Fang, Y.; Yang, G.; Yu, K.; Li, W. Transformer-Based Multi-Modal Data Fusion Method for COPD Classification and Physiological and Biochemical Indicators Identification. Biomolecules 2023, 13, 1391. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, N.; Li, L.; Deng, S.; Tan, C.; Xu, C.; Huang, F.; Si, L.; Chen, H. Hybrid Transformer with Multi-level Fusion for Multimodal Knowledge Graph Completion. In Proceedings of the 45th International ACM SIGIR Conference on Research and Development in Information Retrieval, Madrid, Spain, 11–15 July 2022; pp. 904–915. [Google Scholar]
- Huang, W.; Mao, Y.; Yang, Z.; Zhu, L.; Long, J. Relation classification via knowledge graph enhanced transformer encoder. Knowl.-Based Syst. 2020, 206, 106321. [Google Scholar] [CrossRef]
- Bi, Z.; Cheng, S.; Chen, J.; Liang, X.; Xiong, F.; Zhang, N. Relphormer: Relational Graph Transformer for Knowledge Graph Representations. Neurocomputing 2024, 566, 11. [Google Scholar] [CrossRef]
- Baghershahi, P.; Hosseini, R.; Moradi, H. Self-attention presents low-dimensional knowledge graph embeddings for link prediction. Knowl.-Based Syst. 2023, 260, 8. [Google Scholar] [CrossRef]
- Sun, Z.; Deng, Z.-H.; Nie, J.-Y.; Tang, J. RotatE: Knowledge Graph Embedding by Relational Rotation in Complex Space. arXiv 2019, arXiv:1902.10197. [Google Scholar]
- Wang, L.; You, Z.H.; Li, J.Q.; Huang, Y.A. IMS-CDA: Prediction of CircRNA-Disease Associations From the Integration of Multisource Similarity Information With Deep Stacked Autoencoder Model. IEEE Trans. Cybern. 2020, 51, 5522–5531. [Google Scholar] [CrossRef]
- Lan, W.; Zhang, H.; Dong, Y.; Chen, Q.; Cao, J.; Peng, W.; Liu, J.; Li, M. DRGCNCDA: Predicting circRNA-disease interactions based on knowledge graph and disentangled relational graph convolutional network. Methods 2022, 208, 35–41. [Google Scholar] [CrossRef]
- Yang, X.-H.; Ma, G.-F.; Jin, X.; Long, H.-X.; Xiao, J.; Ye, L. Knowledge graph embedding and completion based on entity community and local importance. Appl. Intell. 2023, 53, 22132–22142. [Google Scholar] [CrossRef]
- Qiu, J.; Tang, J.; Ma, H.; Dong, Y.; Wang, K.; Tang, J. DeepInf: Social Influence Prediction with Deep Learning. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 2110–2119. [Google Scholar]
- Keyulu, X.; Weihua, H.; Jure, L.; Stefanie, J. How Powerful are Graph Neural Networks? In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019; pp. 1–17. [Google Scholar]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive representation learning on large graphs. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 1025–1035. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph neural networks: A review of methods and applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Dai, Q.; Liu, Z.; Wang, Z.; Duan, X.; Guo, M. GraphCDA: A hybrid graph representation learning framework based on GCN and GAT for predicting disease-associated circRNAs. Brief. Bioinform. 2022, 23, bbac379. [Google Scholar] [CrossRef] [PubMed]
- Khwaja, A.; Bjorkholm, M.; Gale, R.E.; Levine, R.L.; Jordan, C.T.; Ehninger, G.; Bloomfield, C.D.; Estey, E.; Burnett, A.; Cornelissen, J.J.; et al. Acute myeloid leukaemia. Nat. Rev. Dis. Primers 2016, 2, 16010. [Google Scholar] [CrossRef]
- Haferlach, T.; Schmidts, I. The power and potential of integrated diagnostics in acute myeloid leukaemia. Br. J. Haematol. 2020, 188, 36–48. [Google Scholar] [CrossRef]
- Chen, H.; Liu, T.; Liu, J.; Feng, Y.; Wang, B.; Wang, J.; Bai, J.; Zhao, W.; Shen, Y.; Wang, X.; et al. Circ-ANAPC7 is Upregulated in Acute Myeloid Leukemia and Appears to Target the MiR-181 Family. Cell. Physiol. Biochem. 2018, 47, 1998–2007. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Jia, Y.; Zhang, R.; Chen, H.; Feng, Y.; Li, F.; Wang, T.; Bai, J.; He, A.; Yang, Y. Using Circ-ANAPC7 as a Novel Type of Biomarker in the Monitoring of Acute Myeloid Leukemia. Acta Haematol. 2022, 145, 176–183. [Google Scholar] [CrossRef]
- Shang, J.; Chen, W.M.; Liu, S.; Wang, Z.H.; Wei, T.N.; Chen, Z.Z.; Wu, W.B. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk. Res. 2019, 85, 106198. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Liu, J.; Jin, Y.; Wang, W.A.-O. Circular RNA circ_0004277 Inhibits Acute Myeloid Leukemia Progression Through MicroRNA-134-5p/Single stranded DNA binding protein 2. Bioengineered 2022, 13, 9662–9673. [Google Scholar] [CrossRef]
- Bray, F.A.-O.; Laversanne, M.; Sung, H.A.-O.; Ferlay, J.; Siegel, R.A.-O.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Gao, S.; Yu, Y.; Liu, L.; Meng, J.; Li, G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019, 233, 116692. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Bahranian, A.; Taheri, M.; Hallajnejad, M. CircMTO1: A circular RNA with roles in the carcinogenesis. Biomed. Pharmacother. 2021, 142, 112025. [Google Scholar] [CrossRef]
- Sui, M.H.; Zhang, W.W.; Geng, D.M.; Sun, D.J. CircPRKCI regulates proliferation, migration and cycle of lung adenocarcinoma cells by targeting miR-219a-5p-regulated CAMK1D. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1899–1909. [Google Scholar]





| Dataset | Databases | circRNA– Disease | circRNA– miRNA | miRNA– Disease | lncRNA– miRNA | lncRNA– Disease | Total |
|---|---|---|---|---|---|---|---|
| Dataset 1 | Circad [22], CircRNADisease [23], LncRNASNP2 [24], LncRNADisease [25] | 346 | 146 | 106 | 202 | 527 | 1327 |
| Dataset 2 | CircR2Cancer [26], LncRNASNP2 [24], LncRNADisease [25] | 647 | 756 | 732 | 308 | 1066 | 3509 |
| Dataset 3 | Circad [22], MNDR [27], Lnc2Cnacer [28], LncRNADisease [29], CircRNADisease [23], Circ2Disease [30], CircR2Cancer [26], HMDD [31], StarBase [32] | 1399 | 1129 | 10,154 | 9506 | 3280 | 25,468 |
| Categories | Model | Advantages | Disadvantages |
|---|---|---|---|
| Information Propagation | KATZHCDA | Simple and efficient, suitable for capturing local associations. | Struggles with sparse data and global association capture. |
| RWR | |||
| CD-LNLP | |||
| Traditional Machine Learning | RWR-KNN | Robust to noise and leverages classic algorithms for classification. | Requires feature engineering and is computationally expensive for large data. |
| ICIRCDA | |||
| RNMFLP | |||
| Deep Learning | DMFCDA | Automatic extraction of higher-order features. | Difficulty in mining and distinguishing higher-order associations. |
| GMNN2CD | |||
| KGETCDA |
| Model | Dataset 1 | Dataset 2 | Dataset 3 | |||
|---|---|---|---|---|---|---|
| AUC | AUPR | AUC | AUPR | AUC | AUPR | |
| KATZHCDA | 0.8033 | 0.0103 | 0.7539 | 0.0129 | 0.5608 | 0.0056 |
| RWR | 0.8302 | 0.0101 | 0.7153 | 0.0089 | 0.6087 | 0.0044 |
| CD-LNLP | 0.7956 | 0.0085 | 0.6956 | 0.0083 | 0.6404 | 0.0059 |
| RWR-KNN | 0.5021 | 0.0028 | 0.5033 | 0.0041 | 0.5046 | 0.0027 |
| ICIRCDA | 0.5662 | 0.0036 | 0.5726 | 0.0051 | 0.6358 | 0.0046 |
| RNMFLP | 0.8726 | 0.0113 | 0.7333 | 0.0130 | 0.5216 | 0.0050 |
| DMFCDA | 0.6367 | 0.0034 | 0.5883 | 0.0047 | 0.6258 | 0.0050 |
| GMNN2CD | 0.8705 | 0.0106 | 0.8109 | 0.0145 | 0.7084 | 0.0069 |
| KGETCDA | 0.9101 | 0.0238 | 0.8483 | 0.0509 | 0.7122 | 0.0070 |
| GIT-DSP(Ours) | 0.9381 | 0.0388 | 0.8728 | 0.0566 | 0.7390 | 0.0108 |
| Improvement (%) | 3.08% | 63.03% | 2.89% | 11.20% | 3.76% | 54.29% |
| Disease | Candidate circRNA | Rank | Evidence (PMID) |
|---|---|---|---|
| acute myeloid leukemia | hsa_circ_100290 | 1 | 30424877 |
| hsa_circ_0000488 | 2 | Unknown | |
| circ-ANAPC7 | 3 | 29969755, 34879367 | |
| circPAN3 | 4 | 30395908, 31401408 | |
| hsa_circ_0035381 | 5 | 28282919, 35917008 | |
| hsa_circ_0001187 | 6 | 28282919, 37280654 | |
| hsa_circ_102533 | 7 | Unknown | |
| hsa_circ_0004277 | 8 | 35412941 | |
| circ_AFF2 | 9 | 28282919 | |
| hsa_circ_0000254 | 10 | 29950198 |
| Disease | Candidate circRNA | Rank | Evidence (PMID) |
|---|---|---|---|
| lung cancer | hsa_circ_0007059 | 1 | 31351967 |
| circ-PRMT5 | 2 | Unknown | |
| circMTO1 | 3 | 30975029 | |
| circ-PRKCI | 4 | 29588350, 33155212, 33660800 | |
| hsa_circ_0046264 | 5 | 29891014 | |
| circPUM1 | 6 | 30528736, 37326964 | |
| hsa_circ_0003028 | 7 | Unknown | |
| CDR1as | 8 | 30841451, 31881486, 36508830 | |
| hsa_circ_0007915 | 9 | Unknown | |
| circ-ERBB2 | 10 | 31109436, 33506582 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Qian, Y.; Sun, Z.; Zhu, H. Prediction of circRNA–Disease Associations via Graph Isomorphism Transformer and Dual-Stream Neural Predictor. Biomolecules 2025, 15, 234. https://doi.org/10.3390/biom15020234
Li H, Qian Y, Sun Z, Zhu H. Prediction of circRNA–Disease Associations via Graph Isomorphism Transformer and Dual-Stream Neural Predictor. Biomolecules. 2025; 15(2):234. https://doi.org/10.3390/biom15020234
Chicago/Turabian StyleLi, Hongchan, Yuchao Qian, Zhongchuan Sun, and Haodong Zhu. 2025. "Prediction of circRNA–Disease Associations via Graph Isomorphism Transformer and Dual-Stream Neural Predictor" Biomolecules 15, no. 2: 234. https://doi.org/10.3390/biom15020234
APA StyleLi, H., Qian, Y., Sun, Z., & Zhu, H. (2025). Prediction of circRNA–Disease Associations via Graph Isomorphism Transformer and Dual-Stream Neural Predictor. Biomolecules, 15(2), 234. https://doi.org/10.3390/biom15020234






