Advances in Computational Drug Repurposing, Driver Genes, and Therapeutics in Lung Adenocarcinoma
Abstract
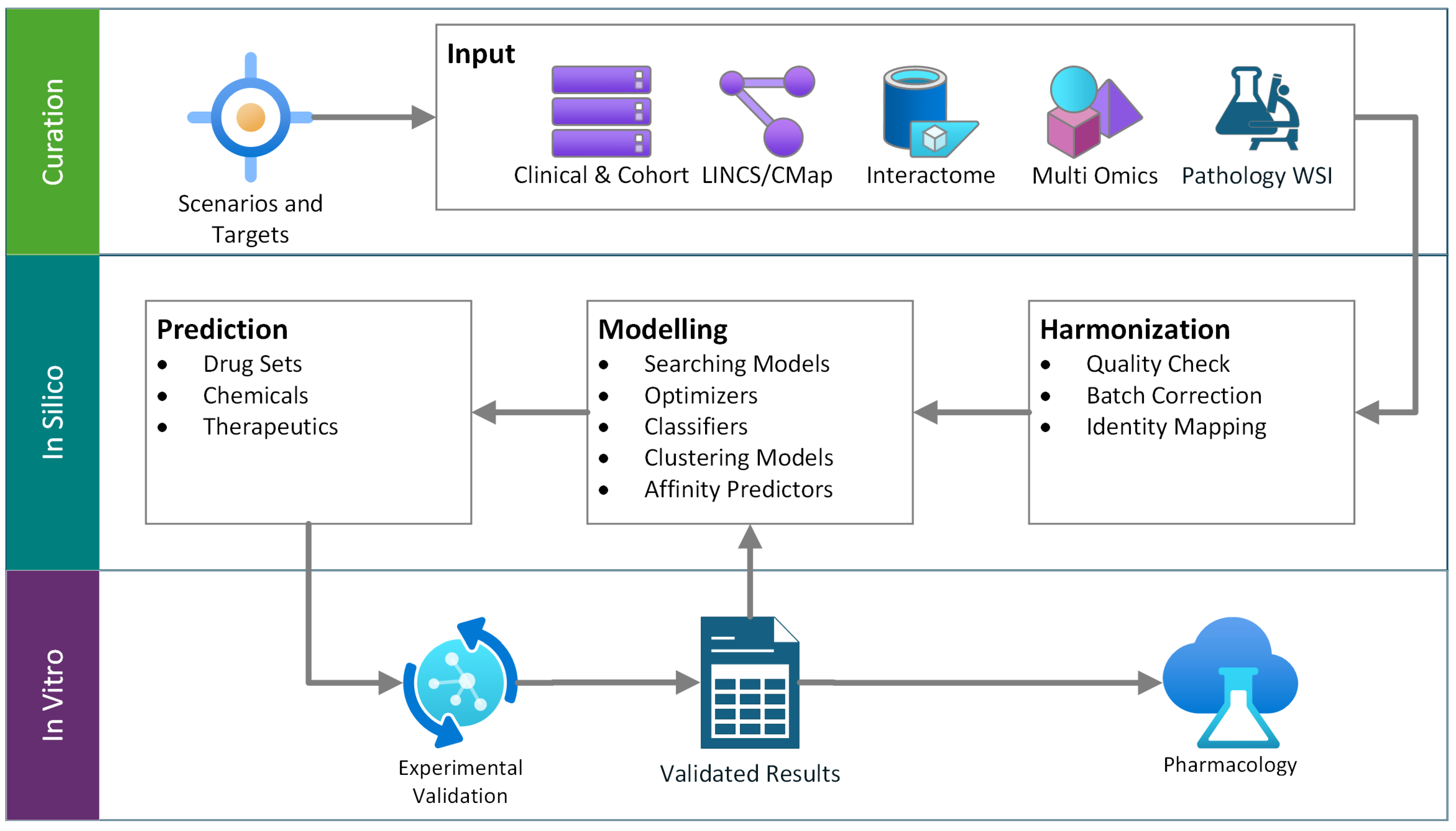
1. Drug Repurposing Review
2. Cancer, Lung Adenocarcinoma, and Recent Therapeutics
3. Gene Dysregulation in LUAD and Therapeutic Counteractions
4. Review on Drug Repurposing in LUAD
4.1. Connectivity Map/LINCS
4.2. Network Medicine/Interactome Proximity
4.3. Knowledge-Graph (KG) Methods
4.4. Ligand-Based Similarity/Chemogenomics
4.5. Side-Effect (Phenotypic) Similarity Mining
4.6. Structure-Based Virtual Screening & Inverse Docking
4.7. Machine Learning/Deep Learning
4.8. Pathway- & Enrichment-Based Approaches
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, M.A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; Carpenter, A.E. Cell Painting, a High-Content Image-Based Assay for Morphological Profiling Using Multiplexed Fluorescent Dyes. Nat. Protoc. 2016, 11, 1757–1774. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Vincent, F.; Lee, J.A.; Eder, J.; Prunotto, M. Opportunities and Challenges in Phenotypic Drug Discovery: An Industry Perspective. Nat. Rev. Drug Discov. 2017, 16, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabási, A.-L.; Loscalzo, J. Network-Based Approach to Prediction and Population-Based Validation of in Silico Drug Repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Ali, R.; Cheng, F. Drug Repurposing Using FDA Adverse Event Reporting System (FAERS) Database. Curr. Drug Targets 2024, 25, 454–464. [Google Scholar] [CrossRef]
- Nelson, M.R.; Tipney, H.; Painter, J.L.; Shen, J.; Nicoletti, P.; Shen, Y.; Floratos, A.; Sham, P.C.; Li, M.J.; Wang, J.; et al. The Support of Human Genetic Evidence for Approved Drug Indications. Nat. Genet. 2015, 47, 856–860. [Google Scholar] [CrossRef]
- Lin, Z.; Pan, W. A Robust Cis-Mendelian Randomization Method with Application to Drug Target Discovery. Nat. Commun. 2024, 15, 6072. [Google Scholar] [CrossRef]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef]
- Morselli Gysi, D.; do Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.J.; Davey, R.A.; Loscalzo, J.; et al. Network Medicine Framework for Identifying Drug-Repurposing Opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118. [Google Scholar] [CrossRef]
- Himmelstein, D.S.; Lizee, A.; Hessler, C.; Brueggeman, L.; Chen, S.L.; Hadley, D.; Green, A.; Khankhanian, P.; Baranzini, S.E. Systematic Integration of Biomedical Knowledge Prioritizes Drugs for Repurposing. eLife 2017, 6, e26726. [Google Scholar] [CrossRef]
- Keiser, M.J.; Setola, V.; Irwin, J.J.; Laggner, C.; Abbas, A.I.; Hufeisen, S.J.; Jensen, N.H.; Kuijer, M.B.; Matos, R.C.; Tran, T.B.; et al. Predicting New Molecular Targets for Known Drugs. Nature 2009, 462, 175–181. [Google Scholar] [CrossRef]
- Campillos, M.; Kuhn, M.; Gavin, A.-C.; Jensen, L.J.; Bork, P. Drug Target Identification Using Side-Effect Similarity. Science 2008, 321, 263–266. [Google Scholar] [CrossRef]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 481382. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, H.; Özgür, A.; Ozkirimli, E. DeepDTA: Deep Drug–Target Binding Affinity Prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef] [PubMed]
- Bang, D.; Lim, S.; Lee, S.; Kim, S. Biomedical Knowledge Graph Learning for Drug Repurposing by Extending Guilt-by-Association to Multiple Layers. Nat. Commun. 2023, 14, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Plans, J.; Piñero, J.; Menche, J.; Sanz, F.; Furlong, L.I.; Schmidt, H.H.H.W.; Oliva, B.; Guney, E. Proximal Pathway Enrichment Analysis for Targeting Comorbid Diseases via Network Endopharmacology. Pharmaceuticals 2018, 11, 61. [Google Scholar] [CrossRef]
- Brown, J.S.; Amend, S.R.; Austin, R.H.; Gatenby, R.A.; Hammarlund, E.U.; Pienta, K.J. Updating the Definition of Cancer. Mol. Cancer Res. 2023, 21, 1142–1147. [Google Scholar] [CrossRef]
- Han, G.; Sinjab, A.; Rahal, Z.; Lynch, A.M.; Treekitkarnmongkol, W.; Liu, Y.; Serrano, A.G.; Feng, J.; Liang, K.; Khan, K.; et al. An Atlas of Epithelial Cell States and Plasticity in Lung Adenocarcinoma. Nature 2024, 627, 656–663. [Google Scholar] [CrossRef]
- Yang, M.; Shen, H.; Flodby, P.; Koss, M.D.; Bassiouni, R.; Liu, Y.; Jashashvili, T.; Neely, A.; Ogbolu, E.; Castillo, J.; et al. Alveolar Type I Cells Can Give Rise to KRAS-Induced Lung Adenocarcinoma. Cell Rep. 2023, 42, 113286. [Google Scholar] [CrossRef]
- Seguin, L.; Durandy, M.; Feral, C.C. Lung Adenocarcinoma Tumor Origin: A Guide for Personalized Medicine. Cancers 2022, 14, 1759. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Ahmedin, J. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Zahed, H.; Feng, X.; Sheikh, M.; Bray, F.; Ferlay, J.; Ginsburg, O.; Shiels, M.S.; Robbins, H.A. Age at Diagnosis for Lung, Colon, Breast and Prostate Cancers: An International Comparative Study. Int. J. Cancer 2024, 154, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Schafer, E.J.; Sung, H.; Bandi, P.; Kratzer, T.; Islami, F.; Siegel, R.L. The Burden of Lung Cancer in Women Compared With Men in the US. JAMA Oncol. 2023, 9, 1727–1728. [Google Scholar] [CrossRef]
- Sisoudiya, S.D.; Houle, A.A.; Fernando, T.; Wilson, T.R.; Schutzman, J.L.; Lee, J.; Schrock, A.; Sokol, E.S.; Sivakumar, S.; Shi, Z.; et al. Ancestry-Associated Co-Alteration Landscape of KRAS and EGFR-Altered Non-Squamous NSCLC. NPJ Precis. Oncol. 2024, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Saka, A.H.; Giaquinto, A.N.; McCullough, L.E.; Tossas, K.Y.; Star, J.; Jemal, A.; Siegel, R.L. Cancer Statistics for African American and Black People, 2025. CA Cancer J. Clin. 2025, 75, 111–140. [Google Scholar] [CrossRef]
- Haga, Y.; Sakamoto, Y.; Kajiya, K.; Kawai, H.; Oka, M.; Motoi, N.; Shirasawa, M.; Yotsukura, M.; Watanabe, S.I.; Arai, M.; et al. Whole-Genome Sequencing Reveals the Molecular Implications of the Stepwise Progression of Lung Adenocarcinoma. Nat. Commun. 2023, 14, 8375. [Google Scholar] [CrossRef]
- Paredes, R.; Borea, R.; Drago, F.; Russo, A.; Nigita, G.; Rolfo, C. Genetic Drivers of Tumor Microenvironment and Immunotherapy Resistance in Non-Small Cell Lung Cancer: The Role of KEAP1, SMARCA4, and PTEN Mutations. J. Immunother. Cancer 2025, 13, 12288. [Google Scholar] [CrossRef]
- Zhou, C.; Tang, K.-J.; Cho, B.C.; Liu, B.; Paz-Ares, L.; Cheng, S.; Kitazono, S.; Thiagarajan, M.; Goldman, J.W.; Sabari, J.K.; et al. Amivantamab plus Chemotherapy in NSCLC with EGFR Exon 20 Insertions. N. Engl. J. Med. 2023, 389, 2039–2051. [Google Scholar] [CrossRef]
- Cho, B.C.; Lu, S.; Felip, E.; Spira, A.I.; Girard, N.; Lee, J.-S.; Lee, S.-H.; Ostapenko, Y.; Danchaivijitr, P.; Liu, B.; et al. Amivantamab plus Lazertinib in Previously Untreated EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2024, 391, 1486–1498. [Google Scholar] [CrossRef]
- Tsuboi, M.; Herbst, R.S.; John, T.; Kato, T.; Majem, M.; Grohé, C.; Wang, J.; Goldman, J.W.; Lu, S.; Su, W.-C.; et al. Overall Survival with Osimertinib in Resected EGFR -Mutated NSCLC. N. Engl. J. Med. 2023, 389, 137–147. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Dziadziuszko, R.; Ahn, J.S.; Barlesi, F.; Nishio, M.; Lee, D.H.; Lee, J.-S.; Zhong, W.; Horinouchi, H.; Mao, W.; et al. Alectinib in Resected ALK -Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2024, 390, 1265–1276. [Google Scholar] [CrossRef]
- Forde, P.M.; Spicer, J.D.; Provencio, M.; Mitsudomi, T.; Awad, M.M.; Wang, C.; Lu, S.; Felip, E.; Swanson, S.J.; Brahmer, J.R.; et al. Overall Survival with Neoadjuvant Nivolumab plus Chemotherapy in Lung Cancer. N. Engl. J. Med. 2025. [Google Scholar] [CrossRef] [PubMed]
- Wakelee, H.; Liberman, M.; Kato, T.; Tsuboi, M.; Lee, S.-H.; Gao, S.; Chen, K.-N.; Dooms, C.; Majem, M.; Eigendorff, E.; et al. Perioperative Pembrolizumab for Early-Stage Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2023, 389, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.K.; Yao, W.; Duruisseaux, M.; Doucet, L.; Azkárate Martínez, A.; Gregorc, V.; Juan-Vidal, O.; Lu, S.; De Bondt, C.; de Marinis, F.; et al. KRYSTAL-12: Phase 3 Study of Adagrasib versus Docetaxel in Patients with Previously Treated Advanced/Metastatic Non-Small Cell Lung Cancer (NSCLC) Harboring a KRASG12C Mutation. J. Clin. Oncol. 2024, 42, LBA8509. [Google Scholar] [CrossRef]
- de Langen, A.J.; Johnson, M.L.; Mazieres, J.; Dingemans, A.M.C.; Mountzios, G.; Pless, M.; Wolf, J.; Schuler, M.; Lena, H.; Skoulidis, F.; et al. Sotorasib versus Docetaxel for Previously Treated Non-Small-Cell Lung Cancer with KRASG12C Mutation: A Randomised, Open-Label, Phase 3 Trial. Lancet 2023, 401, 733–746. [Google Scholar] [CrossRef]
- Li, B.T.; Smit, E.F.; Goto, Y.; Nakagawa, K.; Udagawa, H.; Mazières, J.; Nagasaka, M.; Bazhenova, L.; Saltos, A.N.; Felip, E.; et al. Trastuzumab Deruxtecan in HER2-Mutant Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2022, 386, 241–251. [Google Scholar] [CrossRef]
- Savage, S.R.; Yi, X.; Lei, J.T.; Wen, B.; Zhao, H.; Liao, Y.; Jaehnig, E.J.; Somes, L.K.; Shafer, P.W.; Lee, T.D.; et al. Pan-Cancer Proteogenomics Expands the Landscape of Therapeutic Targets. Cell 2024, 187, 4389–4407.e15. [Google Scholar] [CrossRef]
- Planchard, D.; Jänne, P.A.; Cheng, Y.; Yang, J.C.-H.; Yanagitani, N.; Kim, S.-W.; Sugawara, S.; Yu, Y.; Fan, Y.; Geater, S.L.; et al. Osimertinib with or without Chemotherapy in EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2023, 389, 1935–1948. [Google Scholar] [CrossRef]
- Jänne, P.A.; Planchard, D.; Kobayashi, K.; Cheng, Y.; Lee, C.K.; Valdiviezo, N.; Laktionov, K.; Yang, T.-Y.; Yu, Y.; Kato, T.; et al. CNS Efficacy of Osimertinib With or Without Chemotherapy in Epidermal Growth Factor Receptor–Mutated Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2024, 42, 808–820. [Google Scholar] [CrossRef]
- Moldvay, J.; Tímár, J. KRASG12C Mutant Lung Adenocarcinoma: Unique Biology, Novel Therapies and New Challenges. Pathol. Oncol. Res. 2023, 29, 1611580. [Google Scholar] [CrossRef]
- Jänne, P.A.; Theelen, W.S.M.E.; Garassino, M.C.; Spira, A.I.; Laskin, J.J.; de Marinis, F.; Badin, F.B.; Boom, L.; Aguado De La Rosa, C.; Chmielewska, I.; et al. First-Line Adagrasib (ADA) with Pembrolizumab (PEMBRO) in Patients (Pts) with Advanced/Metastatic KRASG12C-Mutated Non-Small Cell Lung Cancer (NSCLC) from the Phase 2 Portion of the KRYSTAL-7 Study. J. Clin. Oncol. 2025, 43, 8500. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, B.; Wu, M.; Zhang, L.; Ji, M. Current Status of KRAS G12C Inhibitors in NSCLC and the Potential for Combination with Anti-PD-(L)1 Therapy: A Systematic Review. Front. Immunol. 2025, 16, 1509173. [Google Scholar] [CrossRef]
- Ou, S.H.I.; Hagopian, G.G.; Zhang, S.S.; Nagasaka, M. Comprehensive Review of ROS1 Tyrosine Kinase Inhibitors-Classified by Structural Designs and Mutation Spectrum (Solvent Front Mutation [G2032R] and Central β-Sheet 6 [Cβ6] Mutation [L2086F]). J. Thorac. Oncol. 2024, 19, 706–718. [Google Scholar] [CrossRef]
- Peters, S.; Gadgeel, S.M.; Mok, T.; Nadal, E.; Kilickap, S.; Swalduz, A.; Cadranel, J.; Sugawara, S.; Chiu, C.-H.; Yu, C.-J.; et al. Entrectinib in ROS1-Positive Advanced Non-Small Cell Lung Cancer: The Phase 2/3 BFAST Trial. Nat. Med. 2024, 30, 1923–1932. [Google Scholar] [CrossRef]
- Drilon, A.; Camidge, D.R.; Lin, J.J.; Kim, S.-W.; Solomon, B.J.; Dziadziuszko, R.; Besse, B.; Goto, K.; de Langen, A.J.; Wolf, J.; et al. Repotrectinib in ROS1 Fusion–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2024, 390, 118–131. [Google Scholar] [CrossRef]
- Gautschi, O.; Park, K.; Solomon, B.J.; Tomasini, P.; Loong, H.H.; De Braud, F.; Goto, K.; Peterson, P.; Barker, S.; Liming, K.; et al. Selpercatinib in RET Fusion–Positive Non–Small Cell Lung Cancer: Final Safety and Efficacy, Including Overall Survival, From the LIBRETTO-001 Phase I/II Trial. J. Clin. Oncol. 2025, 43, 1758–1764. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non–Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Hochmair, M.; Han, J.Y.; Reguart, N.; Souquet, P.J.; Smit, E.F.; Orlov, S.V.; Vansteenkiste, J.; Nishio, M.; de Jonge, M.; et al. Capmatinib in MET Exon 14-Mutated Non-Small-Cell Lung Cancer: Final Results from the Open-Label, Phase 2 GEOMETRY Mono-1 Trial. Lancet Oncol. 2024, 25, 1357–1370. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Hou, L.; Bai, J.; Sun, C.; Wang, D.; An, G. Trastuzumab Deruxtecan (DS8201) for Advanced Non-Small Cell Lung Cancer with HER2 Exon 20 Insertion Mutation: A Case Report. Anticancer. Drugs 2024, 35, 101–108. [Google Scholar] [CrossRef]
- Smit, E.F.; Felip, E.; Uprety, D.; Nagasaka, M.; Nakagawa, K.; Paz-Ares Rodríguez, L.; Pacheco, J.M.; Li, B.T.; Planchard, D.; Baik, C.; et al. Trastuzumab Deruxtecan in Patients with Metastatic Non-Small-Cell Lung Cancer (DESTINY-Lung01): Primary Results of the HER2-Overexpressing Cohorts from a Single-Arm, Phase 2 Trial. Lancet Oncol. 2024, 25, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Swalduz, A.; Beau-Faller, M.; Planchard, D.; Mazieres, J.; Bayle-Bleuez, S.; Debieuvre, D.; Fallet, V.; Geier, M.; Cortot, A.; Couraud, S.; et al. Real-World Efficacy of the Dabrafenib-Trametinib (D-T) Combination in BRAF V600E-Mutated Metastatic Non-Small Cell Lung Cancer (NSCLC): Results from the IFCT-2004 BLaDE Cohort. Lung Cancer 2025, 199, 108038. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.C.; Chiu, C.H.; Massarelli, E.; Buchschacher, G.L.; Goto, K.; Overbeck, T.R.; Loong, H.H.F.; Chee, C.E.; Garrido, P.; Dong, X.; et al. Updated Efficacy and Safety of Entrectinib in NTRK Fusion-Positive Non-Small Cell Lung Cancer. Lung Cancer 2024, 188, 107442. [Google Scholar] [CrossRef] [PubMed]
- Rodon, J.; Rothe, M.; Mangat, P.K.; Garrett-Mayer, E.; Cannon, T.L.; Hobbs, E.; Kalemkerian, G.P.; Hinshaw, D.C.; Gregory, A.; Grantham, G.N.; et al. Afatinib in Patients with Solid Tumors with Neuregulin 1 (NRG1) Fusions: A Case Series from the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. ESMO Open 2025, 10, 104545. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Shim, B.Y.; Lee, S.-B.; Kim, H.; Park, H.S.; An, H.J. Preclinical Evidence for Synergistic Effects of Autophagy Inhibitor and Alpelisib in PI3KCA Mutated Non-Small Cell Lung Cancer: Implications for Future Clinical Trials. J. Clin. Oncol. 2023, 41, e21022. [Google Scholar] [CrossRef]
- Elkamhawy, A.; Lu, Q.; Nada, H.; Woo, J.; Quan, G.; Lee, K. The Journey of DDR1 and DDR2 Kinase Inhibitors as Rising Stars in the Fight Against Cancer. Int. J. Mol. Sci. 2021, 22, 6535. [Google Scholar] [CrossRef]
- Chen, Q.; Zheng, X.; Cheng, W.; Li, J. Landscape of Targeted Therapies for Lung Squamous Cell Carcinoma. Front. Oncol. 2024, 14, 1467898. [Google Scholar] [CrossRef]
- Payne, L.S.; Huang, P.H. Discoidin Domain Receptor 2 Signaling Networks and Therapy in Lung Cancer. J. Thorac. Oncol. 2014, 9, 900–904. [Google Scholar] [CrossRef]
- Day, E.; Waters, B.; Spiegel, K.; Alnadaf, T.; Manley, P.W.; Buchdunger, E.; Walker, C.; Jarai, G. Inhibition of Collagen-Induced Discoidin Domain Receptor 1 and 2 Activation by Imatinib, Nilotinib and Dasatinib. Eur. J. Pharmacol. 2008, 599, 44–53. [Google Scholar] [CrossRef]
- Haura, E.B.; Hicks, J.K.; Boyle, T.A. Erdafitinib Overcomes FGFR3-TACC3–Mediated Resistance to Osimertinib. J. Thorac. Oncol. 2020, 15, e154–e156. [Google Scholar] [CrossRef]
- Tan, X.; Wu, Z.; Chen, M. MAP2K1 K57N Conferred an Acquired Resistance to Furmonertinib, Dabrafenib and Trametinib Combined Therapy in Advanced Lung Adenocarcinoma with EGFR Mutation and BRAF V600E. Onco Targets Ther. 2024, 17, 307–312. [Google Scholar] [CrossRef]
- DiMarco, A.V.; Ravichandran, M.; Lau, J.; Lima, A.; Lacap, J.; Saenz-Lopez Larrocha, P.; Lin, E.; Weng, J.; Gerosa, L.; Hunsaker, T.; et al. RIT1M90I Is a Driver of Lung Adenocarcinoma Tumorigenesis and Resistance to Targeted Therapy. Cancer Res. 2025. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Kwiatkowski, D.J.; Desai, J.; Dagogo-Jack, I.; Millward, M.; Kindler, H.L.; Tolcher, A.W.; Frentzas, S.; Thurston, A.W.; Post, L.; et al. Abstract CT006: First-in-Class, First-in-Human Phase 1 Trial of VT3989, an Inhibitor of Yes-Associated Protein (YAP)/Transcriptional Enhancer Activator Domain (TEAD), in Patients (Pts) with Advanced Solid Tumors Enriched for Malignant Mesothelioma and Other Tumors with Neurofibromatosis 2 (NF2) Mutations. Cancer Res. 2023, 83, CT006. [Google Scholar] [CrossRef]
- Park, J.; Joo, M.S.; Kim, M.J.; Oh, S.; Tran, P.T.; Kwon, M.; Choi, Y.J.; Lee, J.; Kim, E.-J.; Ki, D.H.; et al. High Cereblon Expression in Neuroendocrine Cancer Confers Vulnerability to GSPT1 Molecular Glue Degrader. Exp. Hematol. Oncol. 2025, 14, 89. [Google Scholar] [CrossRef] [PubMed]
- Chapeau, E.A.; Sansregret, L.; Galli, G.G.; Chène, P.; Wartmann, M.; Mourikis, T.P.; Jaaks, P.; Baltschukat, S.; Barbosa, I.A.M.; Bauer, D.; et al. Direct and Selective Pharmacological Disruption of the YAP–TEAD Interface by IAG933 Inhibits Hippo-Dependent and RAS–MAPK-Altered Cancers. Nat. Cancer 2024, 5, 1102–1120. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Lakhani, N.J.; McKean, M.; Lingaraj, T.; Victor, L.; Sanchez-Martin, M.; Kacena, K.; Malek, K.S.; Santillana, S. A Phase 1, First-in-Human Study of IK-930, an Oral TEAD Inhibitor Targeting the Hippo Pathway in Subjects with Advanced Solid Tumors. J. Clin. Oncol. 2022, 40, TPS3168. [Google Scholar] [CrossRef]
- Galan-Cobo, A.; Vokes, N.I.; Qian, Y.; Molkentine, D.; Ramkumar, K.; Paula, A.G.; Pisegna, M.; McGrail, D.J.; Poteete, A.; Cho, S.; et al. KEAP1 and STK11/LKB1 Alterations Enhance Vulnerability to ATR Inhibition in KRAS Mutant Non-Small Cell Lung Cancer. Cancer Cell 2025, 43, 1530–1548.e9. [Google Scholar] [CrossRef]
- Chen, T.; Ashwood, L.M.; Kondrashova, O.; Strasser, A.; Kelly, G.; Sutherland, K.D. Breathing New Insights into the Role of Mutant P53 in Lung Cancer. Oncogene 2024, 44, 115–129. [Google Scholar] [CrossRef]
- Park, H.; Shapiro, G.I.; Gao, X.; Mahipal, A.; Starr, J.; Furqan, M.; Singh, P.; Ahrorov, A.; Gandhi, L.; Ghosh, A.; et al. Phase Ib Study of Eprenetapopt (APR-246) in Combination with Pembrolizumab in Patients with Advanced or Metastatic Solid Tumors. ESMO Open 2022, 7, 100573. [Google Scholar] [CrossRef]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E.; Gay, L.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef]
- Feng, D.; Jiang, H.; Chen, G.; Guan, W.; Yi, L.; Zhu, Y.; Li, Y.; Huang, G.; He, B.; Tang, J.; et al. Case Report: Therapeutic Response of Front-Line Cadonilimab plus Chemotherapy on Patient with Advanced Lung Adenocarcinoma Harboring STK11 Genetic Aberration. Front. Immunol. 2024, 15, 1485358. [Google Scholar] [CrossRef]
- Na, B.; Shah, S.R.; Vasudevan, H.N. Past, Present, and Future Therapeutic Strategies for NF-1-Associated Tumors. Curr. Oncol. Rep. 2024, 26, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.P.; Coyne, G.O.S.; Wolters, P.L.; Martin, S.; Farschtschi, S.; Blanco, I.; Chen, Z.; Darrigo, L.G.; Eoli, M.; Whittle, J.R.; et al. Efficacy and Safety of Selumetinib in Adults with Neurofibromatosis Type 1 and Symptomatic, Inoperable Plexiform Neurofibromas (KOMET): A Multicentre, International, Randomised, Placebo-Controlled, Parallel, Double-Blind, Phase 3 Study. Lancet 2025, 405, 2217–2230. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Wang, Y.; Bouley, S.J.; Mandigo, T.R.; Sharma, A.; Sengupta, S.; Housden, A.; Perrimon, N.; Walker, J.A.; Housden, B.E. Inhibition of Autophagy as a Novel Treatment for Neurofibromatosis Type 1 Tumors. Mol. Oncol. 2025, 19, 825–851. [Google Scholar] [CrossRef] [PubMed]
- Sait, S.F.; Tang, K.H.; Angus, S.P.; Brown, R.; Sun, D.; Xie, X.; Iltis, C.; Lien, M.; Socci, N.D.; Bale, T.A.; et al. Hydroxychloroquine Prevents Resistance and Potentiates the Antitumor Effect of SHP2 Inhibition in NF1-Associated Malignant Peripheral Nerve Sheath Tumors. Proc. Natl. Acad. Sci. USA 2025, 122, e2407745121. [Google Scholar] [CrossRef]
- Dunne, V.L.; Ghita-Pettigrew, M.; Redmond, K.M.; Small, D.M.; Weldon, S.; Taggart, C.C.; Prise, K.M.; Hanna, G.G.; Butterworth, K.T. PTEN Depletion Increases Radiosensitivity in Response to Ataxia Telangiectasia-Related-3 (ATR) Inhibition in Non-Small Cell Lung Cancer (NSCLC). Int. J. Mol. Sci. 2024, 25, 7817. [Google Scholar] [CrossRef]
- Lv, S.; Yang, J.; Lin, J.; Huang, X.; Zhao, H.; Zhao, C.; Yang, L. CDK4/6 Inhibitors in Lung Cancer: Current Practice and Future Directions. Eur. Respir. Rev. 2024, 33. [Google Scholar] [CrossRef]
- Huang, M.F.; Wang, Y.X.; Chou, Y.T.; Lee, D.F. Therapeutic Strategies for RB1-Deficient Cancers: Intersecting Gene Regulation and Targeted Therapy. Cancers 2024, 16, 1558. [Google Scholar] [CrossRef]
- Wang, G.; Zhou, G.; Han, W.; Jiang, H. The Role of SMARCA4 in Lung Cancer. Sci. Rep. 2025, 15, 28605. [Google Scholar] [CrossRef]
- Lei, X.; Li, Z.; Huang, M.; Huang, L.; Huang, Y.; Lv, S.; Zhang, W.; Chen, Z.; Ke, Y.; Li, S.; et al. Gli1-Mediated Tumor Cell-Derived BFGF Promotes Tumor Angiogenesis and Pericyte Coverage in Non-Small Cell Lung Cancer. J. Exp. Clin. Cancer Res. 2024, 43, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Shi, X.; Zhang, Z.; Chen, Y.; Wu, J.I. Dual Role of Brg Chromatin Remodeling Factor in Sonic Hedgehog Signaling during Neural Development. Proc. Natl. Acad. Sci. USA 2011, 108, 12758–12763. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Y.; Ding, V.; Yan, X.; Lv, Y.; Zhong, M.; Zhu, F.; Zhao, P.; He, C.; Ding, F.; et al. GLI1 Activation Is a Key Mechanism of Erlotinib Resistance in Human Non-Small Cell Lung Cancer. Oncol. Lett. 2020, 20. [Google Scholar] [CrossRef]
- Wu, X.-T.; Wang, Y.-H.; Cai, X.-Y.; Dong, Y.; Cui, Q.; Zhou, Y.-N.; Yang, X.-W.; Lu, W.-F.; Zhang, M. RNF115 Promotes Lung Adenocarcinoma through Wnt/β-Catenin Pathway Activation by Mediating APC Ubiquitination. Cancer Metab. 2021, 9, 7. [Google Scholar] [CrossRef]
- Sarne, V.; Huter, S.; Braunmueller, S.; Rakob, L.; Jacobi, N.; Kitzwögerer, M.; Wiesner, C.; Obrist, P.; Seeboeck, R. Promoter Methylation of Selected Genes in Non-Small-Cell Lung Cancer Patients and Cell Lines. Int. J. Mol. Sci. 2020, 21, 4595. [Google Scholar] [CrossRef]
- Qian, H.; Ali, H.; Karri, V.; Low, J.T.; Ashley, D.M.; Heimberger, A.B.; Godley, L.A.; Sonabend, A.M.; Dmello, C.; Qian, H.; et al. Beyond DNA Damage Response: Immunomodulatory Attributes of CHEK2 in Solid Tumors. Oncotarget 2025, 16, 445–453. [Google Scholar] [CrossRef]
- Xu, P.; Gao, Y.; Jiang, S.; Cui, Y.; Xie, Y.; Kang, Z.; Chen, Y.X.; Sun, D.; Fang, J.Y. CHEK2 Deficiency Increase the Response to PD-1 Inhibitors by Affecting the Tumor Immune Microenvironment. Cancer Lett. 2024, 588, 216595. [Google Scholar] [CrossRef]
- Song, X.; Fan, P.-D.; Bantikassegn, A.; Guha, U.; Threadgill, D.W.; Varmus, H.; Politi, K. ERBB3-Independent Activation of the PI3K Pathway in EGFR-Mutant Lung Adenocarcinomas. Cancer Res. 2015, 75, 1035–1045. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, N.; Wang, Y.; Han, K.; Gao, T.; Li, X. FOXA1, Induced by RC48, Regulates HER2 Transcription to Enhance the Tumorigenic Capacity of Lung Cancer through PI3K/AKT Pathway. J. Cancer 2024, 15, 5863–5875. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; Wang, Q.; Yang, J.; Peng, W.; Li, X.; Shi, M.; Lu, K. Efficacy of Disitamab Vedotin in Non-Small Cell Lung Cancer with HER2 Alterations: A Multicenter, Retrospective Real-World Study. Front. Oncol. 2024, 14, 1441025. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.F.; De Marchi, F.; Lokhandwala, P.M.; Belchis, D.; Xian, R.; Gocke, C.D.; Eshleman, J.R.; Illei, P.; Li, M.T. IDH1 and IDH2 Mutations in Lung Adenocarcinomas: Evidences of Subclonal Evolution. Cancer Med. 2020, 9, 4386–4394. [Google Scholar] [CrossRef] [PubMed]
- Toth, L.N.; de Abreu, F.B.; Tafe, L.J. Non–Small Cell Lung Cancers with Isocitrate Dehydrogenase 1 or 2 (IDH1/2) Mutations. Hum. Pathol. 2018, 78, 138–143. [Google Scholar] [CrossRef]
- Carosi, F.; Broseghini, E.; Fabbri, L.; Corradi, G.; Gili, R.; Forte, V.; Roncarati, R.; Filippini, D.M.; Ferracin, M. Targeting Isocitrate Dehydrogenase (IDH) in Solid Tumors: Current Evidence and Future Perspectives. Cancers 2024, 16, 2752. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Lu, W.; Yang, J.; Xia, Y.; Huang, P. Targeting Mitochondrial IDH2 Enhances Antitumor Activity of Cisplatin in Lung Cancer via ROS-Mediated Mechanism. Biomedicines 2023, 11, 475. [Google Scholar] [CrossRef]
- Du, J.; Qian, J.; Zheng, B.; Xu, G.; Chen, H.; Chen, C. MiR-21-5p Is a Biomarker for Predicting Prognosis of Lung Adenocarcinoma by Regulating PIK3R1 Expression. Int. J. Gen. Med. 2021, 14, 8873–8880. [Google Scholar] [CrossRef]
- Sanaei, M.J.; Razi, S.; Pourbagheri-Sigaroodi, A.; Bashash, D. The PI3K/Akt/MTOR Pathway in Lung Cancer; Oncogenic Alterations, Therapeutic Opportunities, Challenges, and a Glance at the Application of Nanoparticles. Transl. Oncol. 2022, 18, 101364. [Google Scholar] [CrossRef]
- Chen, C.; Liu, H.; Li, Y.; Xu, Q.; Liu, J. Downregulation of PTPRT Elevates the Expression of Survivin and Promotes the Proliferation, Migration, and Invasion of Lung Adenocarcinoma. BMC Cancer 2024, 24, 63. [Google Scholar] [CrossRef]
- Chen, Z.; Ji, W.; Feng, W.; Cui, J.; Wang, Y.; Li, F.; Chen, J.; Guo, Z.; Xia, L.; Zhu, X.; et al. PTPRT Loss Enhances Anti–PD-1 Therapy Efficacy by Regulation of STING Pathway in Non–Small Cell Lung Cancer. Sci. Transl. Med. 2024, 16. [Google Scholar] [CrossRef]
- Pehkonen, H.; de Curtis, I.; Monni, O. Liprins in Oncogenic Signaling and Cancer Cell Adhesion. Oncogene 2021, 40, 6406–6416. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Sugawara, E.; Hatano, S.; Asaka, R.; Okumura, S.; Nakagawa, K.; Mano, H.; Ishikawa, Y. Pulmonary Inflammatory Myofibroblastic Tumor Expressing a Novel Fusion, PPFIBP1–ALK: Reappraisal of Anti-ALK Immunohistochemistry as a Tool for Novel ALK Fusion Identification. Clin. Cancer Res. 2011, 17, 3341–3348. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, X.; Chen, L.; Lai, Z.; Liu, J.; Li, S.; Hu, A.; Lin, Y. Protein Phosphatase 1 Regulatory Subunit 3G (PPP1R3G) Correlates with Poor Prognosis and Immune Infiltration in Lung Adenocarcinoma. Bioengineered 2021, 12, 8336–8346. [Google Scholar] [CrossRef]
- Esfahani, M.S.; Lee, L.J.; Jeon, Y.J.; Flynn, R.A.; Stehr, H.; Hui, A.B.; Ishisoko, N.; Kildebeck, E.; Newman, A.M.; Bratman, S.V.; et al. Functional Significance of U2AF1 S34F Mutations in Lung Adenocarcinomas. Nat. Commun. 2019, 10, 5712. [Google Scholar] [CrossRef]
- Soulette, C.M.; Hrabeta-Robinson, E.; Arevalo, C.; Felton, C.; Tang, A.D.; Marin, M.G.; Brooks, A.N. Full-Length Transcript Alterations in Human Bronchial Epithelial Cells with U2AF1 S34F Mutations. Life Sci. Alliance 2023, 6, e202000641. [Google Scholar] [CrossRef] [PubMed]
- Pulice, J.L.; Meyerson, M. Amplified Dosage of the NKX2-1 Lineage Transcription Factor Controls Its Oncogenic Role in Lung Adenocarcinoma. Mol. Cell 2025, 85, 1311–1329.e16. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, S.; Zhang, X.; Pan, Y.; Yan, Y.; Wang, N.; Ren, Y.; Zuo, J.; Zong, W.-X.; Wang, Z.; et al. RBM10 Loss Promotes EGFR-Driven Lung Cancer and Confers Sensitivity to Spliceosome Inhibition. Cancer Res. 2023, 83, 1490–1502. [Google Scholar] [CrossRef]
- Walter, D.M.; Gladstein, A.C.; Doerig, K.R.; Natesan, R.; Baskaran, S.G.; Gudiel, A.A.; Adler, K.M.; Acosta, J.O.; Wallace, D.C.; Asangani, I.A.; et al. Setd2 Inactivation Sensitizes Lung Adenocarcinoma to Inhibitors of Oxidative Respiration and MTORC1 Signaling. Commun. Biol. 2023, 6, 255. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, J.Q.; Li, X.P. Differences between Lung Adenocarcinoma and Lung Squamous Cell Carcinoma: Driver Genes, Therapeutic Targets, and Clinical Efficacy. Genes. Dis. 2025, 12, 101374. [Google Scholar] [CrossRef]
- Vokes, N.I.; Galan Cobo, A.; Fernandez-Chas, M.; Molkentine, D.; Treviño, S.; Druker, V.; Qian, Y.; Patel, S.; Schmidt, S.; Hong, L.; et al. ATM Mutations Associate with Distinct Co-Mutational Patterns and Therapeutic Vulnerabilities in NSCLC. Clin. Cancer Res. 2023, 29, 4958–4972. [Google Scholar] [CrossRef]
- Wang, J.; Chen, B.; Pu, X.; Li, J.; Xu, Y.; Xu, L.; Xu, F.; Li, K.; Kong, Y.; Liu, L.; et al. Potential Biomarkers of Primary Resistance to First- and Second-Generation EGFR-TKIs in Non-Small-Cell Lung Cancer: A Real-World Study. Ther. Adv. Med. Oncol. 2025, 17. [Google Scholar] [CrossRef] [PubMed]
- Moorthi, S.; Paguirigan, A.; Itagi, P.; Ko, M.; Pettinger, M.; Hoge, A.C.H.; Nag, A.; Patel, N.A.; Wu, F.; Sather, C.; et al. The Genomic Landscape of Lung Cancer in Never-Smokers from the Women’s Health Initiative. JCI Insight 2024, 9, e174643. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes-Young, A.; Davies, K.D.; Trevisan, P.; Nijmeh, H.; Haag, M.; Aisner, D.L.; Patil, T. Clinical and Radiographic Benefit of a Patient With Metastatic Non-Small Cell Lung Cancer Harboring an EGFR::ERBB4 Fusion Through Use of EGFR Tyrosine Kinase Inhibitors. JCO Precis. Oncol. 2024, 8. [Google Scholar] [CrossRef] [PubMed]
- Kalinsky, K.; Hong, F.; McCourt, C.K.; Sachdev, J.C.; Mitchell, E.P.; Zwiebel, J.A.; Doyle, L.A.; McShane, L.M.; Li, S.; Gray, R.J.; et al. Effect of Capivasertib in Patients With an AKT1 E17K-Mutated Tumor: NCI-MATCH Subprotocol EAY131-Y Nonrandomized Trial. JAMA Oncol. 2021, 7, 271–278. [Google Scholar] [CrossRef]
- Friedlaender, A.; Perol, M.; Banna, G.L.; Parikh, K.; Addeo, A. Oncogenic Alterations in Advanced NSCLC: A Molecular Super-Highway. Biomark. Res. 2024, 12, 24. [Google Scholar] [CrossRef]
- Stewart, D.J. Wnt Signaling Pathway in Non–Small Cell Lung Cancer. JNCI J. Natl. Cancer Inst. 2014, 106, djt356. [Google Scholar] [CrossRef]
- Huang, Q.; Li, F.; Hu, H.; Fang, Z.; Gao, Z.; Xia, G.; Ng, W.-L.; Khodadadi-Jamayran, A.; Chen, T.; Deng, J.; et al. Loss of TSC1/TSC2 Sensitizes Immune Checkpoint Blockade in Non–Small Cell Lung Cancer. Sci. Adv. 2022, 8, 9533. [Google Scholar] [CrossRef]
- Sun, D.; Qian, H.; Li, J.; Xing, P. Targeting MDM2 in Malignancies Is a Promising Strategy for Overcoming Resistance to Anticancer Immunotherapy. J. Biomed. Sci. 2024, 31, 17. [Google Scholar] [CrossRef]
- Conca, E.; Lorenzini, D.; Minna, E.; Agnelli, L.; Duca, M.; Gentili, M.; Bodini, B.; Polignano, M.; Mantiero, M.; Damian, S.; et al. Genomic Instability and CCNE1 Amplification as Emerging Biomarkers for Stratifying High-Grade Serous Ovarian Cancer. Front. Oncol. 2025, 15, 1633410. [Google Scholar] [CrossRef]
- Elliott, K.; Singh, V.K.; Bäckerholm, A.; Ögren, L.; Lindberg, M.; Soczek, K.M.; Hoberg, E.; Luijts, T.; Van den Eynden, J.; Falkenberg, M.; et al. Mechanistic Basis of Atypical TERT Promoter Mutations. Nat. Commun. 2024, 15, 1–11. [Google Scholar] [CrossRef]
- Chen, G.; Wang, S.; Zhang, Q.; Liu, J.; Zhu, W.; Song, X.; Song, X. Circulating TERT Serves as the Novel Diagnostic and Prognostic Biomarker for the Resectable NSCLC. Cancer Cell Int. 2024, 24, 420. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kasajima, R.; Yokose, T.; Shimizu, E.; Hatakeyama, S.; Yamaguchi, K.; Yokoyama, K.; Katayama, K.; Yamaguchi, R.; Furukawa, Y.; et al. KMT2C Expression and DNA Homologous Recombination Repair Factors in Lung Cancers with a High-Grade Fetal Adenocarcinoma Component. Transl. Lung Cancer Res. 2023, 12, 1738–1751. [Google Scholar] [CrossRef]
- Pan, Y.; Han, H.; Hu, H.; Wang, H.; Song, Y.; Hao, Y.; Tong, X.; Patel, A.S.; Misirlioglu, S.; Tang, S.; et al. KMT2D Deficiency Drives Lung Squamous Cell Carcinoma and Hypersensitivity to RTK-RAS Inhibition. Cancer Cell 2023, 41, 88–105.e8. [Google Scholar] [CrossRef] [PubMed]
- Alam, H.; Tang, M.; Maitituoheti, M.; Dhar, S.S.; Kumar, M.; Han, C.Y.; Ambati, C.R.; Amin, S.B.; Gu, B.; Chen, T.Y.; et al. KMT2D Deficiency Impairs Super-Enhancers to Confer a Glycolytic Vulnerability in Lung Cancer. Cancer Cell 2020, 37, 599–617.e7. [Google Scholar] [CrossRef]
- Liang, K.H.; Luo, Y.H.; Wang, M.L.; Chiou, S.H.; Chen, Y.M.; Hsu, H.S. A Multiomic Investigation of Lung Adenocarcinoma Molecular Subtypes. J. Chin. Med. Assoc. 2024, 87, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Li, J.; Chen, J.; Lu, J.; Hao, Z.; Shi, J.; Chang, Q.; Zeng, Z. Immunoprognostic Model of Lung Adenocarcinoma and Screening of Sensitive Drugs. Sci. Rep. 2022, 12, 7162. [Google Scholar] [CrossRef]
- Shen, Z.; Feng, C.; Chen, X.; Jiang, Y.; Chen, J. Prognostic Model of Lung Adenocarcinoma Based on Immunoprognosis-Related Genes and Related Drug Prediction. J. Thorac. Dis. 2024, 16, 5860–5877. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Xie, Z.; Kropiwnicki, E.; Wojciechowicz, M.L.; Jagodnik, K.M.; Shu, I.; Bailey, A.; Clarke, D.J.B.; Jeon, M.; Evangelista, J.E.; Kuleshov, M.V.; et al. Getting Started with LINCS Datasets and Tools. Curr. Protoc. 2022, 2, e487. [Google Scholar] [CrossRef]
- Way, G.P.; Natoli, T.; Adeboye, A.; Litichevskiy, L.; Yang, A.; Lu, X.; Caicedo, J.C.; Cimini, B.A.; Karhohs, K.; Logan, D.J.; et al. Morphology and Gene Expression Profiling Provide Complementary Information for Mapping Cell State. Cell Syst. 2022, 13, 911–923.e9. [Google Scholar] [CrossRef]
- Chen, B.; Ma, L.; Paik, H.; Sirota, M.; Wei, W.; Chua, M.-S.; So, S.; Butte, A.J. Reversal of Cancer Gene Expression Correlates with Drug Efficacy and Reveals Therapeutic Targets. Nat. Commun. 2017, 8, 16022. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.X.; Siriwanna, D.; Cho, W.C.; Wan, T.K.; Du, Y.R.; Bennett, A.N.; He, Q.E.; Liu, J.D.; Huang, X.T.; Chan, K.H.K. Lung Adenocarcinoma-Related Target Gene Prediction and Drug Repositioning. Front. Pharmacol. 2022, 13, 936758. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Sinha, A.; Yang, D.; Altorki, N.K.; Tandon, R.; Wang, W.; Chavez, D.; Lee, E.; Patel, A.S.; Sato, T.; et al. Integrative Network Analysis of Early-Stage Lung Adenocarcinoma Identifies Aurora Kinase Inhibition as Interceptor of Invasion and Progression. Nat. Commun. 2022, 13, 1592. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. A Comparison of Network-Based Methods for Drug Repurposing along with an Application to Human Complex Diseases. Int. J. Mol. Sci. 2022, 23, 3703. [Google Scholar] [CrossRef]
- Cheng, F.; Lu, W.; Liu, C.; Fang, J.; Hou, Y.; Handy, D.E.; Wang, R.; Zhao, Y.; Yang, Y.; Huang, J.; et al. A Genome-Wide Positioning Systems Network Algorithm for in Silico Drug Repurposing. Nat. Commun. 2019, 10, 3476. [Google Scholar] [CrossRef]
- Ma, M.; Huang, M.; He, Y.; Fang, J.; Li, J.; Li, X.; Liu, M.; Zhou, M.; Cui, G.; Fan, Q. Network Medicine: A Potential Approach for Virtual Drug Screening. Pharmaceuticals 2024, 17, 899. [Google Scholar] [CrossRef]
- Huang, K.; Chandak, P.; Wang, Q.; Havaldar, S.; Vaid, A.; Leskovec, J.; Nadkarni, G.N.; Glicksberg, B.S.; Gehlenborg, N.; Zitnik, M. A Foundation Model for Clinician-Centered Drug Repurposing. Nat. Med. 2024, 30, 3601–3613. [Google Scholar] [CrossRef]
- Zhou, C.; Cai, C.P.; Huang, X.T.; Wu, S.; Yu, J.L.; Wu, J.W.; Fang, J.S.; Li, G.B. TarKG: A Comprehensive Biomedical Knowledge Graph for Target Discovery. Bioinformatics 2024, 40, btae598. [Google Scholar] [CrossRef]
- Perdomo-Quinteiro, P.; Belmonte-Hernández, A. Knowledge Graphs for Drug Repurposing: A Review of Databases and Methods. Brief. Bioinform. 2024, 25, bbae461. [Google Scholar] [CrossRef]
- Su, X.; Hu, P.; Li, D.; Zhao, B.; Niu, Z.; Herget, T.; Yu, P.S.; Hu, L. Interpretable Identification of Cancer Genes across Biological Networks via Transformer-Powered Graph Representation Learning. Nat. Biomed. Eng. 2025, 9, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Xu, X.; Li, M.; Zhang, X.; Cao, F. Afzelin Induces Immunogenic Cell Death against Lung Cancer by Targeting NQO2. BMC Complement. Med. Ther. 2023, 23, 381. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ren, Y.; Qin, S.; Yang, Z. Exploring the Mechanism of 6-Methoxydihydrosanguinarine in the Treatment of Lung Adenocarcinoma Based on Network Pharmacology, Molecular Docking and Experimental Investigation. BMC Complement. Med. Ther. 2024, 24, 202. [Google Scholar] [CrossRef] [PubMed]
- Duran-Frigola, M.; Pauls, E.; Guitart-Pla, O.; Bertoni, M.; Alcalde, V.; Amat, D.; Juan-Blanco, T.; Aloy, P. Extending the Small-Molecule Similarity Principle to All Levels of Biology with the Chemical Checker. Nat. Biotechnol. 2020, 38, 1087–1096. [Google Scholar] [CrossRef]
- Boldini, D.; Ballabio, D.; Consonni, V.; Todeschini, R.; Grisoni, F.; Sieber, S.A. Effectiveness of Molecular Fingerprints for Exploring the Chemical Space of Natural Products. J. Cheminform. 2024, 16, 35. [Google Scholar] [CrossRef]
- Syahdi, R.R.; Jasial, S.; Maeda, I.; Miyao, T. Bridging Structure- and Ligand-Based Virtual Screening through Fragmented Interaction Fingerprint. ACS Omega 2024, 9, 38957–38969. [Google Scholar] [CrossRef]
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The SIDER Database of Drugs and Side Effects. Nucleic Acids Res. 2016, 44, D1075–D1079. [Google Scholar] [CrossRef]
- Fusaroli, M.; Salvo, F.; Khouri, C.; Raschi, E. The Reporting of Disproportionality Analysis in Pharmacovigilance: Spotlight on the READUS-PV Guideline. Front. Pharmacol. 2024, 15, 1488725. [Google Scholar] [CrossRef]
- Soyer, S.M.; Ozbek, P.; Kasavi, C. Lung Adenocarcinoma Systems Biomarker and Drug Candidates Identified by Machine Learning, Gene Expression Data, and Integrative Bioinformatics Pipeline. OMICS 2024, 28, 408–420. [Google Scholar] [CrossRef]
- Mukherjee, A.; Yadav, P.H.; Mukunthan, K.S. Unveiling Potential Targeted Therapeutic Opportunities for Co-Overexpressed Targeting Protein for Xklp2 and Aurora-A Kinase in Lung Adenocarcinoma. Mol. Biotechnol. 2024, 66, 2792–2803. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, N.M.; Sanad, E.F.; Kassab, S.E.; Essam, M.; Guirguis, M.A.; Basalious, E.B.; Sultan, A.S. Treatment of Non-Small Cell Lung Cancer Using Chem-Bioinformatics-Driven Engineering of Exosomal Cargo-Vehicle for Telmisartan and Pioglitazone Targeted-Delivery. Sci. Rep. 2025, 15, 25166. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, J.; Luttens, A. Structure-Based Virtual Screening of Vast Chemical Space as a Starting Point for Drug Discovery. Curr. Opin. Struct. Biol. 2024, 87, 102829. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H. The Science and Art of Structure-Based Virtual Screening. ACS Med. Chem. Lett. 2024, 15, 436–440. [Google Scholar] [CrossRef]
- Zhou, G.; Rusnac, D.V.; Park, H.; Canzani, D.; Nguyen, H.M.; Stewart, L.; Bush, M.F.; Nguyen, P.T.; Wulff, H.; Yarov-Yarovoy, V.; et al. An Artificial Intelligence Accelerated Virtual Screening Platform for Drug Discovery. Nat. Commun. 2024, 15, 7761. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 Update: A Web Server for Potential Drug Target Identification with a Comprehensive Target Pharmacophore Database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]
- Krause, F.; Voigt, K.; Di Ventura, B.; Öztürk, M.A. ReverseDock: A Web Server for Blind Docking of a Single Ligand to Multiple Protein Targets Using AutoDock Vina. Front. Mol. Biosci. 2023, 10, 1243970. [Google Scholar] [CrossRef]
- Li, B.; Dai, C.; Wang, L.; Deng, H.; Li, Y.; Guan, Z.; Ni, H. A Novel Drug Repurposing Approach for Non-Small Cell Lung Cancer Using Deep Learning. PLoS ONE 2020, 15, e0233112. [Google Scholar] [CrossRef]
- Wei, Z.; Zhu, S.; Chen, X.; Zhu, C.; Duan, B.; Liu, Q. DrSim: Similarity Learning for Transcriptional Phenotypic Drug Discovery. Genom. Proteom. Bioinform. 2022, 20, 1028–1036. [Google Scholar] [CrossRef]
- Cai, L.; Chu, J.; Xu, J.; Meng, Y.; Lu, C.; Tang, X.; Wang, G.; Tian, G.; Yang, J. Machine Learning for Drug Repositioning: Recent Advances and Challenges. Curr. Res. Chem. Biol. 2023, 3, 100042. [Google Scholar] [CrossRef]
- Zeng, X.; Li, S.-J.; Lv, S.-Q.; Wen, M.-L.; Li, Y. A Comprehensive Review of the Recent Advances on Predicting Drug-Target Affinity Based on Deep Learning. Front. Pharmacol. 2024, 15, 1375522. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, J.; Marostica, E.; Yuan, W.; Jin, J.; Zhang, J.; Li, R.; Tang, H.; Wang, K.; Li, Y.; et al. A Pathology Foundation Model for Cancer Diagnosis and Prognosis Prediction. Nature 2024, 634, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Liu, R.; Da, Y. Comparison of Tumor Related Signaling Pathways with Known Compounds to Determine Potential Agents for Lung Adenocarcinoma. Thorac. Cancer 2018, 9, 974–988. [Google Scholar] [CrossRef]
- MotieGhader, H.; Tabrizi-Nezhadi, P.; Deldar Abad Paskeh, M.; Baradaran, B.; Mokhtarzadeh, A.; Hashemi, M.; Lanjanian, H.; Jazayeri, S.M.; Maleki, M.; Khodadadi, E.; et al. Drug Repositioning in Non-Small Cell Lung Cancer (NSCLC) Using Gene Co-Expression and Drug–Gene Interaction Networks Analysis. Sci. Rep. 2022, 12, 9417. [Google Scholar] [CrossRef]
- Garana, B.B.; Joly, J.H.; Delfarah, A.; Hong, H.; Graham, N.A. Drug Mechanism Enrichment Analysis Improves Prioritization of Therapeutics for Repurposing. BMC Bioinform. 2023, 24, 215. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, H.; Chen, M.; Wang, S.; Qian, R.; Zhang, L.; Huang, X.; Wang, J.; Liu, Z.; Qin, W.; et al. A Survey of Optimal Strategy for Signature-Based Drug Repositioning and an Application to Liver Cancer. eLife 2022, 11, e71880. [Google Scholar] [CrossRef]
| Method | Description | |
|---|---|---|
| Cell-based phenotypic screening (Cell Painting) [1] | Goal | Unbiased cellular assays; morphology/profiles cluster MoA |
| Pros | Detects multi-target effects; no prior target needed | |
| Cons | Assay artifacts; translation gap; lab infrastructure | |
| Phenotypic drug discovery across screening models [2] | Goal | Screen in complex models for efficacy/toxicity |
| Pros | Higher physiological relevance; emergent effects | |
| Cons | Costly; low throughput; ethical constraints | |
| EHR/claims mining and target-trial emulation [3] | Goal | Estimate the effects of existing drugs on new outcomes |
| Pros | Real-world, human-level effects; diverse outcomes | |
| Cons | Confounding and bias; data access/cleaning burdens | |
| Pharmacovigilance (FAERS) inverse-signal analyses [4] | Goal | Identify protective drug–outcome associations in safety data |
| Pros | Cheap; wide coverage; early human signals | |
| Cons | Reporting/indication bias; noisy; weak causality | |
| Human genetics for indication selection [5] | Goal | Align drug mechanisms with GWAS/OMIM evidence |
| Pros | Higher clinical success; directionality clues | |
| Cons | Limited to genetically mediated disease; small effects | |
| Drug-target Mendelian randomization (cis-MR) [6] | Goal | Use genetic instruments on targets to infer efficacy/safety |
| Pros | Causal on-target prediction; dose–response hints | |
| Cons | Instrument validity/pleiotropy limits; target coverage | |
| Method | Description | |
|---|---|---|
| Connectivity Map/LINCS [7] | Goal | Match disease expression signatures to drugs that invert them |
| Pros | Human-relevant; target-agnostic; scalable; novel hits | |
| Con | Cell-line mismatch; off-target confounding; signature quality dependent | |
| Network medicine/interactome proximity [8] | Goal | Rank drugs whose targets lie near disease modules in the interactome |
| Pros | Mechanistic context; polypharmacology; interpretable | |
| Cons | Incomplete or biased networks; target mapping gaps | |
| Knowledge-graph (KG) methods [9] | Goal | Predict drug–disease links using heterogeneous biomedical graphs |
| Pros | Integrates diverse evidence; handles indirect paths | |
| Cons | Data noise; edge bias; KG engineering required | |
| Ligand-based similarity/chemogenomics [10] | Goal | Infer new targets/indications from chemical/bioactivity similarity |
| Pros | Fast; simple; reveals off-targets | |
| Cons | Limited novelty; activity cliffs; needs high-quality assays | |
| Side-effect (phenotypic) similarity mining [11] | Goal | Use shared adverse-event profiles to infer common targets/uses |
| Pros | Human phenotype signal; orthogonal to chemistry | |
| Cons | Confounding/indication bias; under-reporting; rare events | |
| Structure-based virtual screening & inverse docking [12] | Goal | Dock approved drugs against target panels to find binders |
| Pros | Atomic mechanism; target-specific; repurpose to novel targets | |
| Cons | Scoring errors, protein flexibility; structure availability | |
| Machine learning/deep learning [13,14] | Goal | Learn patterns across chemical, target, disease features |
| Pros | Captures nonlinear patterns; scalable | |
| Cons | Black-box; data leakage risk; needs large labeled sets | |
| Pathway- and enrichment-based [15] | Goal | Prioritize drugs that modulate disease-enriched pathways |
| Pros | Interpretable; mechanism-level view | |
| Cons | Pathway incompleteness; over-representation bias | |
| Role | Genes | Pathways | |
|---|---|---|---|
| Co-receptor | ERBB3 | PI3K/AKT/mTOR, RTK/RAS/MAPK | |
| Lineage TF | FOXA1, NKX2-1/TTF-1 | Chromatin/Epigenetic, PI3K/AKT/mTOR | |
| Oncogene | Oncogene | AKT1, BRAF, CTNNB1, EGFR, ERBB2/HER2, FGFR2/FGFR3, HRAS, KRAS, MET, NRAS, PIK3CA, RIT1, YAP1 | Adhesion/EMT, Hippo/YAP, Immune modulation, PI3K/AKT/mTOR, RTK/RAS/MAPK, WNT/β-catenin |
| activation | NFE2L2/NRF2 | Immune modulation, Redox/NRF2 | |
| amplification | CCNE1, MDM2 | Cell cycle/RB, DDR | |
| contextual | PPFIBP1, RNF115 | Adhesion/EMT | |
| fusion | ALK, NTRK1/2/3, RET, ROS1 | PI3K/AKT/mTOR, RTK/RAS/MAPK | |
| neomorphic | IDH1 | Metabolism/Cell stress | |
| promoter/amp | TERT | Telomere | |
| rare fusion/mutation | ERBB4 | RTK/RAS/MAPK | |
| resistance | MAP2K1/MEK1 | RTK/RAS/MAPK | |
| wt overexpress/rare mutation | IDH2 | Metabolism/Cell stress, Redox/NRF2 | |
| ligand (fusion) | NRG1 | PI3K/AKT/mTOR, RTK/RAS/MAPK | |
| RNA component | TERC | Telomere | |
| RTK (contextual) | DDR2 | Adhesion/EMT | |
| Tumor suppressor | Tumor suppressor | APC, ARID1A, ATM, CDH1, CDKN2A, CHEK2, KEAP1, KMT2C, KMT2D, MGA, PIK3R1, PTEN, PTPRD, PTPRT, RB1, RBM10, SETD2, SMARCA4/BRG1, STK11/LKB1, TP53 | Adhesion/EMT, Cell cycle/RB, Chromatin/Epigenetic, DDR, Immune modulation, Metabolism/Cell stress, PI3K/AKT/mTOR, Redox/NRF2, Splicing, WNT/β-catenin |
| modifier | U2AF1 | Splicing | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nematzadeh, S.; Karaul, A. Advances in Computational Drug Repurposing, Driver Genes, and Therapeutics in Lung Adenocarcinoma. Biomolecules 2025, 15, 1373. https://doi.org/10.3390/biom15101373
Nematzadeh S, Karaul A. Advances in Computational Drug Repurposing, Driver Genes, and Therapeutics in Lung Adenocarcinoma. Biomolecules. 2025; 15(10):1373. https://doi.org/10.3390/biom15101373
Chicago/Turabian StyleNematzadeh, Sajjad, and Arzu Karaul. 2025. "Advances in Computational Drug Repurposing, Driver Genes, and Therapeutics in Lung Adenocarcinoma" Biomolecules 15, no. 10: 1373. https://doi.org/10.3390/biom15101373
APA StyleNematzadeh, S., & Karaul, A. (2025). Advances in Computational Drug Repurposing, Driver Genes, and Therapeutics in Lung Adenocarcinoma. Biomolecules, 15(10), 1373. https://doi.org/10.3390/biom15101373






