Urinary Proteome and Exosome Analysis Protocol for the Discovery of Respiratory Diseases Biomarkers
Abstract
1. Introduction
2. Material and Methods
2.1. Study Population
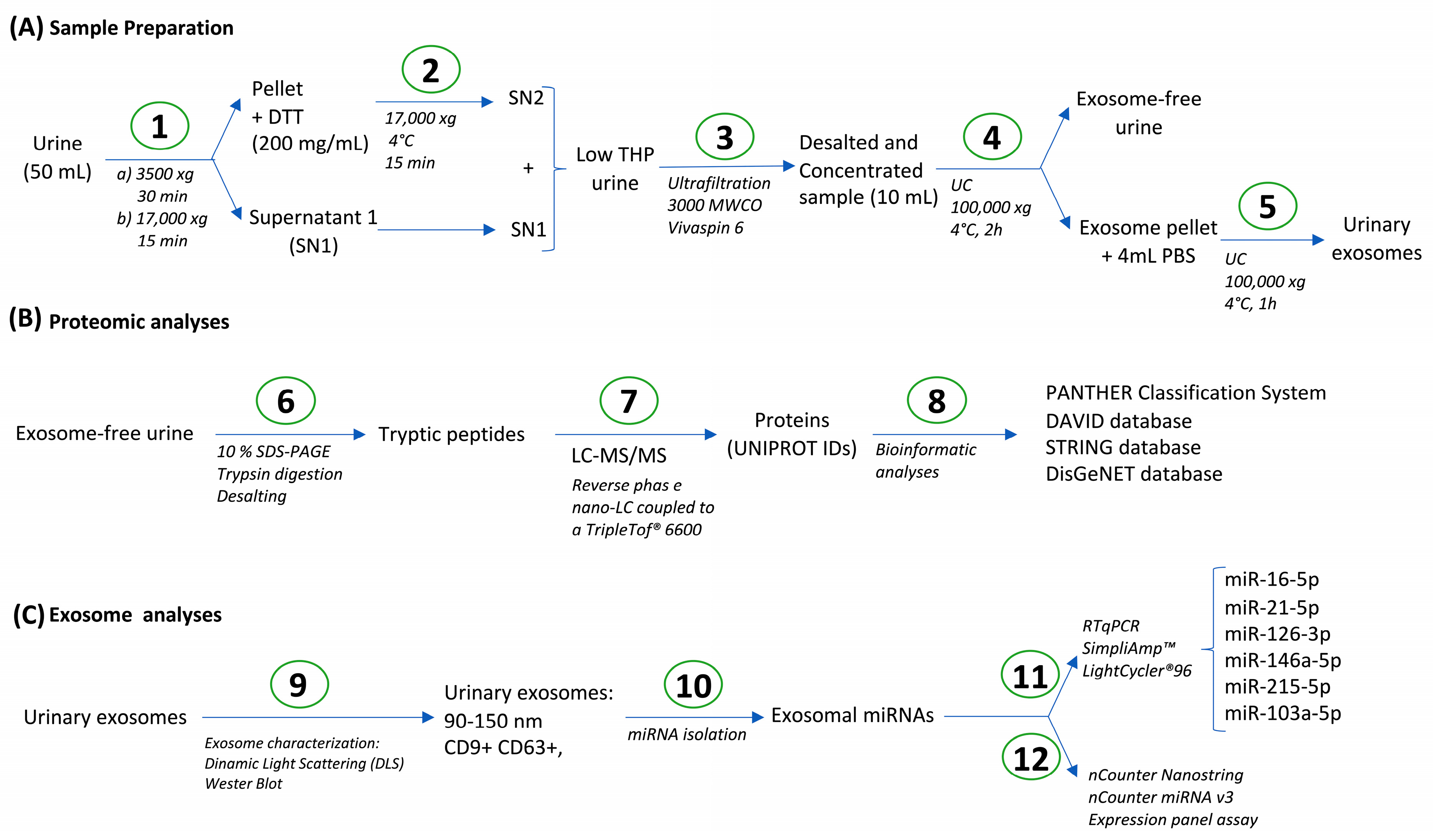
2.2. Urine Processing
2.3. Urinary Proteome Analysis
2.4. Bioinformatic Analysis
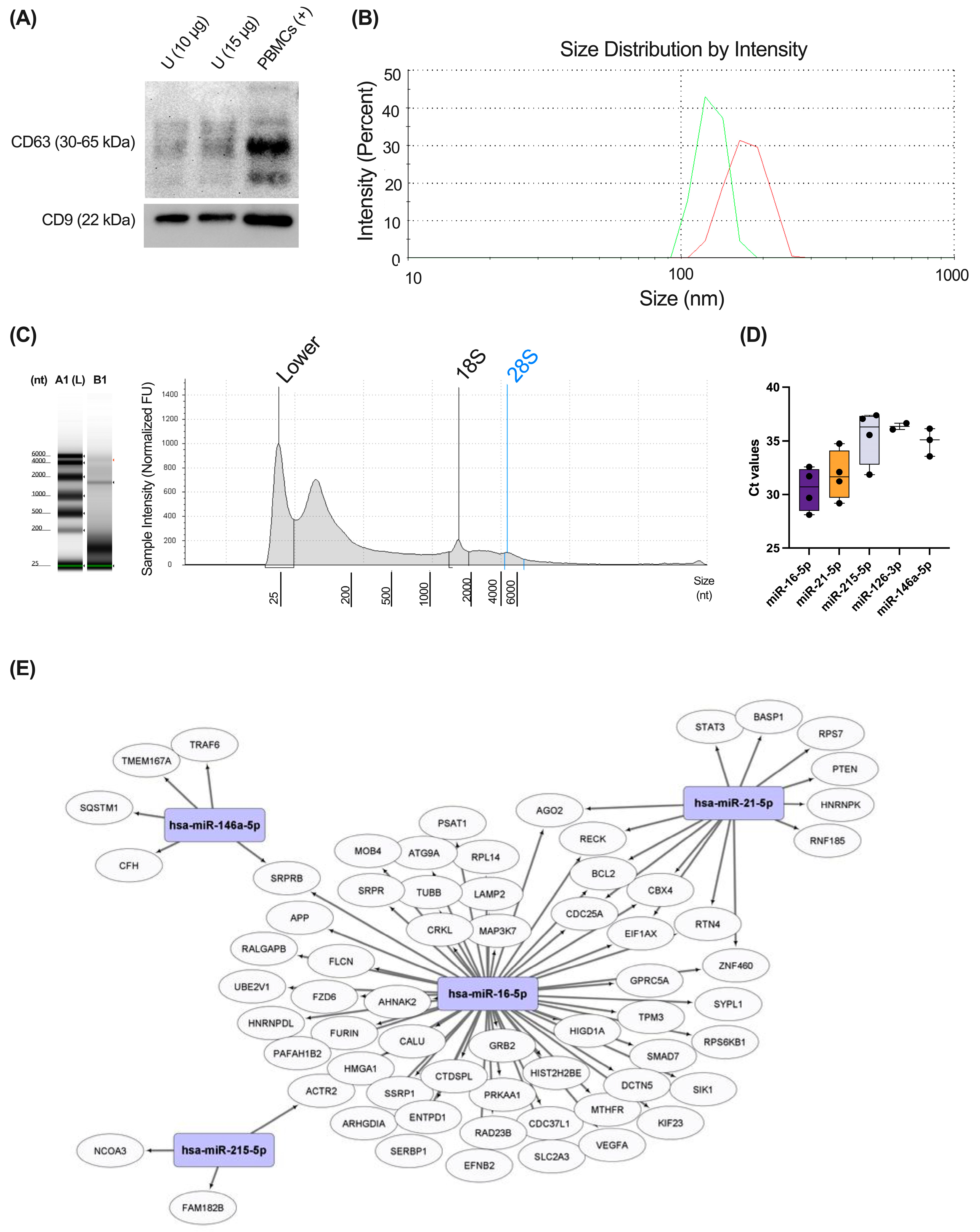
2.5. Exosome Characterization
2.6. microRNA (miRNA) Isolation from Urine Exosomes—RT-qPCR
2.7. Multiplex miRNA Expression Analysis
3. Results
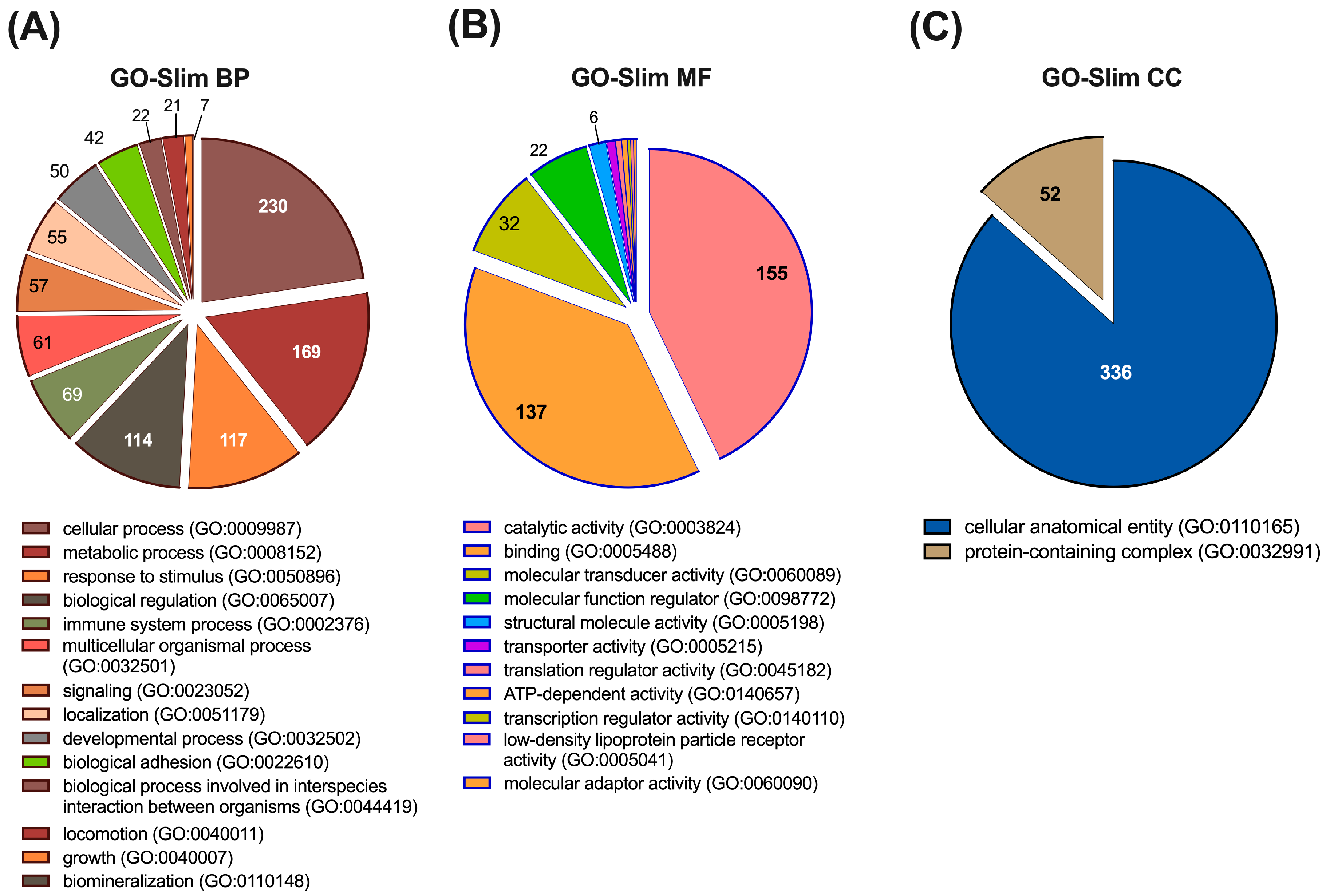
3.1. Tissue Distribution of the Proteins Identified in Exosome-Free Urine Samples
3.2. Functional Characterization of the Proteins Identified in Urine
3.3. Protein–Protein Interaction (PPI) Network Analysis of Proteins Identified in Urine
3.4. Different miRNAs Previously Implicated in Respiratory Pathology Were Detected in Healthy Urinary Exosomes Using RTqPCR
3.5. Several miRNAs from Urinary Exosomes Were Associated with Specific Molecular Phenotypes of Asthma Using a Multiplex miRNA Expression Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chebotareva, N.; Vinogradov, A.; McDonnell, V.; Zakharova, N.V.; Indeykina, M.I.; Moiseev, S.; Nikolaev, E.N.; Kononikhin, A.S. Urinary Protein and Peptide Markers in Chronic Kidney Disease. Int. J. Mol. Sci. 2021, 22, 12123. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Kumar, C.; Zhang, Y.; Olsen, J.V.; Mann, M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006, 7, R80. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Hirao, Y.; Kasuga, K.; Tokutake, T.; Semizu, Y.; Kitamura, K.; Ikeuchi, T.; Nakamura, K.; Yamamoto, T. Molecular Network Analysis of the Urinary Proteome of Alzheimer’s Disease Patients. Dement. Geriatr. Cogn. Dis. Extra 2019, 9, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Virreira Winter, S.; Karayel, O.; Strauss, M.T.; Padmanabhan, S.; Surface, M.; Merchant, K.; Alcalay, R.N.; Mann, M. Urinary proteome profiling for stratifying patients with familial Parkinson’s disease. EMBO Mol. Med. 2021, 13, e13257. [Google Scholar] [CrossRef]
- Sun, H.; Wang, D.; Liu, D.; Guo, Z.; Shao, C.; Sun, W.; Zeng, Y. Differential urinary proteins to diagnose coronary heart disease based on iTRAQ quantitative proteomics. Anal. Bioanal. Chem. 2019, 411, 2273–2282. [Google Scholar] [CrossRef]
- Benabdelkamel, H.; Masood, A.; Okla, M.; Al-Naami, M.Y.; Alfadda, A.A. A Proteomics-Based Approach Reveals Differential Regulation of Urine Proteins between Metabolically Healthy and Unhealthy Obese Patients. Int. J. Mol. Sci. 2019, 20, 4905. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Liu, W.; Xing, S.; Wang, D.; Chen, J.; Sun, L.; Mu, J.; Liu, W.; Xing, B.; et al. Identification of noninvasive diagnostic biomarkers for hepatocellular carcinoma by urinary proteomics. J. Proteomics 2020, 225, 103780. [Google Scholar] [CrossRef]
- Kacirova, M.; Bober, P.; Alexovic, M.; Tomkova, Z.; Tkacikova, S.; Talian, I.; Mederova, L.; Beresova, D.; Toth, R.; Andrasina, I.; et al. Differential Urinary Proteomic Analysis of Endometrial Cancer. Physiol. Res. 2019, 68, S483–S490. [Google Scholar] [CrossRef]
- Kolmert, J.; Gomez, C.; Balgoma, D.; Sjodin, M.; Bood, J.; Konradsen, J.R.; Ericsson, M.; Thorngren, J.O.; James, A.; Mikus, M.; et al. Urinary Leukotriene E(4) and Prostaglandin D(2) Metabolites Increase in Adult and Childhood Severe Asthma Characterized by Type 2 Inflammation. A Clinical Observational Study. Am. J. Respir. Crit. Care Med. 2021, 203, 37–53. [Google Scholar] [CrossRef]
- Tao, J.L.; Chen, Y.Z.; Dai, Q.G.; Tian, M.; Wang, S.C.; Shan, J.J.; Ji, J.J.; Lin, L.L.; Li, W.W.; Yuan, B. Urine metabolic profiles in paediatric asthma. Respirology 2019, 24, 572–581. [Google Scholar] [CrossRef]
- Kelly, R.S.; Dahlin, A.; McGeachie, M.J.; Qiu, W.; Sordillo, J.; Wan, E.S.; Wu, A.C.; Lasky-Su, J. Asthma Metabolomics and the Potential for Integrative Omics in Research and the Clinic. Chest 2017, 151, 262–277. [Google Scholar] [CrossRef] [PubMed]
- Peebles, R.S., Jr. Urine: A Lens for Asthma Pathogenesis and Treatment? Am. J. Respir. Crit. Care Med. 2021, 203, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Diseases, G.B.D.; Injuries, C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Barnes, P.J. Inflammatory endotypes in COPD. Allergy 2019, 74, 1249–1256. [Google Scholar] [CrossRef]
- Breiteneder, H.; Peng, Y.Q.; Agache, I.; Diamant, Z.; Eiwegger, T.; Fokkens, W.J.; Traidl-Hoffmann, C.; Nadeau, K.; O’Hehir, R.E.; O’Mahony, L.; et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy 2020, 75, 3039–3068. [Google Scholar] [CrossRef]
- Kaur, R.; Chupp, G. Phenotypes and endotypes of adult asthma: Moving toward precision medicine. J. Allergy Clin. Immunol. 2019, 144, 1–12. [Google Scholar] [CrossRef]
- Kuruvilla, M.E.; Lee, F.E.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef]
- Agusti, A.; Vogelmeier, C.F.; Halpin, D.M.G. Tackling the global burden of lung disease through prevention and early diagnosis. Lancet Respir. Med. 2022, 10, 1013–1015. [Google Scholar] [CrossRef]
- Elliot, S.; Catanuto, P.; Pereira-Simon, S.; Xia, X.; Shahzeidi, S.; Roberts, E.; Ludlow, J.; Hamdan, S.; Daunert, S.; Parra, J.; et al. Urine-derived exosomes from individuals with IPF carry pro-fibrotic cargo. eLife 2022, 11, e79543. [Google Scholar] [CrossRef]
- Gil-Martinez, M.; Lorente-Sorolla, C.; Rodrigo-Munoz, J.M.; Lendinez, M.A.; Nunez-Moreno, G.; de la Fuente, L.; Minguez, P.; Mahillo-Fernandez, I.; Sastre, J.; Valverde-Monge, M.; et al. Analysis of Differentially Expressed MicroRNAs in Serum and Lung Tissues from Individuals with Severe Asthma Treated with Oral Glucocorticoids. Int. J. Mol. Sci. 2023, 24, 1611. [Google Scholar] [CrossRef]
- Vazquez-Mera, S.; Martelo-Vidal, L.; Miguens-Suarez, P.; Saavedra-Nieves, P.; Arias, P.; Gonzalez-Fernandez, C.; Mosteiro-Anon, M.; Corbacho-Abelaira, M.D.; Blanco-Aparicio, M.; Mendez-Brea, P.; et al. Serum exosome inflamma-miRs are surrogate biomarkers for asthma phenotype and severity. Allergy 2023, 78, 141–155. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, H.E.; Bowman, R.V.; Fong, K.M.; Yang, I.A. Plasma Extracellular Vesicle miRNA Profiles Distinguish Chronic Obstructive Pulmonary Disease Exacerbations and Disease Severity. Int. J. Chron. Obs. Pulmon. Dis. 2022, 17, 2821–2833. [Google Scholar] [CrossRef] [PubMed]
- Gaytan-Pacheco, N.; Ibanez-Salazar, A.; Herrera-Van Oostdam, A.S.; Oropeza-Valdez, J.J.; Magana-Aquino, M.; Adrian Lopez, J.; Monarrez-Espino, J.; Lopez-Hernandez, Y. miR-146a, miR-221, and miR-155 are Involved in Inflammatory Immune Response in Severe COVID-19 Patients. Diagnostics 2022, 13, 133. [Google Scholar] [CrossRef] [PubMed]
- Sangaphunchai, P.; Todd, I.; Fairclough, L.C. Extracellular vesicles and asthma: A review of the literature. Clin. Exp. Allergy 2020, 50, 291–307. [Google Scholar] [CrossRef]
- Gheinani, A.H.; Vogeli, M.; Baumgartner, U.; Vassella, E.; Draeger, A.; Burkhard, F.C.; Monastyrskaya, K. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci. Rep. 2018, 8, 3945. [Google Scholar] [CrossRef]
- Wessel, D.; Flugge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Perez-Hernandez, D.; Gutierrez-Vazquez, C.; Jorge, I.; Lopez-Martin, S.; Ursa, A.; Sanchez-Madrid, F.; Vazquez, J.; Yanez-Mo, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef]
- Bonzon-Kulichenko, E.; Perez-Hernandez, D.; Nunez, E.; Martinez-Acedo, P.; Navarro, P.; Trevisan-Herraz, M.; Ramos Mdel, C.; Sierra, S.; Martinez-Martinez, S.; Ruiz-Meana, M.; et al. A robust method for quantitative high-throughput analysis of proteomes by 18O labeling. Mol. Cell. Proteomics 2011, 10, M110 003335. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Hamilton, A.M.; Furberg, H.; Pietzak, E.; Purdue, M.P.; Troester, M.A.; Hoadley, K.A.; Love, M.I. An approach for normalization and quality control for NanoString RNA expression data. Brief Bioinform. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, C.E.; Goss, V.M.; Balgoma, D.; Nicholas, B.; Brandsma, J.; Skipp, P.J.; Snowden, S.; Burg, D.; D’Amico, A.; Horvath, I.; et al. Application of ’omics technologies to biomarker discovery in inflammatory lung diseases. Eur. Respir. J. 2013, 42, 802–825. [Google Scholar] [CrossRef]
- Chawes, B.L.; Giordano, G.; Pirillo, P.; Rago, D.; Rasmussen, M.A.; Stokholm, J.; Bonnelykke, K.; Bisgaard, H.; Baraldi, E. Neonatal Urine Metabolic Profiling and Development of Childhood Asthma. Metabolites 2019, 9, 185. [Google Scholar] [CrossRef]
- Park, Y.H.; Fitzpatrick, A.M.; Medriano, C.A.; Jones, D.P. High-resolution metabolomics to identify urine biomarkers in corticosteroid-resistant asthmatic children. J. Allergy Clin. Immunol. 2017, 139, 1518–1524.e4. [Google Scholar] [CrossRef][Green Version]
- Mattarucchi, E.; Baraldi, E.; Guillou, C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites. Biomed. Chromatogr. 2012, 26, 89–94. [Google Scholar] [CrossRef]
- Swensen, A.C.; He, J.; Fang, A.C.; Ye, Y.; Nicora, C.D.; Shi, T.; Liu, A.Y.; Sigdel, T.K.; Sarwal, M.M.; Qian, W.J. A Comprehensive Urine Proteome Database Generated From Patients With Various Renal Conditions and Prostate Cancer. Front. Med. 2021, 8, 548212. [Google Scholar] [CrossRef]
- Dubin, R.F.; Rhee, E.P. Proteomics and Metabolomics in Kidney Disease, including Insights into Etiology, Treatment, and Prevention. Clin. J. Am. Soc. Nephrol. 2020, 15, 404–411. [Google Scholar] [CrossRef]
- Bakun, M.; Senatorski, G.; Rubel, T.; Lukasik, A.; Zielenkiewicz, P.; Dadlez, M.; Paczek, L. Urine proteomes of healthy aging humans reveal extracellular matrix (ECM) alterations and immune system dysfunction. Age 2014, 36, 299–311. [Google Scholar] [CrossRef]
- Brown, C.E.; McCarthy, N.S.; Hughes, A.D.; Sever, P.; Stalmach, A.; Mullen, W.; Dominiczak, A.F.; Sattar, N.; Mischak, H.; Thom, S.; et al. Urinary proteomic biomarkers to predict cardiovascular events. Proteomics Clin. Appl. 2015, 9, 610–617. [Google Scholar] [CrossRef]
- Bujold, E.; Fillion, A.; Roux-Dalvai, F.; Scott-Boyer, M.P.; Giguere, Y.; Forest, J.C.; Gotti, C.; Laforest, G.; Guerby, P.; Droit, A. Proteomic Analysis of Maternal Urine for the Early Detection of Preeclampsia and Fetal Growth Restriction. J. Clin. Med. 2021, 10, 4679. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, A.; Macedo-da-Silva, J.; Santiago, V.F.; de Oliveira, G.S.; Guimaraes, T.; Mendonca, C.F.; de Oliveira Branquinho, J.L.; Lucena, C.V.; Osorio, J.; Pernambuco, E.; et al. Urine proteomics as a non-invasive approach to monitor exertional rhabdomyolysis during military training. J. Proteomics 2022, 258, 104498. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, V.; Panarese, A.; di Matteo, F.M.; Favoriti, P.; Favella, L.; Arcieri, S.; Filippini, A. Chievitz’ juxtaparotid organ, free from cancer. Ann. Ital. Chir. 2015, 86, 503–507. [Google Scholar] [PubMed]
- Hao, X.; Guo, Z.; Sun, H.; Liu, X.; Zhang, Y.; Zhang, L.; Sun, W.; Tian, Y. Urinary protein biomarkers for pediatric medulloblastoma. J. Proteomics 2020, 225, 103832. [Google Scholar] [CrossRef]
- Htun, N.M.; Magliano, D.J.; Zhang, Z.Y.; Lyons, J.; Petit, T.; Nkuipou-Kenfack, E.; Ramirez-Torres, A.; von Zur Muhlen, C.; Maahs, D.; Schanstra, J.P.; et al. Prediction of acute coronary syndromes by urinary proteome analysis. PLoS One 2017, 12, e0172036. [Google Scholar] [CrossRef]
- Kang, M.J.; Park, Y.J.; You, S.; Yoo, S.A.; Choi, S.; Kim, D.H.; Cho, C.S.; Yi, E.C.; Hwang, D.; Kim, W.U. Urinary proteome profile predictive of disease activity in rheumatoid arthritis. J. Proteome Res. 2014, 13, 5206–5217. [Google Scholar] [CrossRef]
- Kentsis, A.; Shulman, A.; Ahmed, S.; Brennan, E.; Monuteaux, M.C.; Lee, Y.H.; Lipsett, S.; Paulo, J.A.; Dedeoglu, F.; Fuhlbrigge, R.; et al. Urine proteomics for discovery of improved diagnostic markers of Kawasaki disease. EMBO Mol. Med. 2013, 5, 210–220. [Google Scholar] [CrossRef]
- Meng, W.; Huan, Y.; Gao, Y. Urinary proteome profiling for children with autism using data-independent acquisition proteomics. Transl. Pediatr. 2021, 10, 1765–1778. [Google Scholar] [CrossRef]
- Nielsen, H.H.; Beck, H.C.; Kristensen, L.P.; Burton, M.; Csepany, T.; Simo, M.; Dioszeghy, P.; Sejbaek, T.; Grebing, M.; Heegaard, N.H.; et al. The Urine Proteome Profile Is Different in Neuromyelitis Optica Compared to Multiple Sclerosis: A Clinical Proteome Study. PLoS ONE 2015, 10, e0139659. [Google Scholar] [CrossRef]
- Qian, Y.T.; Liu, X.Y.; Sun, H.D.; Xu, J.Y.; Sun, J.M.; Liu, W.; Chen, T.; Liu, J.W.; Tan, Y.; Sun, W.; et al. Urinary Proteomics Analysis of Active Vitiligo Patients: Biomarkers for Steroid Treatment Efficacy Prediction and Monitoring. Front. Mol. Biosci. 2022, 9, 761562. [Google Scholar] [CrossRef]
- Suganya, V.; Geetha, A.; Sujatha, S. Urine proteome analysis to evaluate protein biomarkers in children with autism. Clin. Chim. Acta 2015, 450, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, M.; Guo, Z.; Song, S.; Liu, S.; Yuan, T.; Fu, Y.; Dong, Y.; Sun, H.; Liu, X.; et al. Urinary proteomic analysis during pregnancy and its potential application in early prediction of gestational diabetes mellitus and spontaneous abortion. Ann. Transl. Med. 2022, 10, 736. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Yu, L.; Zhu, L.; Wu, Z.; Weng, X.; Qiu, G. Urine Proteomics Profiling and Functional Characterization of Knee Osteoarthritis Using iTRAQ Technology. Horm. Metab. Res. 2019, 51, 735–740. [Google Scholar] [CrossRef]
- Yu, Y.; Singh, H.; Kwon, K.; Tsitrin, T.; Petrini, J.; Nelson, K.E.; Pieper, R. Protein signatures from blood plasma and urine suggest changes in vascular function and IL-12 signaling in elderly with a history of chronic diseases compared with an age-matched healthy cohort. Geroscience 2021, 43, 593–606. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Thijs, L.; Petit, T.; Gu, Y.M.; Jacobs, L.; Yang, W.Y.; Liu, Y.P.; Koeck, T.; Zurbig, P.; Jin, Y.; et al. Urinary Proteome and Systolic Blood Pressure as Predictors of 5-Year Cardiovascular and Cardiac Outcomes in a General Population. Hypertension 2015, 66, 52–60. [Google Scholar] [CrossRef]
- Zou, L.; Wang, X.; Guo, Z.; Sun, H.; Shao, C.; Yang, Y.; Sun, W. Differential urinary proteomics analysis of myocardial infarction using iTRAQ quantification. Mol. Med. Rep. 2019, 19, 3972–3988. [Google Scholar] [CrossRef]
- Zhao, M.; Li, M.; Yang, Y.; Guo, Z.; Sun, Y.; Shao, C.; Li, M.; Sun, W.; Gao, Y. A comprehensive analysis and annotation of human normal urinary proteome. Sci. Rep. 2017, 7, 3024. [Google Scholar] [CrossRef]
- Nieto-Fontarigo, J.J.; Salgado, F.J.; San-Jose, M.E.; Cruz, M.J.; Casas-Fernandez, A.; Gomez-Conde, M.J.; Valdes-Cuadrado, L.; Garcia-Gonzalez, M.A.; Arias, P.; Nogueira, M.; et al. The CD14 (-159 C/T) SNP is associated with sCD14 levels and allergic asthma, but not with CD14 expression on monocytes. Sci. Rep. 2018, 8, 4147. [Google Scholar] [CrossRef]
- Nieto-Fontarigo, J.J.; Gonzalez-Barcala, F.J.; Andrade-Bulos, L.J.; San-Jose, M.E.; Cruz, M.J.; Valdes-Cuadrado, L.; Crujeiras, R.M.; Arias, P.; Salgado, F.J. iTRAQ-based proteomic analysis reveals potential serum biomarkers of allergic and nonallergic asthma. Allergy 2020, 75, 3171–3183. [Google Scholar] [CrossRef]
- Nieto-Fontarigo, J.J.; Gonzalez-Barcala, F.J.; San Jose, E.; Arias, P.; Nogueira, M.; Salgado, F.J. CD26 and Asthma: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2019, 56, 139–160. [Google Scholar] [CrossRef]
- Nieto-Fontarigo, J.J.; Gonzalez-Barcala, F.J.; San-Jose, M.E.; Cruz, M.J.; Linares, T.; Soto-Mera, M.T.; Valdes-Cuadrado, L.; Garcia-Gonzalez, M.A.; Andrade-Bulos, L.J.; Arias, P.; et al. Expansion of a CD26low Effector TH Subset and Reduction in Circulating Levels of sCD26 in Stable Allergic Asthma in Adults. J. Investig. Allergol. Clin. Immunol. 2018, 28, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Fontarigo, J.J.; Salgado, F.J.; San-Jose, M.E.; Cruz, M.J.; Valdes, L.; Perez-Diaz, A.; Arias, P.; Nogueira, M.; Gonzalez-Barcala, F.J. Expansion of different subpopulations of CD26(-/low) T cells in allergic and non-allergic asthmatics. Sci. Rep. 2019, 9, 7556. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Wang, T.; Liu, G.; Sun, L.; Han, W.; Gao, Y. Dynamic Urinary Proteome Changes in Ovalbumin-Induced Asthma Mouse Model Using Data-Independent Acquisition Proteomics. J. Asthma Allergy 2021, 14, 1355–1366. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Huang, H.; Dai, Y.; Han, W.; Gao, Y. Proteome analysis of urinary biomarkers in a cigarette smoke-induced COPD rat model. Respir. Res. 2022, 23, 156. [Google Scholar] [CrossRef] [PubMed]
- Airoldi, L.; Magagnotti, C.; Iannuzzi, A.R.; Marelli, C.; Bagnati, R.; Pastorelli, R.; Colombi, A.; Santaguida, S.; Chiabrando, C.; Schiarea, S.; et al. Effects of cigarette smoking on the human urinary proteome. Biochem. Biophys. Res. Commun. 2009, 381, 397–402. [Google Scholar] [CrossRef]
- Brown, J.N.; Brewer, H.M.; Nicora, C.D.; Weitz, K.K.; Morris, M.J.; Skabelund, A.J.; Adkins, J.N.; Smith, R.D.; Cho, J.H.; Gelinas, R. Protein and microRNA biomarkers from lavage, urine, and serum in military personnel evaluated for dyspnea. BMC Med. Genomics 2014, 7, 58. [Google Scholar] [CrossRef]
- Wei, J.; Gao, Y. Early disease biomarkers can be found using animal models urine proteomics. Expert. Rev. Proteomics 2021, 18, 363–378. [Google Scholar] [CrossRef]
- Wu, J.; Li, X.; Zhao, M.; Huang, H.; Sun, W.; Gao, Y. Early Detection of Urinary Proteome Biomarkers for Effective Early Treatment of Pulmonary Fibrosis in a Rat Model. Proteomics Clin. Appl. 2017, 11, 1700103. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, J.; Li, L.; Liu, Y.; Zhao, H.; Li, N.; Li, B.; Zhang, A.; Huang, H.; Chen, S.; et al. Identification of urine protein biomarkers with the potential for early detection of lung cancer. Sci. Rep. 2015, 5, 11805. [Google Scholar] [CrossRef]
- Wei, J.; Ni, N.; Meng, W.; Gao, Y. Early urine proteome changes in the Walker-256 tail-vein injection rat model. Sci. Rep. 2019, 9, 13804. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Qiu, F.; Qiu, Z. Comparative analysis of the human urinary proteome by 1D SDS-PAGE and chip-HPLC-MS/MS identification of the AACT putative urinary biomarker. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010, 878, 3395–3401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Leng, W.; Sun, C.; Lu, T.; Chen, Z.; Men, X.; Wang, Y.; Wang, G.; Zhen, B.; Qin, J. Urine Proteome Profiling Predicts Lung Cancer from Control Cases and Other Tumors. EBioMedicine 2018, 30, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.; Kheirandish-Gozal, L.; Peris, E.; Schoenfelt, K.Q.; Gozal, D. Contextualised urinary biomarker analysis facilitates diagnosis of paediatric obstructive sleep apnoea. Sleep Med. 2014, 15, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Seetho, I.W.; Ramirez-Torres, A.; Albalat, A.; Mullen, W.; Mischak, H.; Parker, R.J.; Craig, S.; Duffy, N.; Hardy, K.J.; Burniston, J.G.; et al. Urinary proteomic profiling in severe obesity and obstructive sleep apnoea with CPAP treatment. Sleep Sci. 2015, 8, 58–67. [Google Scholar] [CrossRef]
- Starodubtseva, N.L.; Kononikhin, A.S.; Bugrova, A.E.; Chagovets, V.; Indeykina, M.; Krokhina, K.N.; Nikitina, I.V.; Kostyukevich, Y.I.; Popov, I.A.; Larina, I.M.; et al. Investigation of urine proteome of preterm newborns with respiratory pathologies. J. Proteomics 2016, 149, 31–37. [Google Scholar] [CrossRef]
- Bi, X.; Liu, W.; Ding, X.; Liang, S.; Zheng, Y.; Zhu, X.; Quan, S.; Yi, X.; Xiang, N.; Du, J.; et al. Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19. Cell. Rep. 2022, 38, 110271. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, N.; Jin, R.; Feng, Y.; Wang, S.; Gao, S.; Gao, R.; Wu, G.; Tian, D.; Tan, W.; et al. Immune suppression in the early stage of COVID-19 disease. Nat. Commun. 2020, 11, 5859. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Liu, H.; Sun, W.; Ding, B.; Zhao, Y.; Chen, P.; Zhu, L.; Li, Z.; Li, N.; et al. Urine proteome of COVID-19 patients. Urine 2020, 2, 1–8. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, X.; Chen, X.; Cai, Y. Proteomics in Biomarker Discovery for Tuberculosis: Current Status and Future Perspectives. Front. Microbiol. 2022, 13, 845229. [Google Scholar] [CrossRef]
- Young, B.L.; Mlamla, Z.; Gqamana, P.P.; Smit, S.; Roberts, T.; Peter, J.; Theron, G.; Govender, U.; Dheda, K.; Blackburn, J. The identification of tuberculosis biomarkers in human urine samples. Eur. Respir. J. 2014, 43, 1719–1729. [Google Scholar] [CrossRef]
- Liu, L.; Deng, J.; Yang, Q.; Wei, C.; Liu, B.; Zhang, H.; Xin, H.; Pan, S.; Liu, Z.; Wang, D.; et al. Urinary proteomic analysis to identify a potential protein biomarker panel for the diagnosis of tuberculosis. IUBMB Life 2021, 73, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Kyyaly, M.A.; Vorobeva, E.V.; Kothalawala, D.M.; Fong, W.C.G.; He, P.; Sones, C.L.; Al-Zahrani, M.; Sanchez-Elsner, T.; Arshad, S.H.; Kurukulaaratchy, R.J. MicroRNAs-A Promising Tool for Asthma Diagnosis and Severity Assessment: A Systematic Review. J. Pers. Med. 2022, 12, 543. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Lee, H.Y.; Choi, J.Y.; Hur, J.; Kim, I.K.; Kim, Y.K.; Kang, J.Y.; Lee, S.Y. Inhibition of MicroRNA-21 by an antagomir ameliorates allergic inflammation in a mouse model of asthma. Exp. Lung Res. 2017, 43, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Hammad Mahmoud Hammad, R.; Hamed, D.; Eldosoky, M.; Ahmad, A.; Osman, H.M.; Abd Elgalil, H.M.; Mahmoud Hassan, M.M. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 2018, 24, 171–179. [Google Scholar] [CrossRef]
- Kim, R.Y.; Horvat, J.C.; Pinkerton, J.W.; Starkey, M.R.; Essilfie, A.T.; Mayall, J.R.; Nair, P.M.; Hansbro, N.G.; Jones, B.; Haw, T.J.; et al. MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase-mediated suppression of histone deacetylase 2. J. Allergy Clin. Immunol. 2017, 139, 519–532. [Google Scholar] [CrossRef]
- Mattes, J.; Collison, A.; Plank, M.; Phipps, S.; Foster, P.S. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA 2009, 106, 18704–18709. [Google Scholar] [CrossRef]
- Olivieri, F.; Prattichizzo, F.; Giuliani, A.; Matacchione, G.; Rippo, M.R.; Sabbatinelli, J.; Bonafe, M. miR-21 and miR-146a: The microRNAs of inflammaging and age-related diseases. Ageing Res. Rev. 2021, 70, 101374. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Kumar, P.; Bhattacharyya, S.; Chattoraj, S.; Srivastava, M.; Pollard, H.B.; Biswas, R. Differential regulation of inflammation by inflammatory mediators in cystic fibrosis lung epithelial cells. J. Interferon. Cytokine Res. 2013, 33, 121–129. [Google Scholar] [CrossRef]
- Zhang, H.H.; Li, C.X.; Tang, L.F. The Differential Expression Profiles of miRNA-let 7a, 7b, and 7c in Bronchoalveolar Lavage Fluid From Infants With Asthma and Airway Foreign Bodies. J. Evid. Based Integr. Med. 2019, 24, 2515690X18821906. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.; Li, P.; Huang, H.; Liu, T.; He, H.; Lin, Z.; Jiang, Y.; Ren, N.; Wu, B.; et al. Circulating microRNA Signatures Associated with Childhood Asthma. Clin. Lab. 2015, 61, 467–474. [Google Scholar] [CrossRef]
- Soccio, P.; Moriondo, G.; Lacedonia, D.; Tondo, P.; Pescatore, D.; Quarato, C.M.I.; Carone, M.; Foschino Barbaro, M.P.; Scioscia, G. MiRNA and Exosomal miRNA as New Biomarkers Useful to Phenotyping Severe Asthma. Biomolecules 2023, 13, 1542. [Google Scholar] [CrossRef] [PubMed]
- Polikepahad, S.; Knight, J.M.; Naghavi, A.O.; Oplt, T.; Creighton, C.J.; Shaw, C.; Benham, A.L.; Kim, J.; Soibam, B.; Harris, R.A.; et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J. Biol. Chem. 2010, 285, 30139–30149. [Google Scholar] [CrossRef] [PubMed]
- Collison, A.; Mattes, J.; Plank, M.; Foster, P.S. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J. Allergy Clin. Immunol. 2011, 128, 160–167.e4. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Yang, G.; Zhang, R.; Xu, G.; Zhang, L.; Wen, W.; Lu, J.; Liu, J.; Yu, Y. Altered microRNA Expression Profiles of Extracellular Vesicles in Nasal Mucus From Patients With Allergic Rhinitis. Allergy Asthma Immunol. Res. 2015, 7, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.; Bao, R.; Roth, M.; Liu, L.; Yang, X.; Tang, X.; Yang, X.; Sun, Q.; Lu, S. microRNA-23a contributes to asthma by targeting BCL2 in airway epithelial cells and CXCL12 in fibroblasts. J. Cell. Physiol. 2019, 234, 21153–21165. [Google Scholar] [CrossRef]
- Tiwari, A.; Hobbs, B.D.; Li, J.; Kho, A.T.; Amr, S.; Celedon, J.C.; Weiss, S.T.; Hersh, C.P.; Tantisira, K.G.; McGeachie, M.J. Blood miRNAs Are Linked to Frequent Asthma Exacerbations in Childhood Asthma and Adult COPD. Noncoding RNA 2022, 8, 27. [Google Scholar] [CrossRef]
- Hu, G.; Du, Y.; Xie, M.; Chen, R.; Shi, F. Circulating miRNAs act as potential biomarkers for asthma. Front. Immunol. 2023, 14, 1296177. [Google Scholar] [CrossRef]

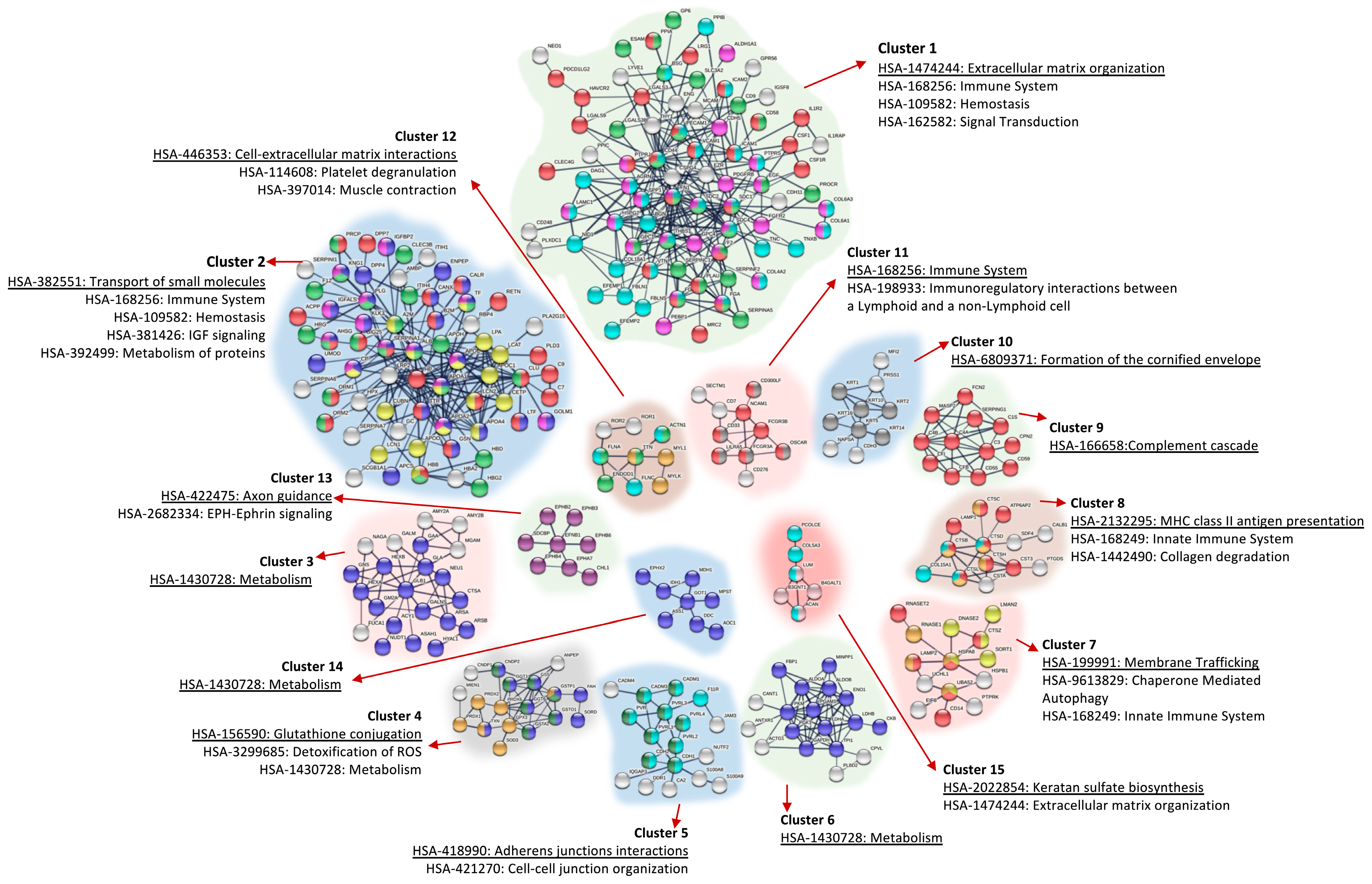

| Cluster | Most Relevant Pathway/Biological Process | N | ES | FE | FDR |
|---|---|---|---|---|---|
| Immune system | |||||
| 1 | R-HSA-168249: Innate immune system | 122 | 33.271 | 3.115 | 3.42 × 10−29 |
| 3 | GO:0002250: Adaptive immune system | 47 | 13.537 | 3.779 | 5.97 × 10−12 |
| 6 | GO:0006958: Complement activation, classical pathway | 31 | 10.222 | 9.066 | 3.46 × 10−17 |
| 10 | R-HSA-166658: Complement cascade | 17 | 5.521 | 7.954 | 7.88 × 10−9 |
| 30 | R-HSA-168898: TLR cascades | 8 | 1.156 | 1.383 | 9.21 × 10−1 |
| 32 | R-HSA-2132295: MHC class II antigen presentation | 8 | 1.075 | 1.765 | 9.08 × 10−1 |
| 37 | R-HSA-447115: IL-12 family signalling | 6 | 0.878 | 2.857 | 4.08 × 10−1 |
| 43 | R-HSA-8953897: Cellular responses to stimuli | 21 | 0.531 | 0.714 | 9.78 × 10−1 |
| 44 | R-HSA-168255: Influenza infection | 4 | 0.356 | 0.696 | 9.30 × 10−1 |
| 53 | R-HSA-9658195: Leishmania infection | 6 | 0.010 | 0.641 | 9.60 × 10−1 |
| Haemostasis | |||||
| 2 | R-HSA-109582: Haemostasis | 70 | 19.675 | 3.059 | 3.70 × 10−15 |
| 9 | GO:0042730: Fibrinolysis | 10 | 6.317 | 19.086 | 9.80 × 10−8 |
| 24 | R-HSA-140837: Intrinsic Pathway of Fibrin Clot Formation | 8 | 2.258 | 9.439 | 2.49 × 10−4 |
| 31 | GO:0072378: Blood coagulation, fibrin clot formation | 4 | 1.104 | 20.722 | 2.16 × 10−2 |
| Extracellular Matrix Organization/Cell adhesion | |||||
| 4 | R-HSA-1500931: Cell–cell communication | 26 | 12.478 | 5.470 | 3.69 × 10−10 |
| 12 | R-HSA-420597: Nectin/Necl trans heterodimerization | 7 | 3.972 | 27.137 | 5.29 × 10−7 |
| 14 | R-HSA-1474228: Degradation of the extracellular matrix | 22 | 3.480 | 4.264 | 1.15 × 10−6 |
| 17 | GO:0098742: Cell–Cell adhesion via plasma-membrane adhesion molecules | 10 | 3.030 | 9.298 | 7.34 × 10−5 |
| 40 | R-HSA-199991: Membrane trafficking | 19 | 0.685 | 0.813 | 9.21 × 10−1 |
| 50 | R-HSA-5619115: Disorders of transmembrane transporters | 5 | 0.037 | 0.543 | 9.84 × 10−1 |
| Signalling | |||||
| 5 | R-HSA-381426: Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | 37 | 10.866 | 8.033 | 1.12 × 10−20 |
| 11 | GO:0007169: Transmembrane receptor PTK signalling pathway | 16 | 4.315 | 4.396 | 2.72 × 10−4 |
| 26 | R-HSA-3928664: Ephrin signalling | 6 | 1.963 | 8.570 | 7.40 × 10−3 |
| 31 | R-HSA-5683057: MAPK family signalling cascades | 13 | 1.104 | 1.085 | 9.21 × 10−1 |
| 38 | R-HSA-6806834: Signalling by MET | 8 | 0.819 | 2.748 | 2.15 × 10−1 |
| 42 | R-HSA-177929: Signalling by EGFR | 4 | 0.586 | 2.171 | 9.21 × 10−1 |
| 48 | R-HSA-1257604: PIP3 activates AKT signalling | 5 | 0.063 | 0.508 | 9.90 × 10−1 |
| 49 | R-HSA-9006931: Signalling by nuclear receptors | 9 | 0.058 | 0.817 | 9.21 × 10−1 |
| 51 | R-HSA-195721: Signalling by WNT | 4 | 0.026 | 0.327 | 1.00 |
| 52 | R-HSA-194315: Signalling by Rho GTPases | 14 | 0.017 | 0.537 | 9.99 × 10−1 |
| 54 | R-HSA-372790: Signalling by GPCR | 7 | 0.005 | 0.268 | 1.00 |
| Metabolism | |||||
| 7 | R-HSA-5663084: Diseases of carbohydrate metabolism | 11 | 6.897 | 8.780 | 6.73 × 10−6 |
| 8 | R-HSA-71387: Metabolism of carbohydrates | 46 | 6.389 | 4.232 | 1.68 × 10−14 |
| 16 | R-HSA-70326: Glucose metabolism | 11 | 3.382 | 3.280 | 2.35 × 10−2 |
| 18 | R-HSA-2022377: Metabolism of Angiotensinogen to Angiotensins | 7 | 2.724 | 10.553 | 5.62 × 10−4 |
| 19 | R-HSA-174824: Plasma lipoprotein assembly, remodelling, and clearance | 12 | 2.604 | 4.401 | 1.29 × 10−3 |
| 20 | GO:0006869: Lipid transport | 10 | 2.529 | 4.075 | 2.38 × 10−2 |
| 21 | R-HSA-1660662: Glycosphingolipid metabolism | 10 | 2.513 | 5.899 | 6.71 × 10−4 |
| 22 | GO:0019915: Lipid storage | 4 | 2.399 | 5.579 | 4.00 × 10−1 |
| 27 | R-HSA-156590: Glutathione conjugation | 8 | 1.839 | 6.030 | 4.52 × 10−3 |
| 28 | R-HSA-189085: Digestion of dietary carbohydrates | 4 | 1.551 | 9.868 | 6.93 × 10−2 |
| 29 | R-HSA-159740: Gamma carboxylation of protein precursors | 5 | 1.278 | 8.141 | 3.64 × 10−1 |
| 35 | R-HSA-3299685: Detoxification of ROS | 7 | 0.956 | 5.134 | 2.65 × 10−2 |
| 36 | R-HSA-3781860: Diseases associated with N-glycosylation of proteins | 4 | 0.918 | 5.427 | 2.72 × 10−1 |
| 41 | R-HSA-8953854: Metabolism of RNA | 5 | 0.594 | 0.201 | 1.00 |
| 45 | R-HSA-70268: Pyruvate metabolism | 3 | 0.241 | 2.626 | 9.21 × 10−1 |
| 56 | R-HSA-74160: Gene expression (Transcription) | 12 | 0.000 | 0.217 | 1.00 |
| Cell death | |||||
| 33, 47 | R-HSA-9612973: Autophagy | 6 | 1.052 | 1.078 | 9.21 × 10−1 |
| 34 | GO:2000352: Negative regulation of endothelial cell apoptotic process | 5 | 1.025 | 5.037 | 2.30 × 10−1 |
| 46 | R-HSA-109581: Apoptosis | 6 | 0.207 | 0.905 | 9.21 × 10−1 |
| Others | |||||
| 13, 23, 25 | R-HSA-1638074: Keratan sulphate/Keratin metabolism | 9 | 3.877 | 7.183 | 4.70 × 10−4 |
| 15 | R-HSA-1480926: O2/CO2 exchange in erythrocytes | 5 | 3.435 | 10.437 | 1.35 × 10−2 |
| 39 | R-HSA-1266738: Developmental Biology | 42 | 0.800 | 1.01 | 9.21 × 10−1 |
| 55 | R-HSA-1640170: Cell cycle | 4 | 0.000 | 0.157 | 1.00 |
| baseMean | Log2FC | lfcSE | Stat | p Value | |
|---|---|---|---|---|---|
| Asthma vs. Healthy | |||||
| hsa-miR-574-5p | 40.5671479 | −0.9148423 | 0.35729808 | −2.5604455 | 0.01045381 |
| hsa-miR-342-3p | 22.7834699 | 0.8300255 | 0.34822973 | 2.38355728 | 0.01714622 |
| hsa-miR-488-3p | 17.6996462 | −0.8831223 | 0.38710618 | −2.2813439 | 0.0225281 |
| hsa-miR-320e | 94.2924488 | 1.26604719 | 0.56756082 | 2.23068109 | 0.02570226 |
| hsa-miR-377-3p | 23.1056833 | −0.641396 | 0.30913106 | −2.0748353 | 0.0380018 |
| hsa-miR-139-3p | 24.4883407 | 0.69054879 | 0.33788118 | 2.04376221 | 0.04097704 |
| hsa-miR-34b-3p | 16.6518662 | 0.80311135 | 0.40940465 | 1.96165662 | 0.04980248 |
| T2high vs. Healthy | |||||
| hsa-miR-4454+hsa-miR-7975 | 1871.43143 | 4.84845886 | 1.09064723 | 4.4454877 | 8.77 × 10−6 |
| hsa-miR-4286 | 95.6039239 | 2.34300078 | 0.58675153 | 3.99317369 | 6.52 × 10−5 |
| hsa-let-7b-5p | 91.2725914 | 1.89394928 | 0.53071736 | 3.56865903 | 0.00035881 |
| hsa-miR-200c-3p | 38.446332 | 1.48357943 | 0.42808608 | 3.46561006 | 0.00052903 |
| hsa-miR-1260a | 30.3256399 | 1.6754408 | 0.51630868 | 3.24503707 | 0.00117435 |
| hsa-let-7a-5p | 60.4218451 | 2.15717229 | 0.66622303 | 3.23791312 | 0.00120407 |
| hsa-miR-23a-3p | 63.1060858 | 1.78852509 | 0.6104412 | 2.92988923 | 0.00339083 |
| hsa-miR-125a-5p | 20.2466154 | 1.51553846 | 0.52845536 | 2.86786468 | 0.00413252 |
| hsa-miR-4516 | 67.6961056 | 2.05065792 | 0.71652204 | 2.8619607 | 0.00421029 |
| hsa-miR-488-3p | 17.6996462 | −1.7776733 | 0.62174119 | −2.8591853 | 0.00424731 |
| hsa-miR-204-5p | 67.847085 | 1.9333541 | 0.71686145 | 2.69697041 | 0.00699735 |
| hsa-miR-10b-5p | 55.5235107 | 1.73778902 | 0.67353223 | 2.58011265 | 0.00987681 |
| hsa-miR-191-5p | 27.4502163 | 1.3938602 | 0.56984399 | 2.44603827 | 0.01444357 |
| hsa-let-7c-5p | 21.9687882 | 1.12353434 | 0.49590837 | 2.26560875 | 0.02347535 |
| hsa-miR-10a-5p | 44.1971719 | 1.14162491 | 0.50780162 | 2.24817107 | 0.02456528 |
| hsa-miR-30d-5p | 74.7454413 | 1.72851576 | 0.77089042 | 2.24223277 | 0.02494633 |
| hsa-miR-630 | 27.9137898 | 1.15203733 | 0.53392982 | 2.15765685 | 0.03095452 |
| hsa-miR-301a-3p | 26.8636724 | −1.0874893 | 0.50811571 | −2.1402394 | 0.03233543 |
| hsa-miR-342-3p | 22.7834699 | 1.0648488 | 0.50082727 | 2.12617975 | 0.0334883 |
| hsa-miR-30e-3p | 24.8830148 | 1.00361175 | 0.48709092 | 2.06041976 | 0.03935843 |
| hsa-miR-30a-3p | 21.0812686 | 0.99494462 | 0.49195509 | 2.02242977 | 0.04313197 |
| T2low vs. Healthy | |||||
| hsa-miR-4454+hsa-miR-7975 | 1871.43143 | −2.5504233 | 0.93497526 | −2.7277976 | 0.00637587 |
| hsa-miR-4286 | 95.6039239 | −1.3041977 | 0.51370779 | −2.538793 | 0.01112356 |
| hsa-miR-574-5p | 40.5671479 | −1.096622 | 0.44726633 | −2.4518322 | 0.01421309 |
| hsa-miR-4516 | 67.6961056 | −1.3061093 | 0.61141963 | −2.1361913 | 0.03266382 |
| T2high vs. T2low | |||||
| hsa-miR-4454+hsa-miR-7975 | 1871.43143 | 7.39888211 | 1.31874659 | 5.61054123 | 2.02 × 10−8 |
| hsa-miR-4286 | 95.6039239 | 3.6471985 | 0.7116184 | 5.12521669 | 2.97 × 10−7 |
| hsa-miR-4516 | 67.6961056 | 3.35676722 | 0.85783089 | 3.91308736 | 9.11 × 10−5 |
| hsa-miR-1260a | 30.3256399 | 2.3236675 | 0.60848414 | 3.8187807 | 0.00013411 |
| hsa-let-7a-5p | 60.4218451 | 2.75193846 | 0.79141495 | 3.47723839 | 0.00050661 |
| hsa-let-7b-5p | 91.2725914 | 2.12931239 | 0.63735641 | 3.34085034 | 0.00083522 |
| hsa-miR-200c-3p | 38.446332 | 1.62529815 | 0.50490333 | 3.21902838 | 0.00128626 |
| hsa-miR-191-5p | 27.4502163 | 1.98309985 | 0.67706047 | 2.92898484 | 0.00340071 |
| hsa-miR-23a-3p | 63.1060858 | 1.98414825 | 0.72983499 | 2.71862581 | 0.00655537 |
| hsa-miR-10b-5p | 55.5235107 | 2.15897028 | 0.80699947 | 2.67530571 | 0.00746611 |
| hsa-let-7c-5p | 21.9687882 | 1.4493829 | 0.59007958 | 2.45624986 | 0.01403955 |
| hsa-miR-125a-5p | 20.2466154 | 1.41552655 | 0.61068218 | 2.3179431 | 0.02045241 |
| hsa-miR-630 | 27.9137898 | 1.35069962 | 0.64138636 | 2.10590637 | 0.03521249 |
| hsa-miR-10a-5p | 44.1971719 | 1.26924038 | 0.6064849 | 2.09278152 | 0.03636866 |
| hsa-miR-204-5p | 67.847085 | 1.75264746 | 0.85172349 | 2.0577658 | 0.03961262 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martelo-Vidal, L.; Vázquez-Mera, S.; Miguéns-Suárez, P.; Bravo-López, S.B.; Makrinioti, H.; Domínguez-Arca, V.; de-Miguel-Díez, J.; Gómez-Carballa, A.; Salas, A.; González-Barcala, F.J.; et al. Urinary Proteome and Exosome Analysis Protocol for the Discovery of Respiratory Diseases Biomarkers. Biomolecules 2025, 15, 60. https://doi.org/10.3390/biom15010060
Martelo-Vidal L, Vázquez-Mera S, Miguéns-Suárez P, Bravo-López SB, Makrinioti H, Domínguez-Arca V, de-Miguel-Díez J, Gómez-Carballa A, Salas A, González-Barcala FJ, et al. Urinary Proteome and Exosome Analysis Protocol for the Discovery of Respiratory Diseases Biomarkers. Biomolecules. 2025; 15(1):60. https://doi.org/10.3390/biom15010060
Chicago/Turabian StyleMartelo-Vidal, Laura, Sara Vázquez-Mera, Pablo Miguéns-Suárez, Susana Belén Bravo-López, Heidi Makrinioti, Vicente Domínguez-Arca, Javier de-Miguel-Díez, Alberto Gómez-Carballa, Antonio Salas, Francisco Javier González-Barcala, and et al. 2025. "Urinary Proteome and Exosome Analysis Protocol for the Discovery of Respiratory Diseases Biomarkers" Biomolecules 15, no. 1: 60. https://doi.org/10.3390/biom15010060
APA StyleMartelo-Vidal, L., Vázquez-Mera, S., Miguéns-Suárez, P., Bravo-López, S. B., Makrinioti, H., Domínguez-Arca, V., de-Miguel-Díez, J., Gómez-Carballa, A., Salas, A., González-Barcala, F. J., Salgado, F. J., & Nieto-Fontarigo, J. J. (2025). Urinary Proteome and Exosome Analysis Protocol for the Discovery of Respiratory Diseases Biomarkers. Biomolecules, 15(1), 60. https://doi.org/10.3390/biom15010060







