Biosynthesis of Arabinoside from Sucrose and Nucleobase via a Novel Multi-Enzymatic Cascade
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Strains and Plasmids
2.3. Protein Expression and Purification
2.4. Enzymatic Activity Assays
2.5. Production of Arabinosides from Sucrose or G6P
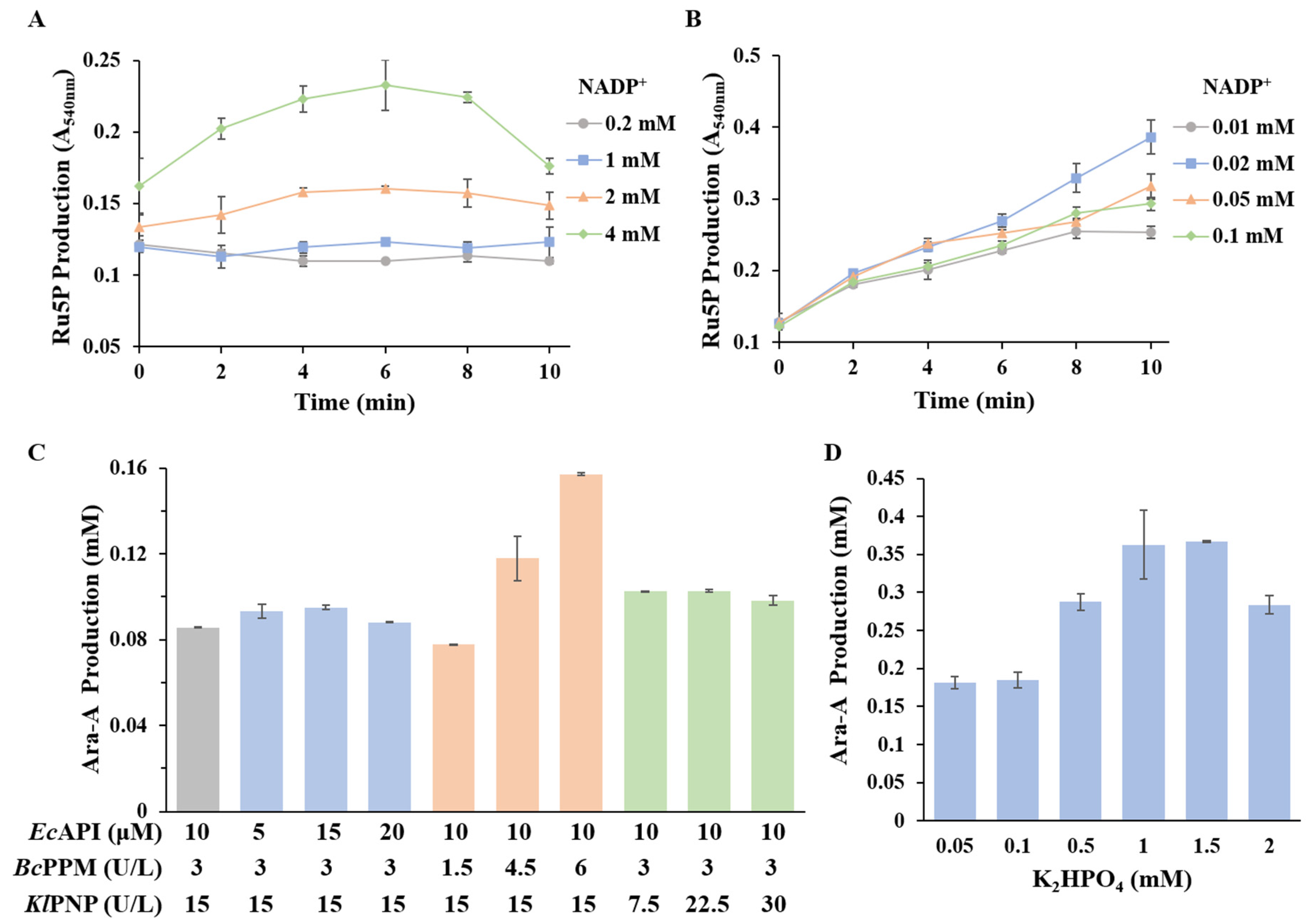
2.6. Effect of NADP+ Concentrates on Ru5P Accumulation
2.7. Effect of Phosphate Concentrations on the Accumulation of Intermediate Metabolite
2.8. Analytical Methods
3. Results and Discussion
3.1. Design of the Multi-Enzymatic Cascade for Arabinoside Production from Sucrose and Nucleobase
3.2. Enzyme Selection of NP
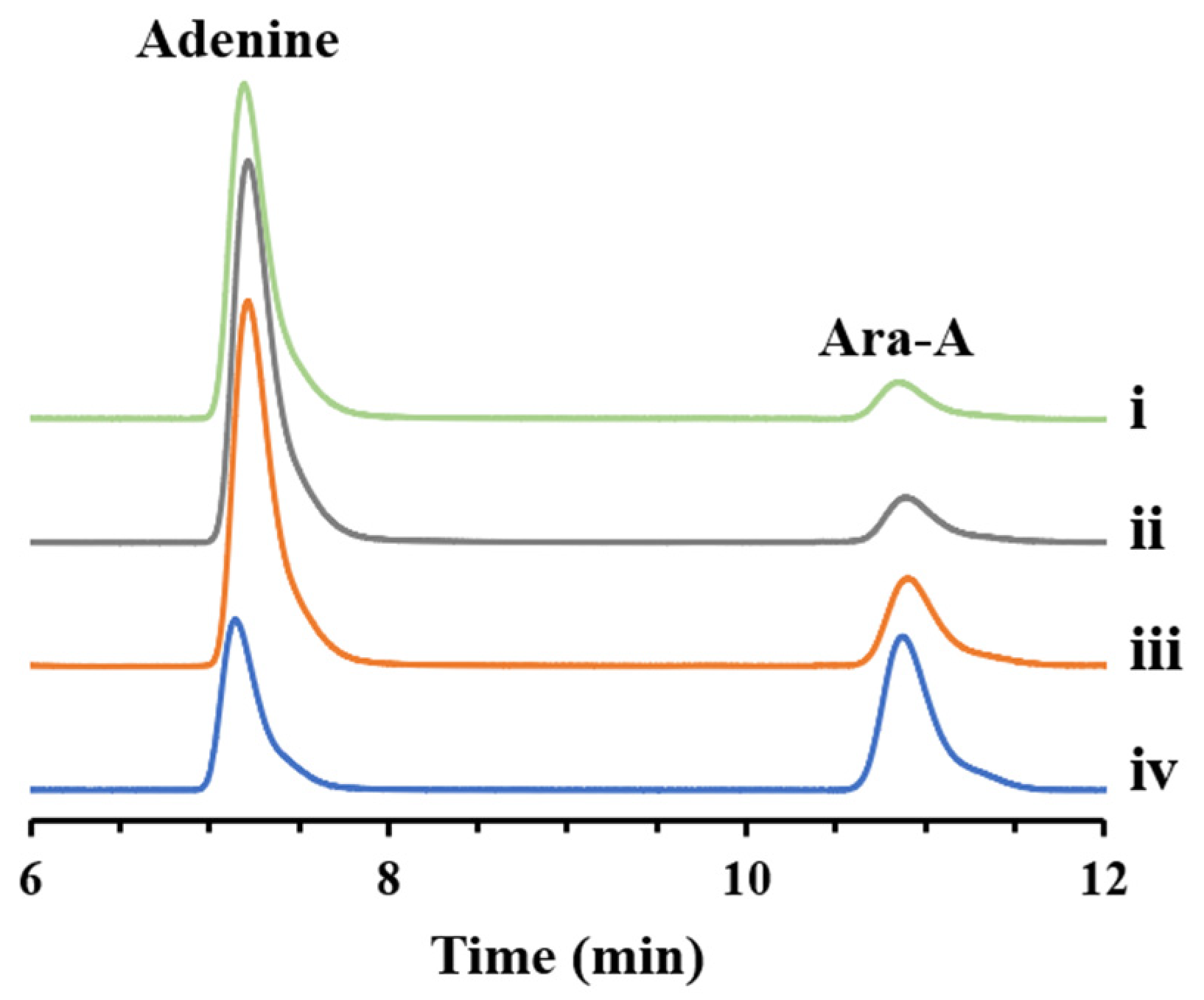
3.3. Feasibility Verification of Ara-A Production from Sucrose and Adenine
3.4. Optimization of Ara-A Production from Sucrose and Adenine
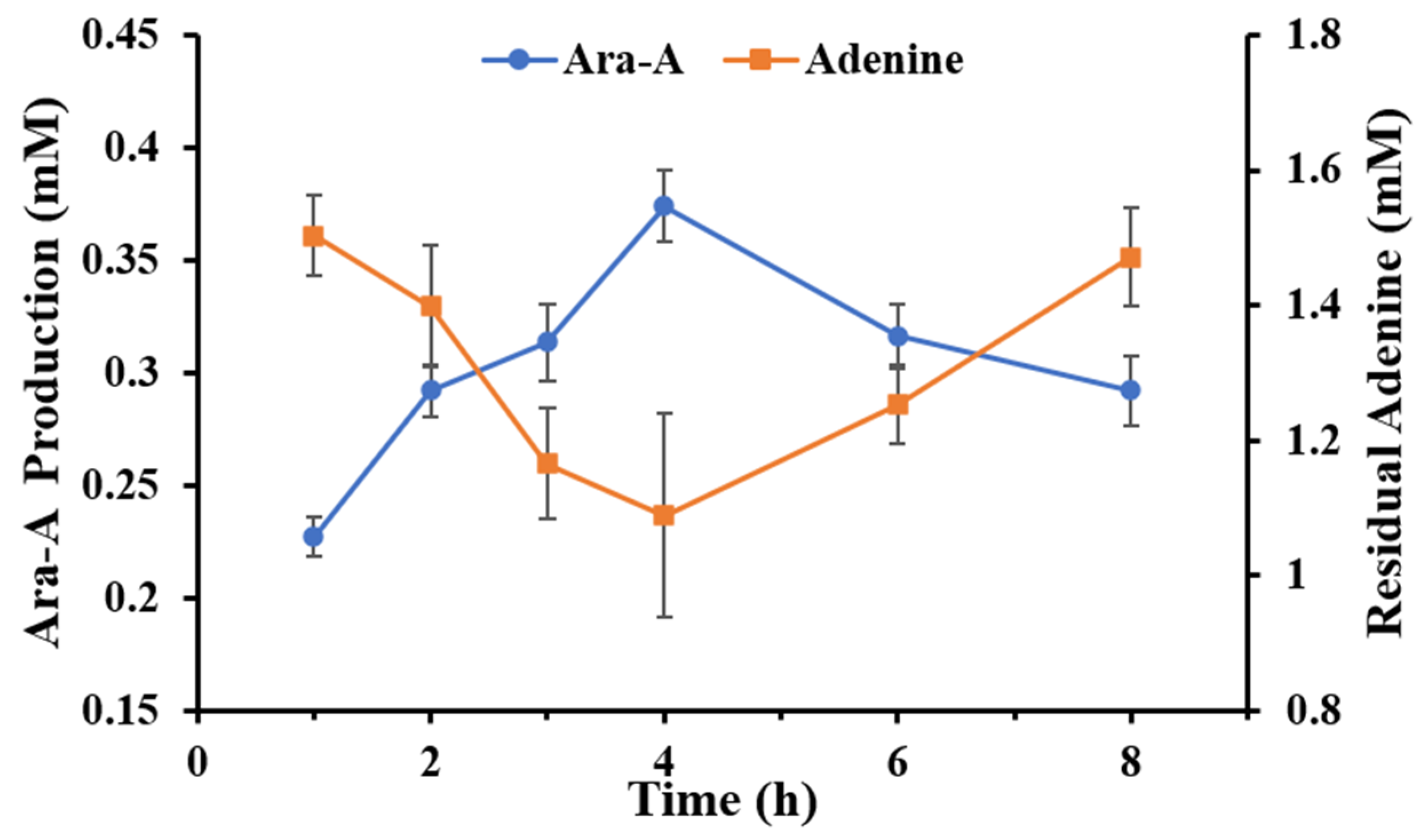
3.5. Production of Ara-A from Sucrose and Adenine under Optimal Conditions
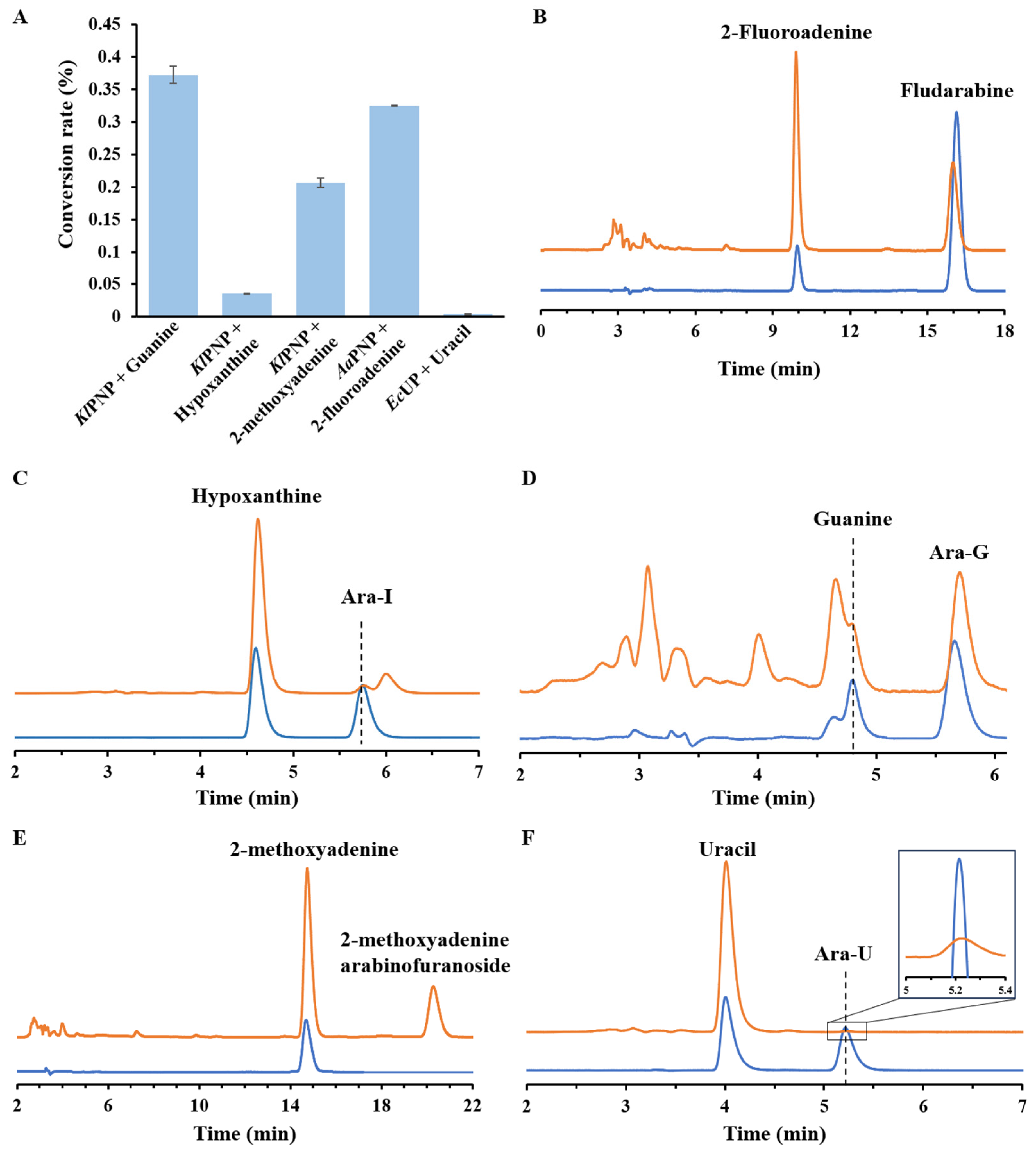
3.6. Production of Other Arabinosides through In Vitro Multi-Enzymatic Cascade Equipped with Different NPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ara-A | Vidarabine |
| Ara-G | Arabinofuranosylguanine |
| Ara-C | Cytarabine |
| Ara-U | Spongouridine |
| Ara-I | Hypoxanthine arabinofuranoside |
| NADP+ | Nicotinamide adenine dinucleotide phosphate |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| MTT | Methylthiazolyldiphenyl-tetrazolium bromide |
| PES | Phenazine ethosulfate |
| SP | Sucrose phosphorylase |
| PGM | Phosphoglucomutase |
| G6PD | Glucose 6-phosphate 1-dehydrogenase |
| 6PGDH | 6-phosphogluconate dehydrogenase |
| API | D-arabinose 5-phosphate isomerase |
| PPM | Phosphopentomutase |
| NP | Nucleoside phosphorylase |
| PNP | Purine nucleoside phosphorylase |
| UP | Uridine phosphorylase |
| TP | Thymidine phosphorylase |
| PyNP | Pyrimidine nucleoside phosphorylase |
| RK | Ribokinase |
| TPNOX | H2O-forming NADPH oxidase |
| G1P | Glucose 1-phosphate |
| G6P | Glucose 6-phosphate |
| GlcA6P | 6-phosphogluconate |
| Ru5P | Ribulose 5-phosphate |
| A5P | Arabinose 5-phosphate |
| A1P | Arabinose 1-phosphate |
| HPLC | High-performance liquid chromatography |
References
- Niu, G.; Tan, H. Nucleoside antibiotics: Biosynthesis, regulation, and biotechnology. Trends Microbiol. 2015, 23, 110–119. [Google Scholar] [CrossRef]
- Seley-Radtke, K.L.; Yates, M.K. The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antivir. Res. 2018, 154, 66–86. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Attar, F.; Bloukh, S.H.; Sharifi, M.; Nabi, F.; Bai, Q.; Khan, R.H.; Falahati, M. A review on the interaction of nucleoside analogues with SARS-CoV-2 RNA dependent RNA polymerase. Int. J. Biol. Macromol. 2021, 181, 605–611. [Google Scholar] [CrossRef]
- Zandi, K.; Amblard, F.; Musall, K.; Downs-Bowen, J.; Kleinbard, R.; Oo, A.; Cao, D.; Liang, B.; Russell, O.O.; McBrayer, T.; et al. Repurposing nucleoside analogs for human coronaviruses. Antimicrob. Agents Chemother. 2021, 65, e01652-20. [Google Scholar] [CrossRef]
- Lapponi, M.J.; Rivero, C.W.; Zinni, M.A.; Britos, C.N.; Trelles, J.A. New developments in nucleoside analogues biosynthesis: A review. J. Mol. Catal. B Enzym. 2016, 133, 218–233. [Google Scholar] [CrossRef]
- Tsesmetzis, N.; Paulin, C.B.J.; Rudd, S.G.; Herold, N. Nucleobase and nucleoside analogues: Resistance and re-sensitisation at the level of pharmacokinetics, pharmacodynamics and metabolism. Cancers 2018, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-M.; Chen, Y.-N.; Zeng, Z.; Gao, C.-H.; Su, X.; Peng, Y. Marine nucleosides: Structure, bioactivity, synthesis and biosynthesis. Mar. Drugs 2014, 12, 5817–5838. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wan, D.; Xu, G.; Wang, G.; Ma, H.; Wang, T.; Gao, Y.; Qi, J.; Chen, X.; Zhu, J.; et al. An unusual protector-protege strategy for the biosynthesis of purine nucleoside antibiotics. Cell Chem. Biol. 2017, 24, 171–181. [Google Scholar] [CrossRef]
- Mehellou, Y.; Valente, R.; Mottram, H.; Walsby, E.; Mills, K.I.; Balzarini, J.; McGuigan, C. Phosphoramidates of 2’-β-D-arabinouridine (AraU) as phosphate prodrugs; design, synthesis, in vitro activity and metabolism. Biorg. Med. Chem. 2010, 18, 2439–2446. [Google Scholar] [CrossRef]
- Benitez-Mateos, A.I.; Padrosa, D.R.; Paradisi, F. Multistep enzyme cascades as a route towards green and sustainable pharmaceutical syntheses. Nat. Chem. 2022, 14, 489–499. [Google Scholar] [CrossRef]
- Naik, K.; Dheeraj, S.; Jeevani, K.; Saravanan, T. Evaluating multienzyme cascade routes for pharmaceutically relevant molecules. Eur. J. Org. Chem. 2024, 27, e202301236. [Google Scholar] [CrossRef]
- Devine, P.N.; Howard, R.M.; Kumar, R.; Thompson, M.P.; Truppo, M.D.; Turner, N.J. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2018, 2, 409–421. [Google Scholar] [CrossRef]
- Li, P.; Jing, R.; Zhou, M.; Jia, P.; Li, Z.; Liu, G.; Wang, Z.; Wang, H. Whole-cell biosynthesis of cytarabine by an unnecessary protein-reduced Escherichia coli that coexpresses purine and uracil phosphorylase. Biotechnol. Bioeng. 2022, 119, 1768–1780. [Google Scholar]
- Del Arco, J.; Acosta, J.; Fernandez-Lucas, J. New trends in the biocatalytic production of nucleosidic active pharmaceutical ingredients using 2’-deoxyribosyltransferases. Biotechnol. Adv. 2021, 51, 107701. [Google Scholar] [CrossRef]
- Kamel, S.; Thiele, I.; Neubauer, P.; Wagner, A. Thermophilic nucleoside phosphorylases: Their properties, characteristics and applications. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140304. [Google Scholar] [CrossRef] [PubMed]
- Robescu, M.S.; Serra, I.; Terreni, M.; Ubiali, D.; Bavaro, T. A multi-enzymatic cascade reaction for the synthesis of vidarabine 5’-monophosphate. Catalysts 2020, 10, 60. [Google Scholar] [CrossRef]
- Serra, I.; Daly, S.; Alcantara, A.R.; Bianchi, D.; Terreni, M.; Ubiali, D. Redesigning the synthesis of vidarabine via a multienzymatic reaction catalyzed by immobilized nucleoside phosphorylases. RSC Adv. 2015, 5, 23569–23577. [Google Scholar] [CrossRef]
- Liu, G.; Wang, J.; Chu, J.; Jiang, T.; Qin, S.; Gao, Z.; He, B. Engineering substrate promiscuity of nucleoside phosphorylase via an insertions-deletions strategy. JACS Au 2024, 4, 454–464. [Google Scholar] [CrossRef]
- Fateev, I.V.; Kostromina, M.A.; Abramchik, Y.A.; Eletskaya, B.Z.; Mikheeva, O.O.; Lukoshin, D.D.; Zayats, E.A.; Berzina, M.Y.; Dorofeeva, E.V.; Paramonov, A.S.; et al. Multi-enzymatic cascades in the synthesis of modified nucleosides: Comparison of the thermophilic and mesophilic pathways. Biomolecules 2021, 11, 586. [Google Scholar] [CrossRef]
- Cracan, V.; Titov, D.V.; Shen, H.; Grabarek, Z.; Mootha, V.K. A genetically encoded tool for manipulation of NADP+/NADPH in living cells. Nat. Chem. Biol. 2017, 13, 1088–1095. [Google Scholar] [CrossRef]
- Meredith, T.C.; Woodard, R.W. Escherichia coli YrbH is a D-arabinose 5-phosphate isomerase. J. Biol. Chem. 2003, 278, 32771–32777. [Google Scholar] [CrossRef] [PubMed]
- Zayats, E.A.; Fateev, I.V.; Kostromina, M.A.; Abramchik, Y.A.; Lykoshin, D.D.; Yurovskaya, D.O.; Timofeev, V.I.; Berzina, M.Y.; Eletskaya, B.Z.; Konstantinova, I.D.; et al. Rational mutagenesis in the lid domain of ribokinase from E. coli results in an order of magnitude increase in activity towards D-arabinose. Int. J. Mol. Sci. 2022, 23, 12540. [Google Scholar] [CrossRef] [PubMed]
- Birmingham, W.R.; Starbird, C.A.; Panosian, T.D.; Nannemann, D.P.; Iverson, T.M.; Bachmann, B.O. Bioretrosynthetic construction of a didanosine biosynthetic pathway. Nat. Chem. Biol. 2014, 10, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Bachosz, K.; Zdarta, J.; Bilal, M.; Meyer, A.S.; Jesionowski, T. Enzymatic cofactor regeneration systems: A new perspective on efficiency assessment. Sci. Total Environ. 2023, 868, 161630. [Google Scholar] [CrossRef]
- Yehia, H.; Kamel, S.; Paulick, K.; Neubauer, P.; Wagner, A. Substrate spectra of nucleoside phosphorylases and their potential in the production of pharmaceutically active compounds. Curr. Pharm. Des. 2017, 23, 6913–6935. [Google Scholar] [CrossRef]
- Joshi, J.G.; Handler, P. Phosphoglucomutase. I. Purification and properties of phosphoglucomutase from Escherichia Coli. J. Biol. Chem. 1964, 239, 2741–2751. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Ko, T.P.; Chen, W.H.; Lo, L.P.; Lin, C.H.; Wang, A.H.J. Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism. J. Struct. Biol. 2010, 169, 25–35. [Google Scholar] [CrossRef]
- Panosian, T.D.; Nannemann, D.P.; Watkins, G.R.; Phelan, V.V.; McDonald, W.H.; Wadzinski, B.E.; Bachmann, B.O.; Iverson, T.M. Bacillus cereus phosphopentomutase is an alkaline phosphatase family member that exhibits an altered entry point into the catalytic cycle. J. Biol. Chem. 2011, 286, 8043–8054. [Google Scholar] [CrossRef]
- Huffman, M.A.; Fryszkowska, A.; Alvizo, O.; Borra-Garske, M.; Campos, K.R.; Canada, K.A.; Devine, P.N.; Duan, D.; Forstater, J.H.; Grosser, S.T.; et al. Design of an invitro biocatalytic cascade for the manufacture of islatravir. Science 2019, 366, 1255–1259. [Google Scholar] [CrossRef]
- Iglesias, L.E.; Lewkowicz, E.S.; Medici, R.; Bianchi, P.; Iribarren, A.M. Biocatalytic approaches applied to the synthesis of nucleoside prodrugs. Biotechnol. Adv. 2015, 33, 412–434. [Google Scholar] [CrossRef]
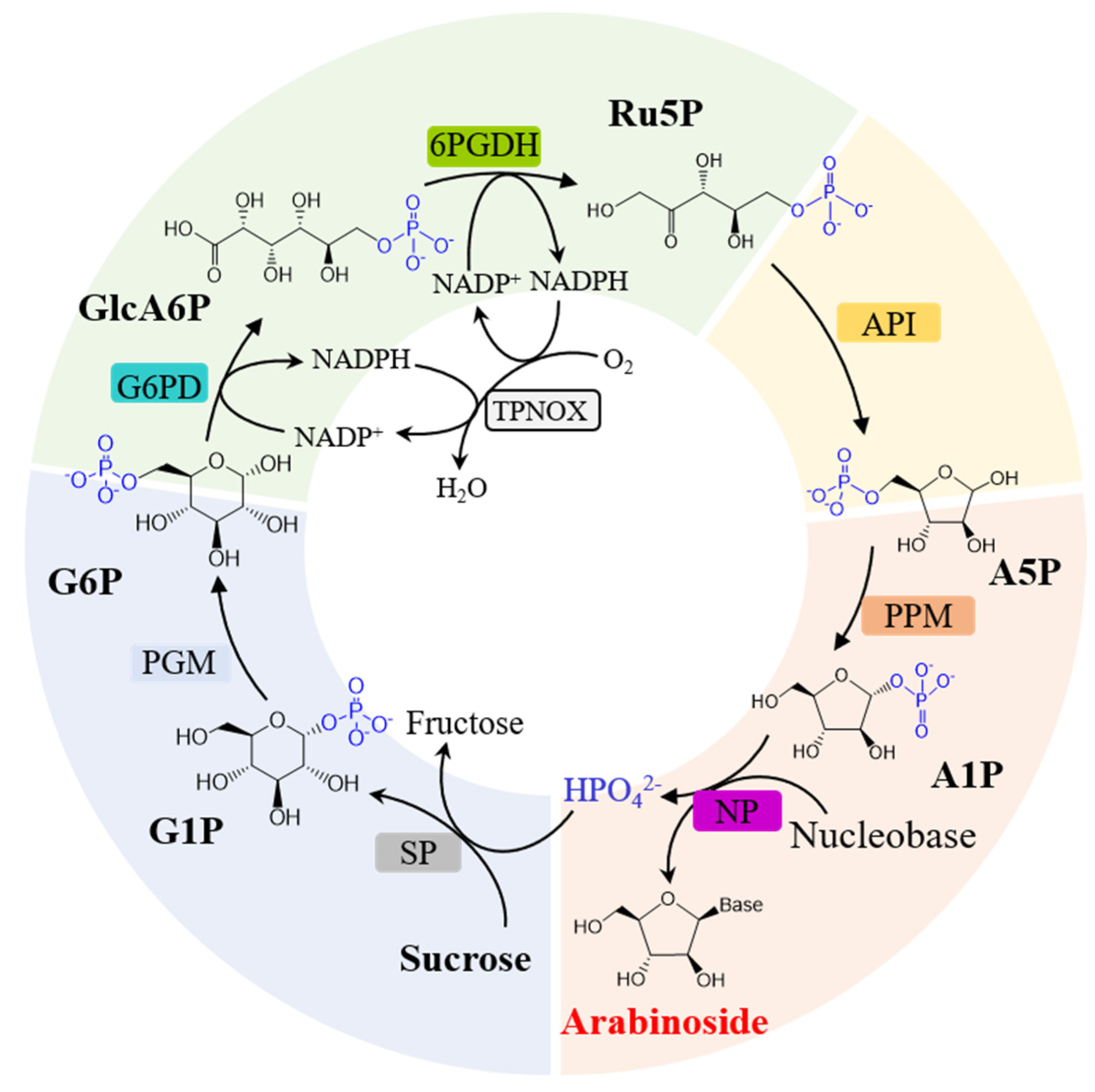
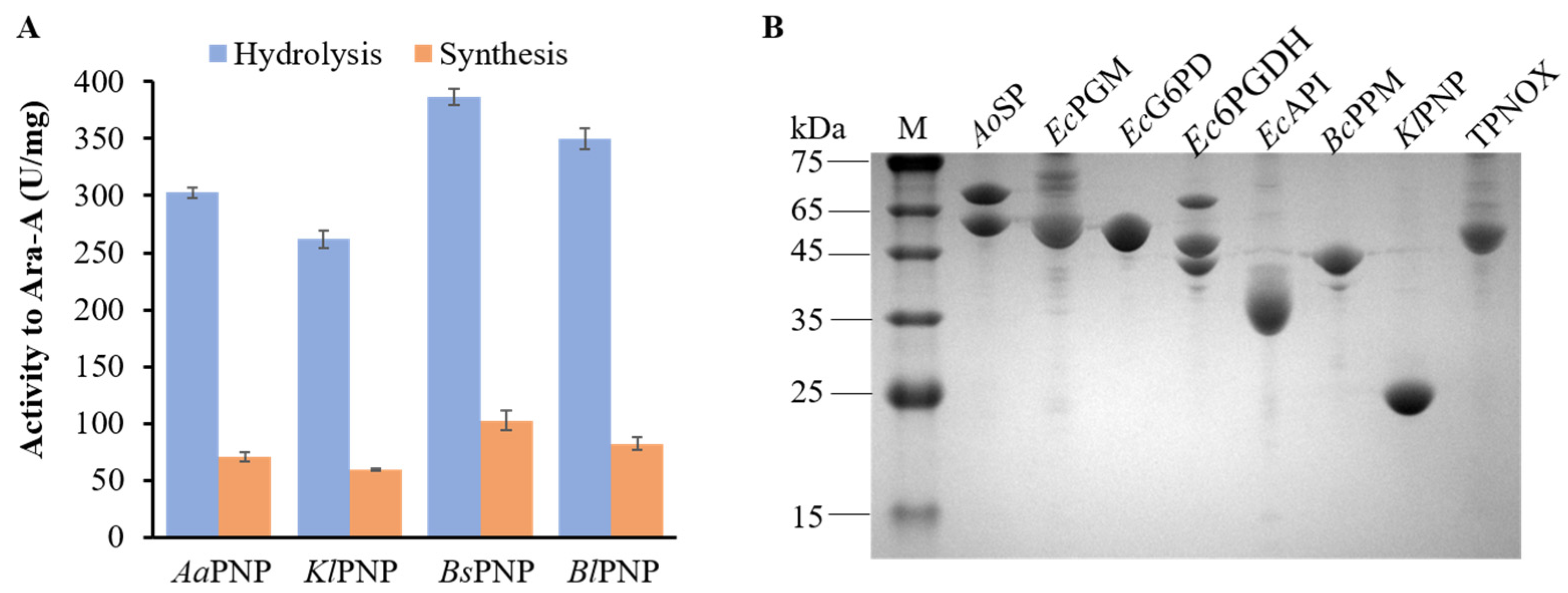
| Name | Enzyme | Source | Substrate Spectra |
|---|---|---|---|
| KlPNP | Purine nucleoside phosphorylase | Klebsiella | Ara-A, Ara-G, and Ara-I |
| AaPNP | Purine nucleoside phosphorylase | Alicyclobacillus acidoterrestris | Ara-A, Ara-G, and Ara-I |
| BsPNP | Purine nucleoside phosphorylase | Bacillus subtilis | Ara-A, Ara-G, and Ara-I |
| BlPNP | Purine nucleoside phosphorylase | Bacillus licheniformis | Ara-A, Ara-G, and Ara-I |
| EcPNP | Purine nucleoside phosphorylase | E. coli | Ara-U |
| EcUP | Uridine phosphorylase | E. coli | Ara-U |
| EcXP | Purine nucleoside phosphorylase 2 | E. coli | Ara-G |
| EcTP | Thymidine phosphorylase | E. coli | Ara-U |
| Name | Enzyme | Source | Conditions Required for Activity | Reference |
|---|---|---|---|---|
| EcPGM | Phosphoglucomutase | E. coli | pH 8.0, Mg2+ | [26] |
| EcG6PD | Glucose-6-phosphate 1-dehydrogenase | E. coli | / | |
| Ec6PGDH | 6-phosphogluconate dehydrogenase | E. coli | pH 8.0, NADP+, Mg2+ | [27] |
| EcAPI | D-arabinose 5-phosphate isomerase | E. coli | pH 8.5; Zn2+ inhibited | [21] |
| BcPPM | Phosphopentomutase | Bacillus cereus | pH 8.0, glucose 1,6-bisphosphate dependent, Mn2+ | [28] |
| TPNOX | NADPH oxidase | Lactobacillus brevis | pH 7.5, NADPH | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Yang, E.; Zhang, X.; Liu, X.; Tang, X.; Wang, Z.; Wang, H. Biosynthesis of Arabinoside from Sucrose and Nucleobase via a Novel Multi-Enzymatic Cascade. Biomolecules 2024, 14, 1107. https://doi.org/10.3390/biom14091107
Liu Y, Yang E, Zhang X, Liu X, Tang X, Wang Z, Wang H. Biosynthesis of Arabinoside from Sucrose and Nucleobase via a Novel Multi-Enzymatic Cascade. Biomolecules. 2024; 14(9):1107. https://doi.org/10.3390/biom14091107
Chicago/Turabian StyleLiu, Yuxue, Erchu Yang, Xiaojing Zhang, Xiaobei Liu, Xiaoting Tang, Zhenyu Wang, and Hailei Wang. 2024. "Biosynthesis of Arabinoside from Sucrose and Nucleobase via a Novel Multi-Enzymatic Cascade" Biomolecules 14, no. 9: 1107. https://doi.org/10.3390/biom14091107
APA StyleLiu, Y., Yang, E., Zhang, X., Liu, X., Tang, X., Wang, Z., & Wang, H. (2024). Biosynthesis of Arabinoside from Sucrose and Nucleobase via a Novel Multi-Enzymatic Cascade. Biomolecules, 14(9), 1107. https://doi.org/10.3390/biom14091107






