The Emerging Role of Ubiquitin-Specific Protease 36 (USP36) in Cancer and Beyond
Abstract
1. Introduction
2. The Structure of USP36
3. Biological Function of USP36
3.1. Classical Deubiquitination Activity of USP36
3.2. The Functions of USP36 in Nucleolar Protein SUMOylation
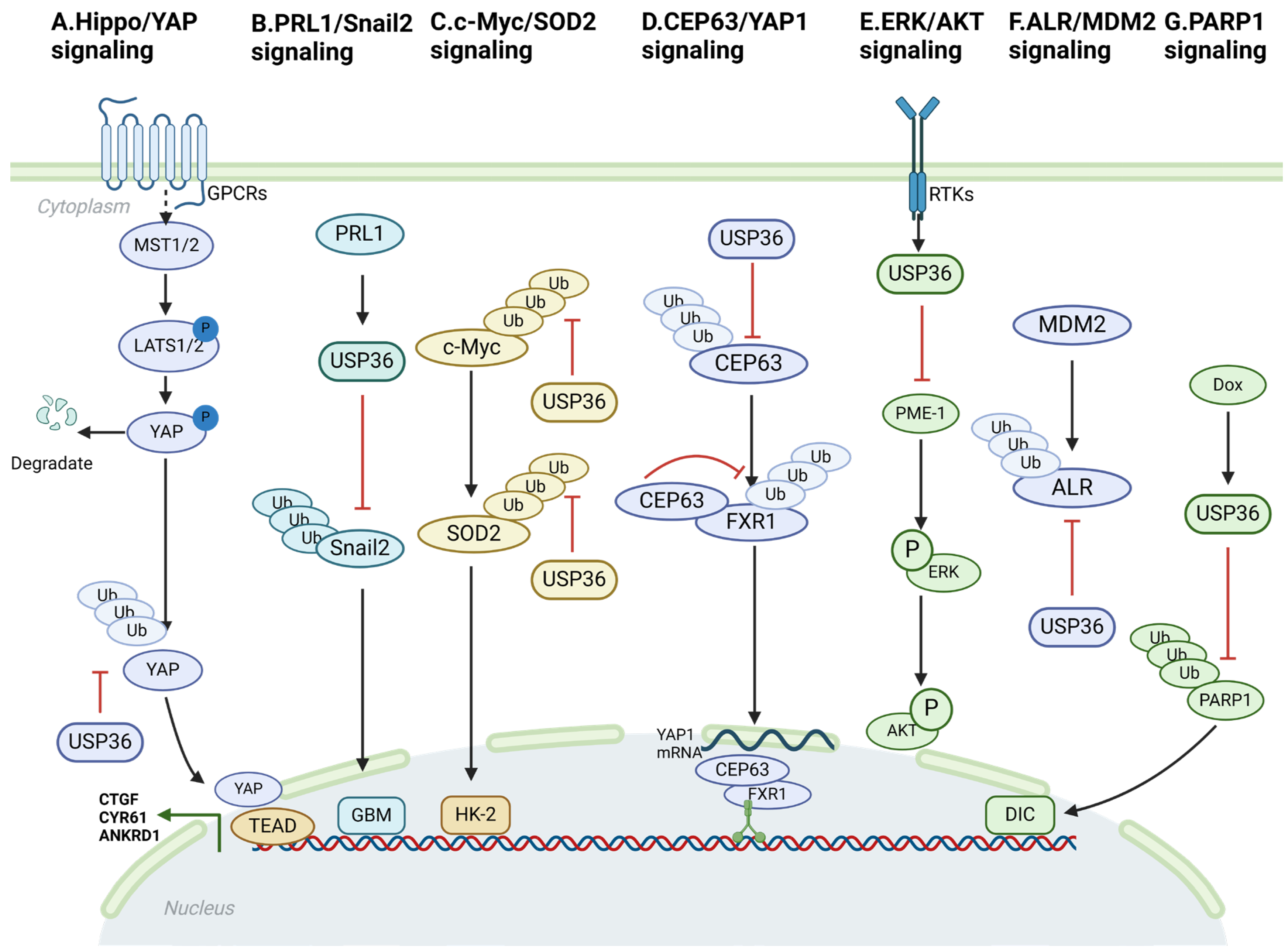
4. USP36-Mediated Signaling Pathway
4.1. Hippo/YAP Signaling
4.2. PRL1/Snail2 Signaling
4.3. c-Myc/SOD2 and c-Myc/Fbw7g Signaling
4.4. CEP63/YAP1 Signaling
4.5. ERK/AKT Signaling
4.6. ALR/MDM2 Signaling
4.7. USP36/PARP1 Signaling
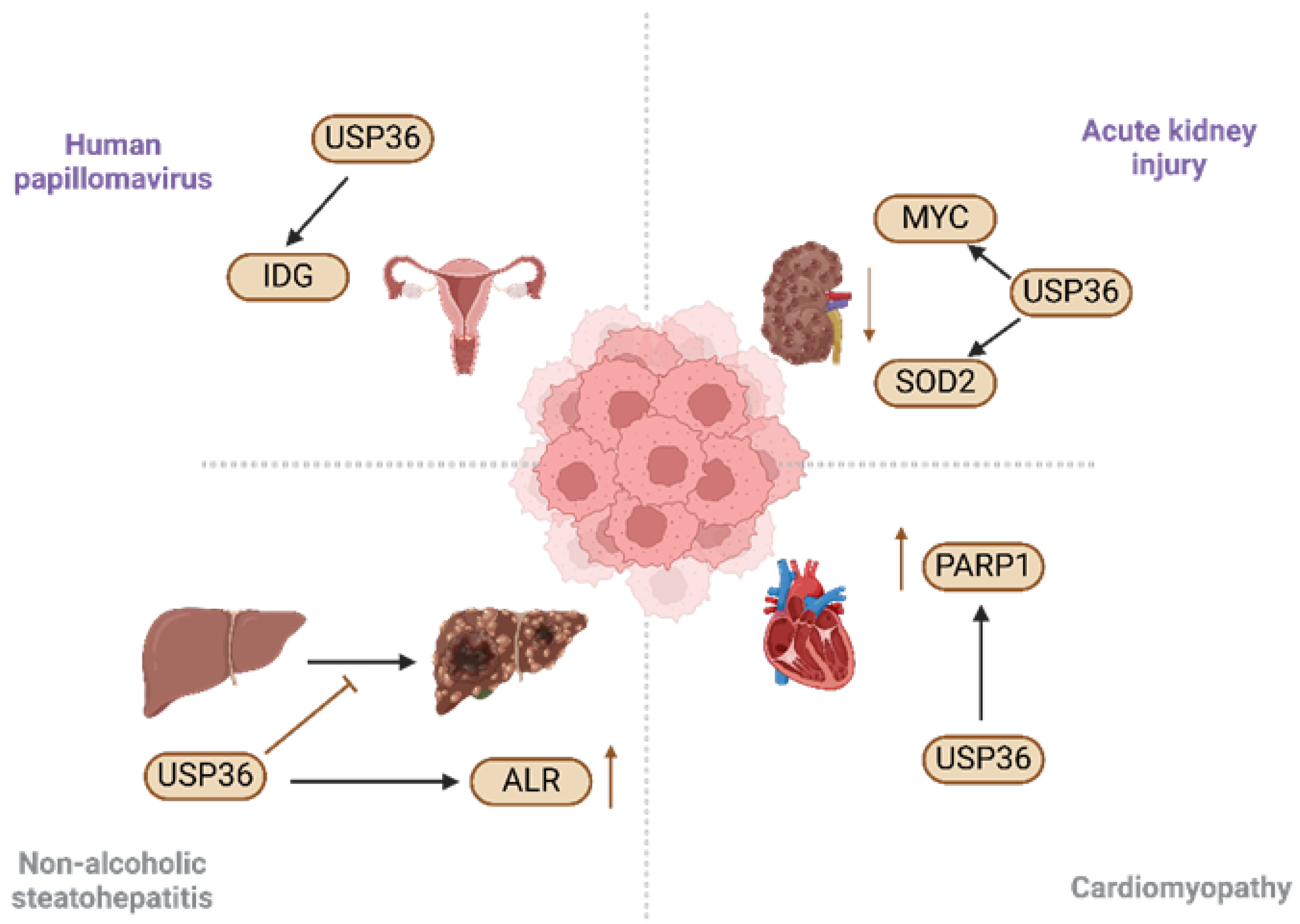
5. Role of USP36 in Diseases
5.1. Acute Kidney Injury
5.2. Non-Alcoholic Steatohepatitis
5.3. Human Papillomavirus
5.4. Cardiomyopathy
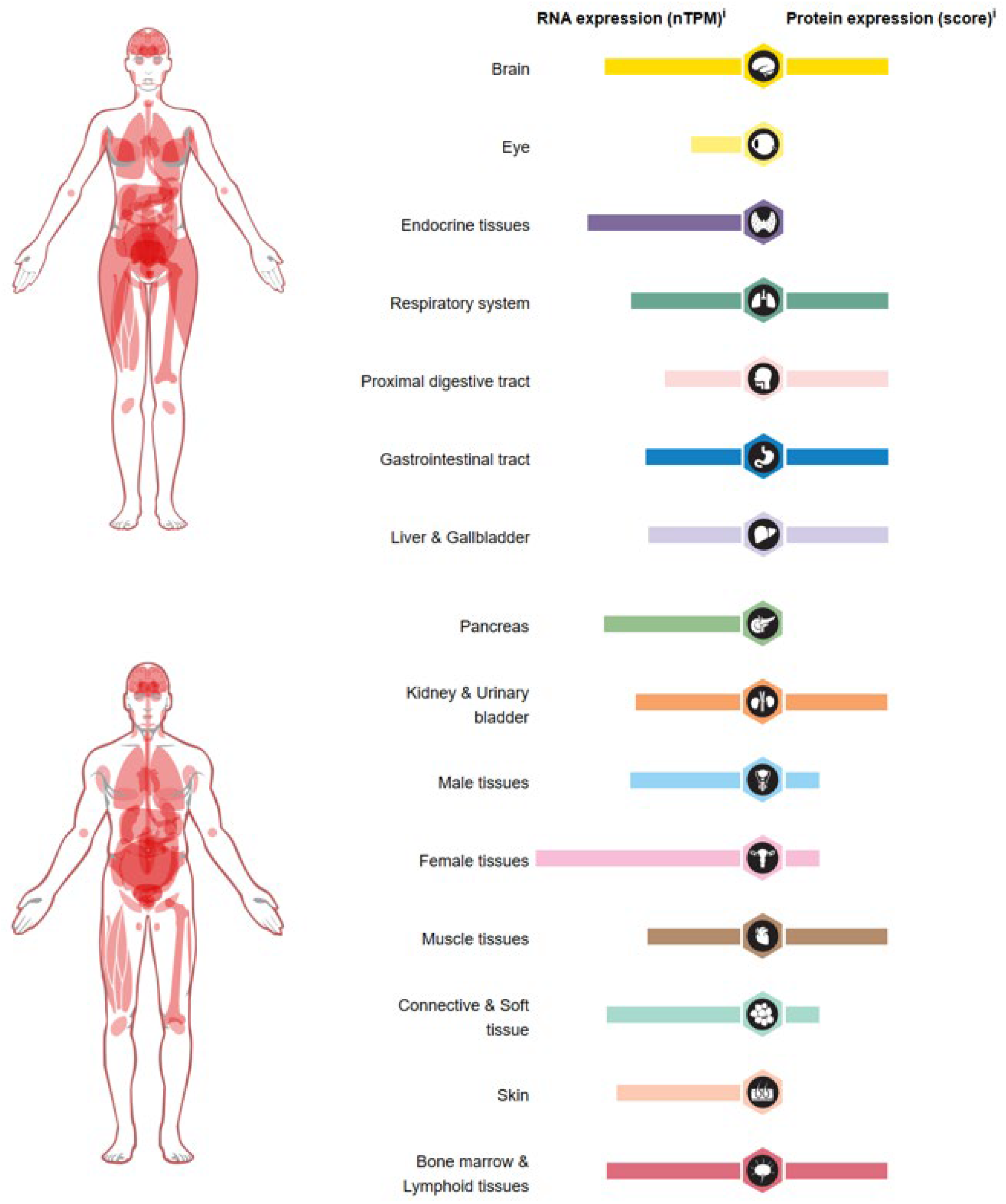
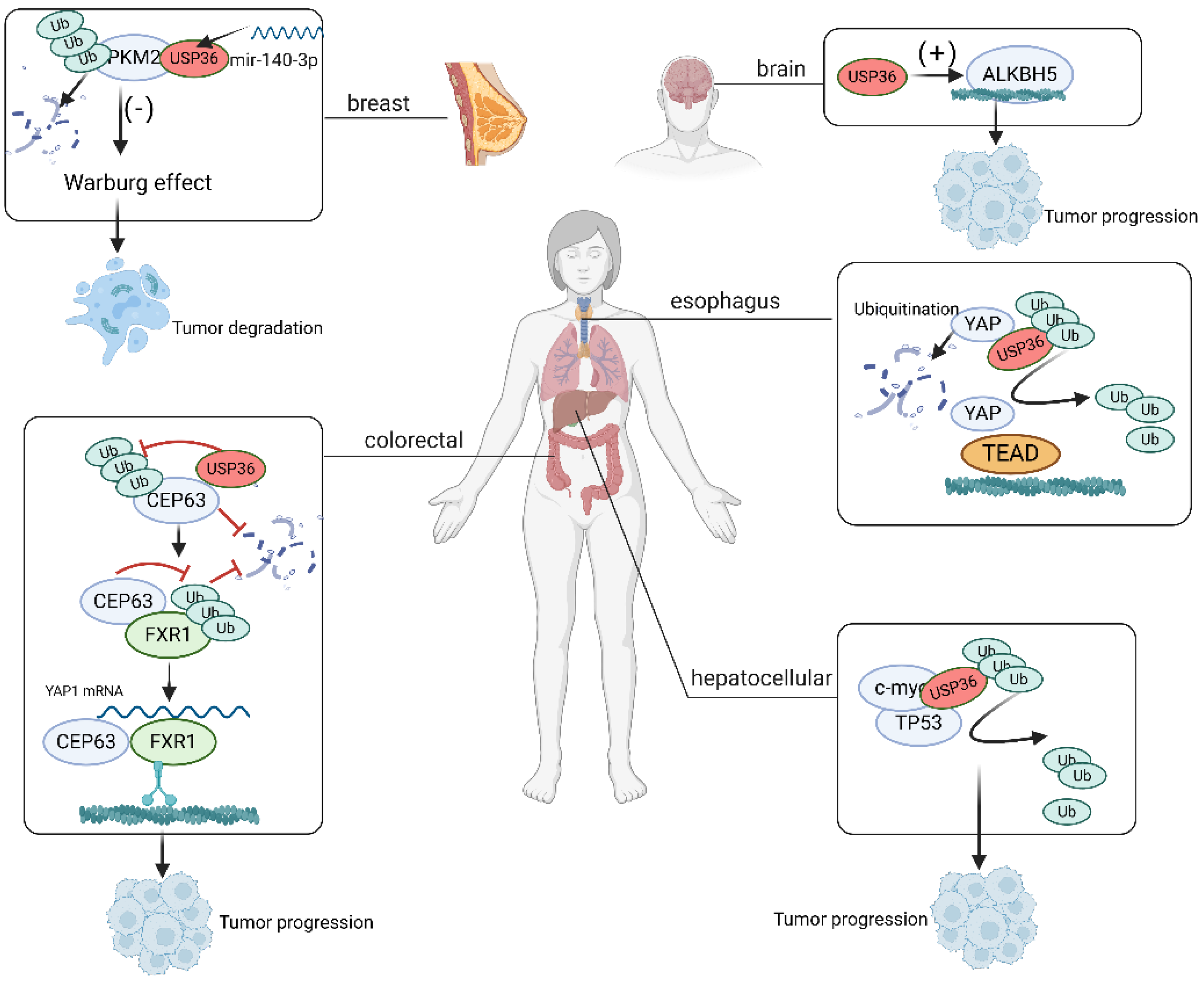
6. Role of USP36 in Cancer
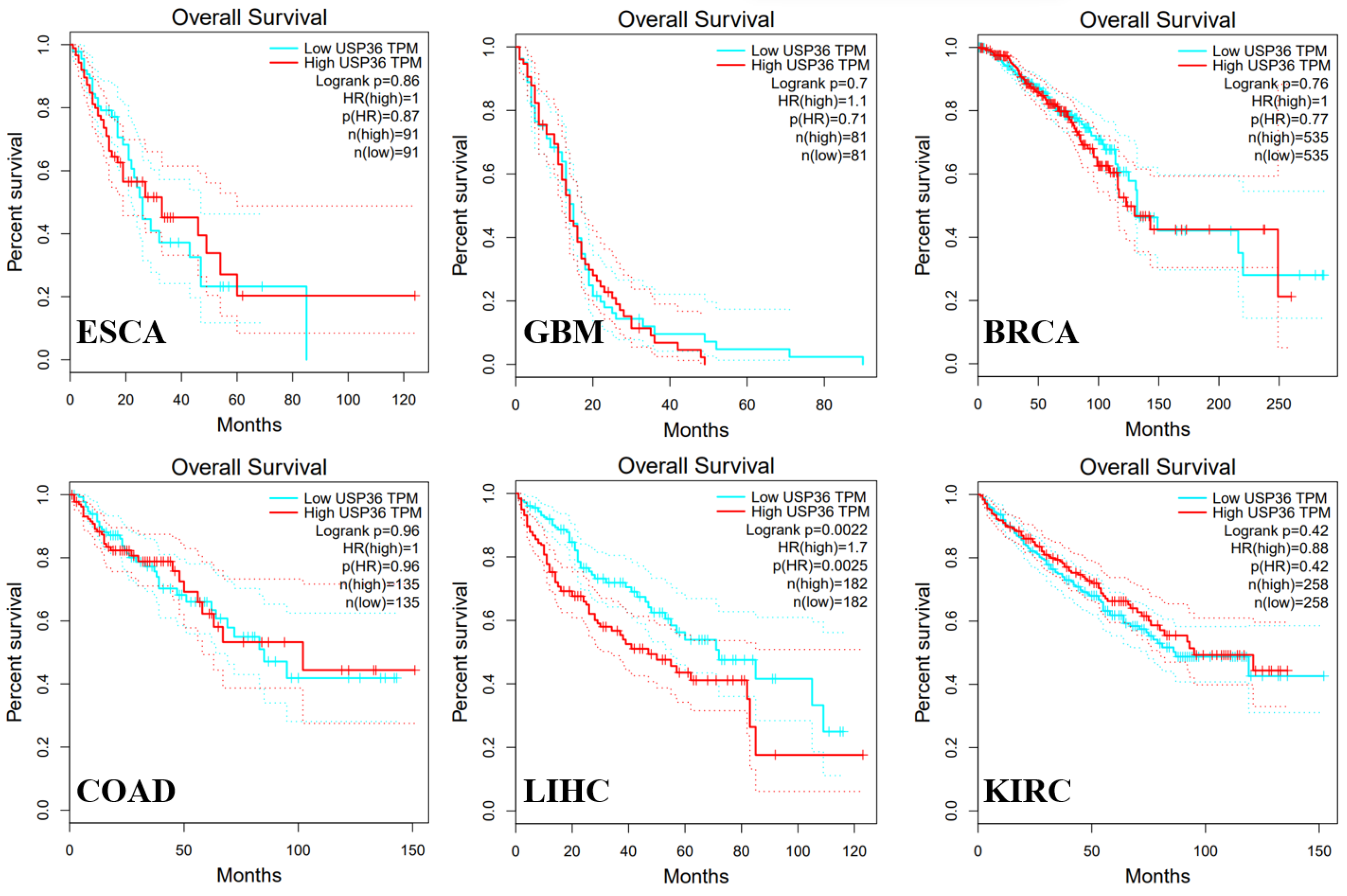
6.1. Esophageal Carcinoma
6.2. Glioblastoma
6.3. Hepatocellular Carcinoma
6.4. Colorectal Cancer
6.5. Breast Cancer
6.6. T Cell Lymphoma
7. USP36 as a Target for Cancer Therapy
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dewson, G.; Eichhorn, P.J.A.; Komander, D. Deubiquitinases in cancer. Nat. Rev. Cancer 2023, 23, 842–862. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.M.; Armstrong, L.A.; Kulathu, Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol. Cell 2022, 82, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Y.; Li, C.Y.; Chen, R.Y.; Shi, J.J.; Liu, Y.J.; Lu, J.F.; Yang, G.J.; Chen, J. The emerging role of deubiquitylating enzyme USP21 as a potential therapeutic target in cancer. Bioorg. Chem. 2024, 10, 107400. [Google Scholar] [CrossRef]
- Ashton-Beaucage, D.; Lemieux, C.; Udell, C.M.; Sahmi, M.; Rochette, S.; Therrien, M. The Deubiquitinase USP47 Stabilizes MAPK by Counteracting the Function of the N-end Rule ligase POE/UBR4 in Drosophila. PLoS Biol. 2016, 14, e1002539. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.A.; Kumar, R.; Penzes, P.; Piper, M.; Gecz, J. The DUB Club: Deubiquitinating Enzymes and Neurodevelopmental Disorders. Biol. Psychiatry 2022, 92, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Kitamura, N.; Komada, M. Nucleophosmin/B23 regulates ubiquitin dynamics in nucleoli by recruiting deubiquitylating enzyme USP36. J. Biol. Chem. 2009, 284, 27918–27923. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Matsumoto, M.; Inada, T.; Yamamoto, A.; Nakayama, K.I.; Kitamura, N.; Komada, M. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J. Cell Sci. 2009, 122, 678–686. [Google Scholar] [CrossRef]
- Taillebourg, E.; Gregoire, I.; Viargues, P.; Jacomin, A.-C.; Thevenon, D.; Faure, M.; Fauvarque, M.-O. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy 2012, 8, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-M.; Zhang, Y.Z.; Ramakrishna, S.; Lim, C.T. Electrospinning and mechanical characterization of gelatin nanofibers. Polymer 2004, 45, 5361–5368. [Google Scholar] [CrossRef]
- Richardson, L.A.; Reed, B.J.; Charette, J.M.; Freed, E.F.; Fredrickson, E.K.; Locke, M.N.; Baserga, S.J.; Gardner, R.G. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep. 2012, 2, 372–385. [Google Scholar] [CrossRef]
- Peltonen, K.; Colis, L.; Liu, H.; Trivedi, R.; Moubarek, M.S.; Moore, H.M.; Bai, B.; Rudek, M.A.; Bieberich, C.J.; Laiho, M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 2014, 25, 77–90. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Sun, X.X.; Chen, Y.; Li, Y.; Wang, X.; Dai, R.S.; Zhu, H.M.; Klimek, J.; David, L.; Fedorov, L.M.; et al. The deubiquitinase USP36 promotes snoRNP group SUMOylation and is essential for ribosome biogenesis. EMBO Rep. 2021, 22, e50684. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; Vertegaal, A.C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595. [Google Scholar] [CrossRef]
- Panse, V.G.; Kressler, D.; Pauli, A.; Petfalski, E.; Gnädig, M.; Tollervey, D.; Hurt, E. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 2006, 7, 1311–1321. [Google Scholar] [CrossRef]
- Finkbeiner, E.; Haindl, M.; Muller, S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. Embo J 2011, 30, 1067–1078. [Google Scholar] [CrossRef]
- Raman, N.; Weir, E.; Müller, S. The AAA ATPase MDN1 Acts as a SUMO-Targeted Regulator in Mammalian Pre-ribosome Remodeling. Mol. Cell 2016, 64, 607–615. [Google Scholar] [CrossRef]
- Filippopoulou, C.; Thomé, C.C.; Perdikari, S.; Ntini, E.; Simos, G.; Bohnsack, K.E.; Chachami, G. Hypoxia-driven deSUMOylation of EXOSC10 promotes adaptive changes in the transcriptome profile. Cell Mol. Life Sci. 2024, 81, 58. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Dai, R.S.; Savage, J.C.; Shinde, U.; Klimek, J.; David, L.L.; Young, E.A.; Hafner, M.; Sears, R.C.; et al. The ubiquitin-specific protease USP36 SUMOylates EXOSC10 and promotes the nucleolar RNA exosome function in rRNA processing. Nucleic Acids Res. 2023, 51, 3934–3949. [Google Scholar] [CrossRef]
- Li, Y.; Carey, T.S.; Feng, C.H.; Zhu, H.-M.; Sun, X.-X.; Dai, M.-S. The Ubiquitin-specific Protease USP36 Associates with the Microprocessor Complex and Regulates miRNA Biogenesis by SUMOylating DGCR8. Cancer Res. Commun. 2023, 3, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Moroishi, T.; Mottier-Pavie, V.; Plouffe, S.W.; Hansen, C.G.; Hong, A.W.; Park, H.W.; Mo, J.S.; Lu, W.; Lu, S.; et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015, 6, 8357. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Varelas, X.; Guan, K.L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes. Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Park, H.W.; Kim, Y.C.; Yu, B.; Moroishi, T.; Mo, J.S.; Plouffe, S.W.; Meng, Z.; Lin, K.C.; Yu, F.X.; Alexander, C.M.; et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell 2015, 162, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Choi, S.S.; Michelotti, G.A.; Chan, I.S.; Swiderska-Syn, M.; Karaca, G.F.; Xie, G.; Moylan, C.A.; Garibaldi, F.; Premont, R.; et al. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology 2012, 143, 1319–1329.e1311. [Google Scholar] [CrossRef]
- Plouffe, S.W.; Hong, A.W.; Guan, K.L. Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 2015, 21, 212–222. [Google Scholar] [CrossRef]
- Zhou, A.; Yu, H.; Liu, J.; Zheng, J.; Jia, Y.; Wu, B.; Xiang, L. Role of Hippo-YAP Signaling in Osseointegration by Regulating Osteogenesis, Angiogenesis, and Osteoimmunology. Front. Cell Dev. Biol. 2020, 8, 780. [Google Scholar] [CrossRef]
- Zhang, W.; Luo, J.; Xiao, Z.; Zang, Y.; Li, X.; Zhou, Y.; Zhou, J.; Tian, Z.; Zhu, J.; Zhao, X. USP36 facilitates esophageal squamous carcinoma progression via stabilizing YAP. Cell Death Dis. 2022, 13, 1021. [Google Scholar] [CrossRef]
- Bai, Y.; Zhou, H.-M.; Zhang, L.; Dong, Y.; Zeng, Q.; Shou, W.; Zhang, Z.-Y. Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis. Sci. Rep. 2016, 6, 34211. [Google Scholar] [CrossRef]
- Bai, Y.; Luo, Y.; Liu, S.; Zhang, L.; Shen, K.; Dong, Y.; Walls, C.D.; Quilliam, L.A.; Wells, C.D.; Cao, Y.; et al. PRL-1 Protein Promotes ERK1/2 and RhoA Protein Activation through a Non-canonical Interaction with the Src Homology 3 Domain of p115 Rho GTPase-activating Protein*. J. Biol. Chem. 2011, 286, 42316–42324. [Google Scholar] [CrossRef]
- Assani, G.; Zhou, Y. Effect of modulation of epithelial-mesenchymal transition regulators Snail1 and Snail2 on cancer cell radiosensitivity by targeting of the cell cycle, cell apoptosis and cell migration/invasion. Oncol. Lett. 2019, 17, 23–30. [Google Scholar] [CrossRef]
- Meng, J.; Ai, X.; Lei, Y.; Zhong, W.; Qian, B.; Qiao, K.; Wang, X.; Zhou, B.; Wang, H.; Huai, L.; et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics 2019, 9, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Min, L.; Liu, C.; Chen, S.; Gao, C.; Ma, J.; Wu, X.; Li, W.; Wu, L.; Zhu, L. RNF8 Promotes Epithelial-Mesenchymal Transition in Lung Cancer Cells via Stabilization of Slug. Mol. Cancer Res. 2020, 18, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Cai, X.; Xu, K.; Song, S.; Xiao, Z.; Hou, Y.; Qi, X.; Liu, F.; Chen, Y.; Yang, H.; et al. PRL1 Promotes Glioblastoma Invasion and Tumorigenesis via Activating USP36-Mediated Snail2 Deubiquitination. Front. Oncol. 2021, 11, 795633. [Google Scholar] [CrossRef] [PubMed]
- van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Blackwood, E.M.; Eisenman, R.N. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 1991, 251, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.Y.; Watson, J.D.; Yan, P.S.; Barsyte-Lovejoy, D.; Khosravi, F.; Wong, W.W.; Farnham, P.J.; Huang, T.H.; Penn, L.Z. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003, 13, 882–886. [Google Scholar] [CrossRef]
- Bieda, M.; Xu, X.; Singer, M.A.; Green, R.; Farnham, P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006, 16, 595–605. [Google Scholar] [CrossRef]
- Liu, Q.; Sheng, W.; Ma, Y.; Zhen, J.; Roy, S.; Alvira Jafar, C.; Xin, W.; Wan, Q. USP36 protects proximal tubule cells from ischemic injury by stabilizing c-Myc and SOD2. Biochem. Biophys. Res. Commun. 2019, 513, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Welcker, M.; Clurman, B.E. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 2008, 8, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Welcker, M.; Orian, A.; Grim, J.E.; Eisenman, R.N.; Clurman, B.E. A Nucleolar Isoform of the Fbw7 Ubiquitin Ligase Regulates c-Myc and Cell Size. Curr. Biol. 2004, 14, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin. Cancer Biol. 2006, 16, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Lutterbach, B.; Hann, S.R. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol. Cell Biol. 1994, 14, 5510–5522. [Google Scholar] [CrossRef]
- Sears, R.; Nuckolls, F.; Haura, E.; Taya, Y.; Tamai, K.; Nevins, J.R. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes. Dev. 2000, 14, 2501–2514. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-X.; Sears, R.C.; Dai, M.-S. Deubiquitinating c-Myc: USP36 steps up in the nucleolus. Cell Cycle 2015, 14, 3786–3793. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.X.; He, X.; Yin, L.; Komada, M.; Sears, R.C.; Dai, M.S. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc. Natl. Acad. Sci. USA 2015, 112, 3734–3739. [Google Scholar] [CrossRef]
- Fraile, J.M.; Campos-Iglesias, D.; Rodríguez, F.; Astudillo, A.; Vilarrasa-Blasi, R.; Verdaguer-Dot, N.; Prado, M.A.; Paulo, J.A.; Gygi, S.P.; Martín-Subero, J.I.; et al. Loss of the deubiquitinase USP36 destabilizes the RNA helicase DHX33 and causes preimplantation lethality in mice. J. Biol. Chem. 2018, 293, 2183–2194. [Google Scholar] [CrossRef]
- Thevenon, D.; Engel, E.; Avet-Rochex, A.; Gottar, M.; Bergeret, E.; Tricoire, H.; Benaud, C.; Baudier, J.; Taillebourg, E.; Fauvarque, M.-O. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe 2009, 64, 309–320. [Google Scholar] [CrossRef]
- Löffler, H.; Fechter, A.; Matuszewska, M.; Saffrich, R.; Mistrik, M.; Marhold, J.; Hornung, C.; Westermann, F.; Bartek, J.; Krämer, A. Cep63 recruits Cdk1 to the centrosome: Implications for regulation of mitotic entry, centrosome amplification, and genome maintenance. Cancer Res. 2011, 71, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Sir, J.H.; Barr, A.R.; Nicholas, A.K.; Carvalho, O.P.; Khurshid, M.; Sossick, A.; Reichelt, S.; D’Santos, C.; Woods, C.G.; Gergely, F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 2011, 43, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, D.; Kodani, A.; Gonzalez, D.M.; Mancias, J.D.; Mochida, G.H.; Vagnoni, C.; Johnson, J.; Krogan, N.; Harper, J.W.; Reiter, J.F.; et al. Microcephaly Proteins Wdr62 and Aspm Define a Mother Centriole Complex Regulating Centriole Biogenesis, Apical Complex, and Cell Fate. Neuron 2016, 92, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Madiraju, C.; Novack, J.P.; Reed, J.C.; Matsuzawa, S.I. K63 ubiquitination in immune signaling. Trends Immunol. 2022, 43, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Perrotti, D.; Neviani, P. Protein phosphatase 2A: A target for anticancer therapy. Lancet Oncol. 2013, 14, e229–e238. [Google Scholar] [CrossRef] [PubMed]
- Arriazu, E.; Pippa, R.; Odero, M.D. Protein Phosphatase 2A as a Therapeutic Target in Acute Myeloid Leukemia. Front. Oncol. 2016, 6, 78. [Google Scholar] [CrossRef]
- Kim, S.Y.; Choi, J.; Lee, D.H.; Park, J.H.; Hwang, Y.J.; Baek, K.H. PME-1 is regulated by USP36 in ERK and Akt signaling pathways. FEBS Lett. 2018, 592, 1575–1588. [Google Scholar] [CrossRef]
- Zheng, M.; Ai, Z.; Guo, Y.; Chen, Y.; Xie, P.; An, W. Imbalance in ALR ubiquitination accelerates the progression of nonalcoholic steatohepatitis to hepatocellular carcinoma. Oncogene 2023, 42, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Tewey, K.M.; Rowe, T.C.; Yang, L.; Halligan, B.D.; Liu, L.F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 1984, 226, 466–468. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Bawa-Khalfe, T.; Lu, L.S.; Lyu, Y.L.; Liu, L.F.; Yeh, E.T. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012, 18, 1639–1642. [Google Scholar] [CrossRef]
- Wu, L.; Wang, L.; Du, Y.; Zhang, Y.; Ren, J. Mitochondrial quality control mechanisms as therapeutic targets in doxorubicin-induced cardiotoxicity. Trends Pharmacol. Sci. 2023, 44, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Jiang, Z.; Kan, J.; Jiang, X.; Pan, C.; You, S.; Chang, R.; Zhang, J.; Yang, H.; Zhu, L.; et al. USP36-mediated PARP1 deubiquitination in doxorubicin-induced cardiomyopathy. Cell Signal 2024, 117, 111070. [Google Scholar] [CrossRef] [PubMed]
- Iden, M.; Tsaih, S.W.; Huang, Y.W.; Liu, P.; Xiao, M.; Flister, M.J.; Rader, J.S. Multi-omics mapping of human papillomavirus integration sites illuminates novel cervical cancer target genes. Br. J. Cancer 2021, 125, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Ympa, Y.P.; Sakr, Y.; Reinhart, K.; Vincent, J.L. Has mortality from acute renal failure decreased? A systematic review of the literature. Am. J. Med. 2005, 118, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Chawla, L.S.; Amdur, R.L.; Amodeo, S.; Kimmel, P.L.; Palant, C.E. The severity of acute kidney injury predicts progression to chronic kidney disease. Kidney Int. 2011, 79, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Ronco, C.; Bellomo, R.; Kellum, J.A. Acute kidney injury. Lancet 2019, 394, 1949–1964. [Google Scholar] [CrossRef] [PubMed]
- Hultström, M.; Becirovic-Agic, M.; Jönsson, S. Comparison of acute kidney injury of different etiology reveals in-common mechanisms of tissue damage. Physiol. Genom. 2018, 50, 127–141. [Google Scholar] [CrossRef]
- Baffy, G.; Brunt, E.M.; Caldwell, S.H. Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J. Hepatol. 2012, 56, 1384–1391. [Google Scholar] [CrossRef]
- LaBrecque, D.R.; Pesch, L.A. Preparation and partial characterization of hepatic regenerative stimulator substance (SS) from rat liver. J. Physiol. 1975, 248, 273–284. [Google Scholar] [CrossRef]
- Lane, D.P.; Hall, P.A. MDM2--arbiter of p53’s destruction. Trends Biochem. Sci. 1997, 22, 372–374. [Google Scholar] [CrossRef]
- Pett, M.; Coleman, N. Integration of high-risk human papillomavirus: A key event in cervical carcinogenesis? J. Pathol. 2007, 212, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shen, J.; Liu, J.; Han, K.; Liang, L.; Gao, Y. Gene Signature and Prognostic Value of Ubiquitin-Specific Proteases Members in Hepatocellular Carcinoma and Explored the Immunological Role of USP36. Front. Biosci. 2022, 27, 190. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Cao, C.H.; Han, K.; Lv, Y.R.; Ma, X.D.; Cao, J.H.; Chen, J.W.; Li, S.; Lin, J.L.; Fang, Y.J.; et al. CEP63 upregulates YAP1 to promote colorectal cancer progression through stabilizing RNA binding protein FXR1. Oncogene 2022, 41, 4433–4445. [Google Scholar] [CrossRef]
- Wu, H.; Jiao, Y.; Zhou, C.; Guo, X.; Wu, Z.; Lv, Q. miR-140-3p/usp36 axis mediates ubiquitination to regulate PKM2 and suppressed the malignant biological behavior of breast cancer through Warburg effect. Cell Cycle 2023, 22, 680–692. [Google Scholar] [CrossRef]
- Li, B.; Yan, J.; Phyu, T.; Fan, S.; Chung, T.H.; Mustafa, N.; Lin, B.; Wang, L.; Eichhorn, P.J.A.; Goh, B.C.; et al. MELK mediates the stability of EZH2 through site-specific phosphorylation in extranodal natural killer/T-cell lymphoma. Blood 2019, 134, 2046–2058. [Google Scholar] [CrossRef]
- Chang, G.; Xie, G.S.; Ma, L.; Li, P.; Li, L.; Richard, H.T. USP36 promotes tumorigenesis and drug sensitivity of glioblastoma by deubiquitinating and stabilizing ALKBH5. Neuro Oncol. 2023, 25, 841–853. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chen, W.; Zheng, R.; Zeng, H.; Zhang, S.; He, J. Annual report on status of cancer in China, 2011. Chin. J. Cancer Res. 2015, 27, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.D.; Liao, X.Y.; Chen, Y.B.; Huang, S.Y.; Xue, W.Q.; Li, F.F.; Ge, X.S.; Liu, D.Q.; Cai, Q.; Long, J.; et al. Genomic Characterization of Esophageal Squamous Cell Carcinoma Reveals Critical Genes Underlying Tumorigenesis and Poor Prognosis. Am. J. Hum. Genet. 2016, 98, 709–727. [Google Scholar] [CrossRef]
- Gao, Y.B.; Chen, Z.L.; Li, J.G.; Hu, X.D.; Shi, X.J.; Sun, Z.M.; Zhang, F.; Zhao, Z.R.; Li, Z.T.; Liu, Z.Y.; et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 1097–1102. [Google Scholar] [CrossRef]
- Lo Sardo, F.; Canu, V.; Maugeri-Saccà, M.; Strano, S.; Blandino, G. YAP and TAZ: Monocorial and bicorial transcriptional co-activators in human cancers. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188756. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Wang, W.; Wang, S.; Hou, J.; Zhang, A.; Lv, B.; Gao, C.; Yan, Z.; Pang, D.; et al. Regulation of Hippo/YAP signaling and Esophageal Squamous Carcinoma progression by an E3 ubiquitin ligase PARK2. Theranostics 2020, 10, 9443–9457. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, H.W.; Wang, S.; Fan, L.; Feng, S.; Cai, X.; Peng, C.; Wu, X.; Lu, J.; Chen, D.; et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J. Clin. Invest. 2019, 129, 2043–2055. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Smith-Cohn, M.; Cohen, A.L.; Colman, H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics 2017, 14, 284–297. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Patel, N.P.; Lyon, K.A.; Huang, J.H. The effect of race on the prognosis of the glioblastoma patient: A brief review. Neurol. Res. 2019, 41, 967–971. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e596. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Cho, J.; Song, E.J. Ubiquitin-proteasome system (UPS) as a target for anticancer treatment. Arch. Pharm. Res. 2020, 43, 1144–1161. [Google Scholar] [CrossRef]
- Shachaf, C.M.; Kopelman, A.M.; Arvanitis, C.; Karlsson, A.; Beer, S.; Mandl, S.; Bachmann, M.H.; Borowsky, A.D.; Ruebner, B.; Cardiff, R.D.; et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004, 431, 1112–1117. [Google Scholar] [CrossRef]
- Gao, Q.; Zhu, H.; Dong, L.; Shi, W.; Chen, R.; Song, Z.; Huang, C.; Li, J.; Dong, X.; Zhou, Y.; et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019, 179, 561–577.e522. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Cao, J.; Lei, X.; Shi, Y.; Wu, L. Multi-omics data identified TP53 and LRP1B as key regulatory gene related to immune phenotypes via EPCAM in HCC. Cancer Med. 2022, 11, 2145–2158. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Shan, Q.; Zhan, Q.; Ye, Q.; Liu, P.; Xu, S.; He, X.; Ma, J.; Xiang, J.; Jiang, G.; et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut 2020, 69, 1322–1334. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, F.; Ma, R.; Zhang, L.; Du, G.; Niu, D.; Yin, D. Cep63 knockout inhibits the malignant phenotypes of papillary thyroid cancer cell line TPC-1. Oncol. Rep. 2021, 46, 8150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ren, P.; Xu, D.; Liu, X.; Liu, Z.; Zhang, C.; Li, Y.; Wang, L.; Du, X.; Xing, B. Human UTP14a promotes colorectal cancer progression by forming a positive regulation loop with c-Myc. Cancer Lett. 2019, 440–441, 106–115. [Google Scholar] [CrossRef]
- Zhong, X.D.; Chen, L.J.; Xu, X.Y.; Liu, Y.J.; Tao, F.; Zhu, M.H.; Li, C.Y.; Zhao, D.; Yang, G.J.; Chen, J. Berberine as a potential agent for breast cancer therapy. Front. Oncol. 2022, 12, 993775. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, P.; Yarani, R.; Valipour, E.; Kiani, S.; Hoseinkhani, Z.; Mansouri, K. Cell line-directed breast cancer research based on glucose metabolism status. Biomed. Pharmacother. 2022, 146, 112526. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.J.; Tao, F.; Zhong, H.J.; Yang, C.; Chen, J. Targeting PGAM1 in cancer: An emerging therapeutic opportunity. Eur. J. Med. Chem. 2022, 244, 114798. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Biol. 2019, 95, 912–919. [Google Scholar] [CrossRef]
- Yan, J.; Ng, S.B.; Tay, J.L.; Lin, B.; Koh, T.L.; Tan, J.; Selvarajan, V.; Liu, S.C.; Bi, C.; Wang, S.; et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 2013, 121, 4512–4520. [Google Scholar] [CrossRef] [PubMed]
- Adelaiye-Ogala, R.; Budka, J.; Damayanti, N.P.; Arrington, J.; Ferris, M.; Hsu, C.C.; Chintala, S.; Orillion, A.; Miles, K.M.; Shen, L.; et al. EZH2 Modifies Sunitinib Resistance in Renal Cell Carcinoma by Kinome Reprogramming. Cancer Res. 2017, 77, 6651–6666. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, Z.; Cenciarini, M.E.; Proietti, C.J.; Amasino, M.; Hong, T.; Yang, M.; Liao, Y.; Chiang, H.C.; Kaklamani, V.G.; et al. Tamoxifen Resistance in Breast Cancer Is Regulated by the EZH2-ERα-GREB1 Transcriptional Axis. Cancer Res. 2018, 78, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Gardner, E.E.; Lok, B.H.; Schneeberger, V.E.; Desmeules, P.; Miles, L.A.; Arnold, P.K.; Ni, A.; Khodos, I.; de Stanchina, E.; Nguyen, T.; et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017, 31, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Banasavadi-Siddegowda, Y.; Mo, X.; Kim, S.H.; Mao, P.; Kig, C.; Nardini, D.; Sobol, R.W.; Chow, L.M.; Kornblum, H.I.; et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells 2013, 31, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cai, S.; Gu, T.; Song, F.; Xue, Y.; Sun, D. MiR-140-3p Impedes Gastric Cancer Progression and Metastasis by Regulating BCL2/BECN1-Mediated Autophagy. Onco Targets Ther. 2021, 14, 2879–2892. [Google Scholar] [CrossRef]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X.; Zhao, S. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Mol. Oncol. 2021, 15, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Yang, G.J.; Wang, W.; Ma, D.L.; Leung, C.H. Discovery of a tetrahydroisoquinoline-based CDK9-cyclin T1 protein-protein interaction inhibitor as an anti-proliferative and anti-migration agent against triple-negative breast cancer cells. Genes. Dis. 2022, 9, 1674–1688. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.S.; Yang, G.J.; Wang, W.; Leung, C.H.; Ma, D.L. Correction to: The design and development of covalent protein-protein interaction inhibitors for cancer treatment. J. Hematol. Oncol. 2020, 13, 102. [Google Scholar] [CrossRef]
- Jiang, H.; He, H.; Chen, Y.; Huang, W.; Cheng, J.; Ye, J.; Wang, A.; Tao, J.; Wang, C.; Liu, Q.; et al. Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J. Exp. Med. 2017, 214, 3219–3238. [Google Scholar] [CrossRef]
- Leung, C.-H.; Zhang, J.-T.; Yang, G.-J.; Liu, H.; Han, Q.-B.; Ma, D.-L. Emerging Screening Approaches in the Development of Nrf2–Keap1 Protein–Protein Interaction Inhibitors. Int. J. Mol. Sci. 2019, 20, 4445. [Google Scholar] [CrossRef]

| References | Functions | Substrates | Cancer Type |
|---|---|---|---|
| [31] | Facilitating ESCC progression via the Hippo/YAP axis | Hippo/YAP | Esophageal carcinoma |
| [77] | Deubiquitinating Snail2 and thus promoting glioblastoma invasion and tumorigenesis | Snail2 | Glioblastoma invasion |
| [73] | Synergizing with TP53 and promoting HCC progression | TP53 | Hepatocellular carcinoma |
| [74] | Stabilizing CEP63 by reducing its K48 ubiquitination and promoting colorectal cancer progression | CEP63 | Colorectal cancer |
| [75] | Deubiquitinating PKM2 to suppress the malignant phenotypes through the Warburg effect | PKM2 | Breast cancer |
| [76] | USP36 helps mediate the stabilization of EZH2 | MELK | T cell lymphoma |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, M.-Y.; Liu, Y.-J.; Shi, J.-J.; Chen, R.-Y.; Zhang, S.; Li, C.-Y.; Cao, J.-F.; Yang, G.-J.; Chen, J. The Emerging Role of Ubiquitin-Specific Protease 36 (USP36) in Cancer and Beyond. Biomolecules 2024, 14, 572. https://doi.org/10.3390/biom14050572
Niu M-Y, Liu Y-J, Shi J-J, Chen R-Y, Zhang S, Li C-Y, Cao J-F, Yang G-J, Chen J. The Emerging Role of Ubiquitin-Specific Protease 36 (USP36) in Cancer and Beyond. Biomolecules. 2024; 14(5):572. https://doi.org/10.3390/biom14050572
Chicago/Turabian StyleNiu, Meng-Yao, Yan-Jun Liu, Jin-Jin Shi, Ru-Yi Chen, Shun Zhang, Chang-Yun Li, Jia-Feng Cao, Guan-Jun Yang, and Jiong Chen. 2024. "The Emerging Role of Ubiquitin-Specific Protease 36 (USP36) in Cancer and Beyond" Biomolecules 14, no. 5: 572. https://doi.org/10.3390/biom14050572
APA StyleNiu, M.-Y., Liu, Y.-J., Shi, J.-J., Chen, R.-Y., Zhang, S., Li, C.-Y., Cao, J.-F., Yang, G.-J., & Chen, J. (2024). The Emerging Role of Ubiquitin-Specific Protease 36 (USP36) in Cancer and Beyond. Biomolecules, 14(5), 572. https://doi.org/10.3390/biom14050572









