Serum EphA2 as a Promising Biomarker for the Early Detection and Diagnosis of Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Patient Enrollment and Sample Collection
2.3. Monoclonal Antibodies to Human EphA2
2.4. In-House ELISA for Detecting of Human EphA2
2.5. Western Blotting
2.6. Flow Cytometry Analysis
2.7. Statistical Analysis
3. Results
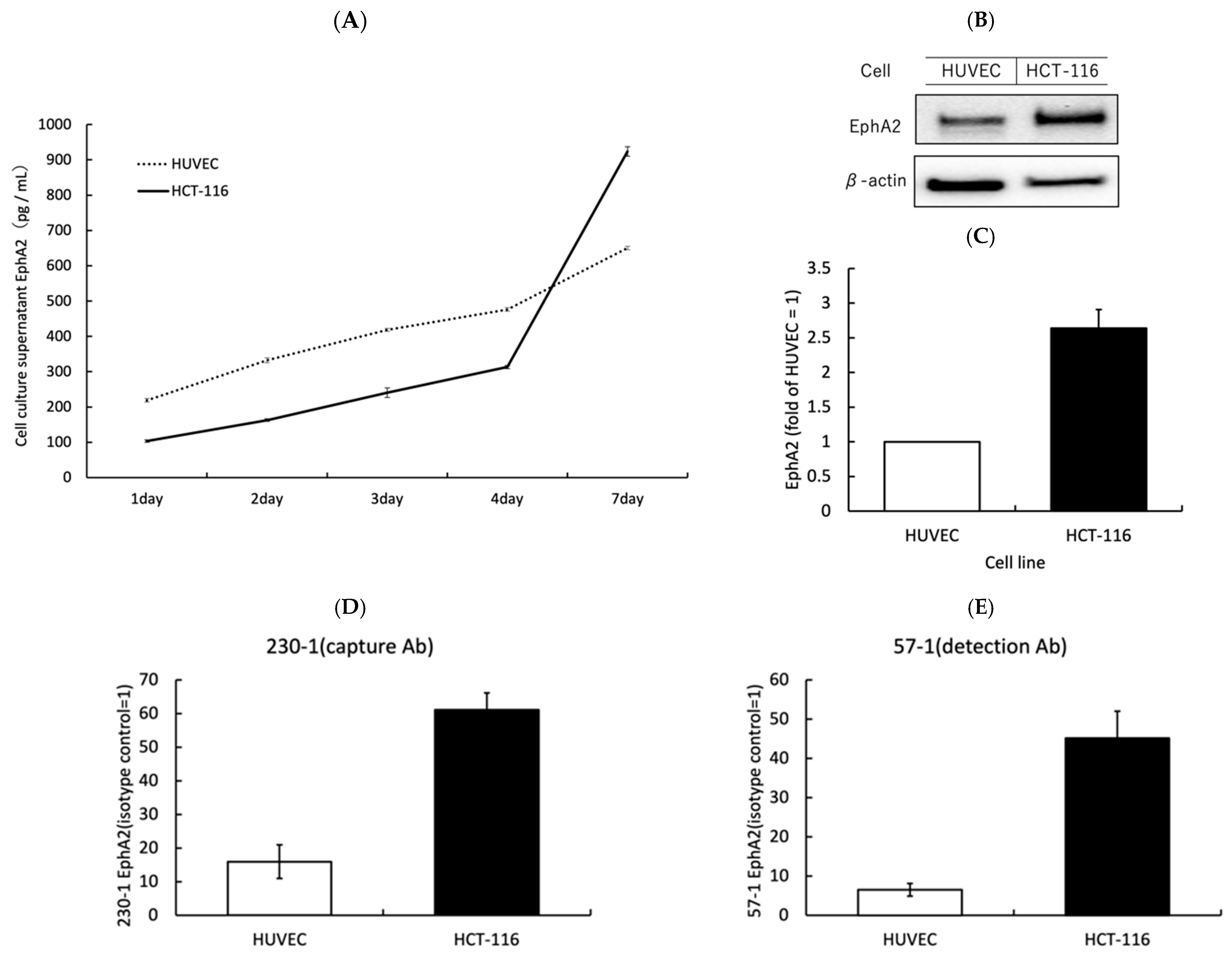
3.1. EphA2 Is Strongly Expressed on the Surface of CRC Cells and Is Released from CRC Cells
3.2. Soluble EphA2 in Serum Samples from CRC Patients and Its Clinical Correlations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Xiao, Y.; Wang, W.; Tang, Y.Y.; Xiao, Z.; Su, M. Targeting EphA2 in cancer. J. Hematol. Oncol. 2020, 13, 114. [Google Scholar] [CrossRef]
- Kurose, H.; Ueda, K.; Kondo, R.; Ogasawara, S.; Kusano, H.; Sanada, S.; Naito, Y.; Nakiri, M.; Nishihara, K.; Kakuma, T.; et al. Elevated Expression of EPHA2 Is Associated With Poor Prognosis After Radical Prostatectomy in Prostate Cancer. Anticancer. Res. 2019, 39, 6249–6257. [Google Scholar] [CrossRef]
- Amato, K.R.; Wang, S.; Tan, L.; Hastings, A.K.; Song, W.; Lovly, C.M.; Meador, C.B.; Ye, F.; Lu, P.; Balko, J.M.; et al. EPHA2 Blockade Overcomes Acquired Resistance to EGFR Kinase Inhibitors in Lung Cancer. Cancer Res. 2016, 76, 305–318. [Google Scholar] [CrossRef]
- Miyazaki, T.; Kato, H.; Fukuchi, M.; Nakajima, M.; Kuwano, H. EphA2 overexpression correlates with poor prognosis in esophageal squamous cell carcinoma. Int. J. Cancer 2003, 103, 657–663. [Google Scholar] [CrossRef]
- Martini, G.; Cardone, C.; Vitiello, P.P.; Belli, V.; Napolitano, S.; Troiani, T.; Ciardiello, D.; Della Corte, C.M.; Morgillo, F.; Matrone, N.; et al. EPHA2 Is a Predictive Biomarker of Resistance and a Potential Therapeutic Target for Improving Antiepidermal Growth Factor Receptor Therapy in Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 845–855. [Google Scholar] [CrossRef]
- Wu, D.; Suo, Z.; Kristensen, G.B.; Li, S.; Troen, G.; Holm, R.; Nesland, J.M. Prognostic value of EphA2 and EphrinA-1 in squamous cell cervical carcinoma. Gynecol. Oncol. 2004, 94, 312–319. [Google Scholar] [CrossRef]
- Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Merritt, W.M.; Landen, C.N.; Deavers, M.T.; Fletcher, M.S.; Urbauer, D.L.; Kinch, M.S.; Sood, A.K. EphA2 overexpression is associated with angiogenesis in ovarian cancer. Cancer 2007, 109, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, V.M.; Kim, L.C.; Edwards, D.N.; Hwang, Y.; Santapuram, P.R.; Stirdivant, S.M.; Lu, P.; Ye, F.; Brantley-Sieders, D.M.; Chen, J. The Ephrin-A1/EPHA2 Signaling Axis Regulates Glutamine Metabolism in HER2-Positive Breast Cancer. Cancer Res. 2016, 76, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Zhao, X.; Dong, X.; Liu, T.; Zhao, N.; Zhang, D.; Wang, W.; Zhang, Y.; Sun, B. Effect of EphA2 knockdown on melanoma metastasis depends on intrinsic ephrinA1 level. Cell. Oncol. 2020, 43, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Torlot, L.; Jarzab, A.; Albert, J.; Pók-Udvari, Á.; Stahler, A.; Holch, J.W.; Gerlinger, M.; Heinemann, V.; Klauschen, F.; Kirchner, T.; et al. Proteomics uncover EPHA2 as a potential novel therapeutic target in colorectal cancer cell lines with acquired cetuximab resistance. J. Cancer Res. Clin. Oncol. 2023, 149, 669–682. [Google Scholar] [CrossRef]
- Nehal, M.; Khatoon, J.; Akhtar, S.; Khan, M.K.A. Exploring the potential of EphA2 receptor signaling pathway: A comprehensive review in cancer treatment. Mol. Biol. Rep. 2024, 51, 337. [Google Scholar] [CrossRef]
- Cantor, D.I.; Cheruku, H.R.; Westacott, J.; Shin, J.S.; Mohamedali, A.; Ahn, S.B. Proteomic investigations into resistance in colorectal cancer. Expert Rev. Proteomics 2020, 17, 49–65. [Google Scholar] [CrossRef]
- Lei, X.; He, Q.; Li, Z.; Zou, Q.; Xu, P.; Yu, H.; Ding, Y.; Zhu, W. Cancer stem cells in colorectal cancer and the association with chemotherapy resistance. Med. Oncol. 2021, 38, 43. [Google Scholar] [CrossRef]
- Medina, G.G.; McQueen, A.; Greisinger, A.J.; Bartholomew, L.K.; Vernon, S.W. What would make getting colorectal cancer screening easier? Perspectives from screeners and nonscreeners. Gastroenterol. Res. Pract. 2012, 2012, 895807. [Google Scholar] [CrossRef]
- Meissner, H.I.; Klabunde, C.N.; Breen, N.; Zapka, J.M. Breast and colorectal cancer screening: U.S. primary care physicians’ reports of barriers. Am. J. Prev. Med. 2012, 43, 584–589. [Google Scholar] [CrossRef]
- Dunne, P.D.; Dasgupta, S.; Blayney, J.K.; McArt, D.G.; Redmond, K.L.; Weir, J.A.; Bradley, C.A.; Sasazuki, T.; Shirasawa, S.; Wang, T.; et al. EphA2 Expression Is a Key Driver of Migration and Invasion and a Poor Prognostic Marker in Colorectal Cancer. Clin. Cancer Res. 2016, 22, 230–242. [Google Scholar] [CrossRef]
- Saito, T.; Masuda, N.; Miyazaki, T.; Kanoh, K.; Suzuki, H.; Shimura, T.; Asao, T.; Kuwano, H. Expression of EphA2 and E-cadherin in colorectal cancer: Correlation with cancer metastasis. Oncol. Rep. 2004, 11, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Lakemeyer, L.; Sander, S.; Wittau, M.; Henne-Bruns, D.; Kornmann, M.; Lemke, J. Diagnostic and Prognostic Value of CEA and CA19-9 in Colorectal Cancer. Diseases 2021, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Miyake, H.; Nagai, H.; Yoshioka, Y.; Shibata, K.; Asai, S.; Takamizawa, J.; Yuasa, N. Optimal cutoff value of preoperative CEA and CA19-9 for prognostic significance in patients with stage II/III colon cancer. Langenbecks Arch. Surg. 2021, 406, 1987–1997. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef]
- Herath, N.I.; Spanevello, M.D.; Doecke, J.D.; Smith, F.M.; Pouponnot, C.; Boyd, A.W. Complex expression patterns of Eph receptor tyrosine kinases and their ephrin ligands in colorectal carcinogenesis. Eur. J. Cancer 2012, 48, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Strimpakos, A.; Pentheroudakis, G.; Kotoula, V.; De Roock, W.; Kouvatseas, G.; Papakostas, P.; Makatsoris, T.; Papamichael, D.; Andreadou, A.; Sgouros, J.; et al. The prognostic role of ephrin A2 and endothelial growth factor receptor pathway mediators in patients with advanced colorectal cancer treated with cetuximab. Clin. Color. Cancer 2013, 12, 267–274.e262. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Yang, X.; Zhang, Y.; Lu, Y.; Li, Y. The expression and diagnostic value of serum levels of EphA2 and VEGF-A in patients with colorectal cancer. Cancer Biomark. 2021, 31, 399–408. [Google Scholar] [CrossRef]
- Koshikawa, N.; Hoshino, D.; Taniguchi, H.; Minegishi, T.; Tomari, T.; Nam, S.O.; Aoki, M.; Sueta, T.; Nakagawa, T.; Miyamoto, S.; et al. Proteolysis of EphA2 Converts It from a Tumor Suppressor to an Oncoprotein. Cancer Res. 2015, 75, 3327–3339. [Google Scholar] [CrossRef]
- Tröster, A.; Jores, N.; Mineev, K.S.; Sreeramulu, S.; DiPrima, M.; Tosato, G.; Schwalbe, H. Targeting EPHA2 with Kinase Inhibitors in Colorectal Cancer. ChemMedChem 2023, 18, e202300420. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xiao, T.; Lu, S.S.; Hung, X.P.; Yi, H.; He, Q.Y.; Huang, W.; Tang, Y.Y.; Xiao, Z.Q. ANXA1-derived peptides suppress gastric and colon cancer cell growth by targeting EphA2 degradation. Int. J. Oncol. 2020, 57, 1203–1213. [Google Scholar] [CrossRef]


| Category | CRC n = 65 |
|---|---|
| Age, Median (IQR) | 73 (66–79) |
| Sex Male:Female | 42:23 |
| Location Right:Left | 25:40 |
| Stage I:II:III:IV | 10:30:19:6 |
| Category | CRC (n = 65) | EphA2 (pg/mL) (Range) | p Value | |
|---|---|---|---|---|
| Tumor location | Right | 25 | 641 (329–3145) | N.S. |
| Left | 40 | 692 (359–2325) | ||
| Tumor size | ≤30 mm | 18 | 636 (329–1117) | 0.0346 |
| >30 mm | 46 | 730 (359–3145) | ||
| Depth of invasion | pT1–2 | 12 | 540 (449–812) | 0.0311 |
| pT3–4 | 53 | 714 (329–3145) | ||
| Lymph node metastasis | Negative | 43 | 689 (329–3145) | N.S. |
| Positive | 22 | 607 (368–1465) | ||
| Distant metastasis | Negative | 59 | 672 (329–3145) | N.S. |
| Positive | 6 | 856 (522–1465) | ||
| Vascular invasion | Negative | 14 | 657 (449–2325) | N.S. |
| Positive | 48 | 689 (329–3145) | ||
| Lymphatic invasion | Negative | 13 | 812 (449–2325) | 0.0431 |
| Positive | 49 | 631 (329–3145) | ||
| Stage | I–II | 40 | 686 (329–3145) | N.S. |
| III–IV | 25 | 656 (368–1465) | ||
| CEA (ng/mL) | ≤5 | 39 | 642 (329–1565) | N.S. |
| >5 | 25 | 782 (359–3145) | ||
| CA19–9 (U/mL) | ≤37 | 54 | 649 (329–3145) | N.S. |
| >37 | 10 | 712 (359–1465) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakuraba, S.; Koizumi, A.; Iwasawa, T.; Ito, T.; Kato, K. Serum EphA2 as a Promising Biomarker for the Early Detection and Diagnosis of Colorectal Cancer. Biomolecules 2024, 14, 1504. https://doi.org/10.3390/biom14121504
Sakuraba S, Koizumi A, Iwasawa T, Ito T, Kato K. Serum EphA2 as a Promising Biomarker for the Early Detection and Diagnosis of Colorectal Cancer. Biomolecules. 2024; 14(12):1504. https://doi.org/10.3390/biom14121504
Chicago/Turabian StyleSakuraba, Shunsuke, Akihiro Koizumi, Takumi Iwasawa, Tomoaki Ito, and Kazunori Kato. 2024. "Serum EphA2 as a Promising Biomarker for the Early Detection and Diagnosis of Colorectal Cancer" Biomolecules 14, no. 12: 1504. https://doi.org/10.3390/biom14121504
APA StyleSakuraba, S., Koizumi, A., Iwasawa, T., Ito, T., & Kato, K. (2024). Serum EphA2 as a Promising Biomarker for the Early Detection and Diagnosis of Colorectal Cancer. Biomolecules, 14(12), 1504. https://doi.org/10.3390/biom14121504






