Impact of De Novo Cholesterol Biosynthesis on the Initiation and Progression of Breast Cancer
Abstract
1. Introduction
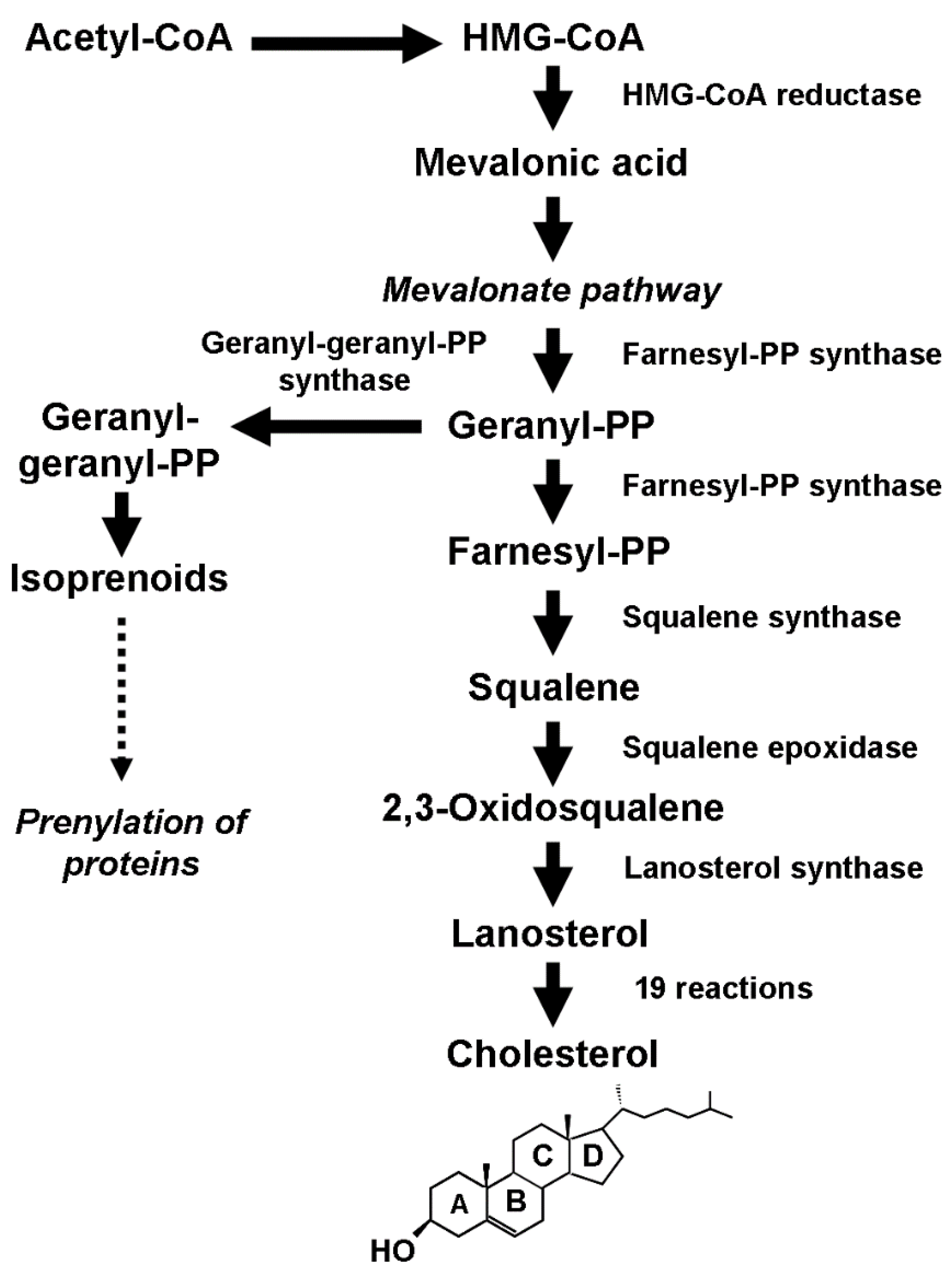
2. Cholesterol Biology
2.1. Cholesterol and Membrane Dynamics
2.2. Cholesterol Direct Activity
2.3. Cholesterol Derivatives
3. Cholesterol Biosynthesis and Breast Cancer Initiation and Development
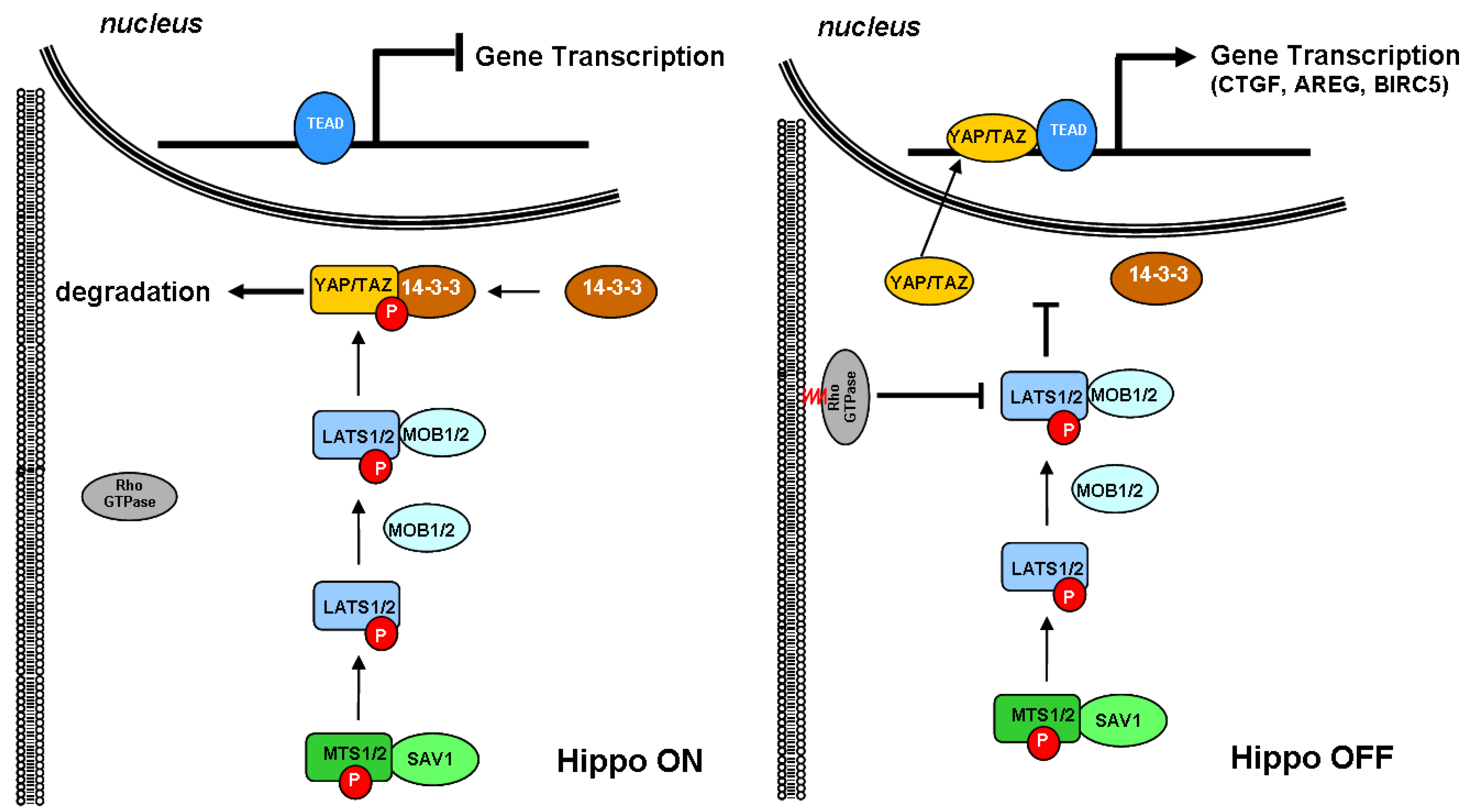
3.1. Cholesterol Biosynthesis and the Hippo Signaling Pathway
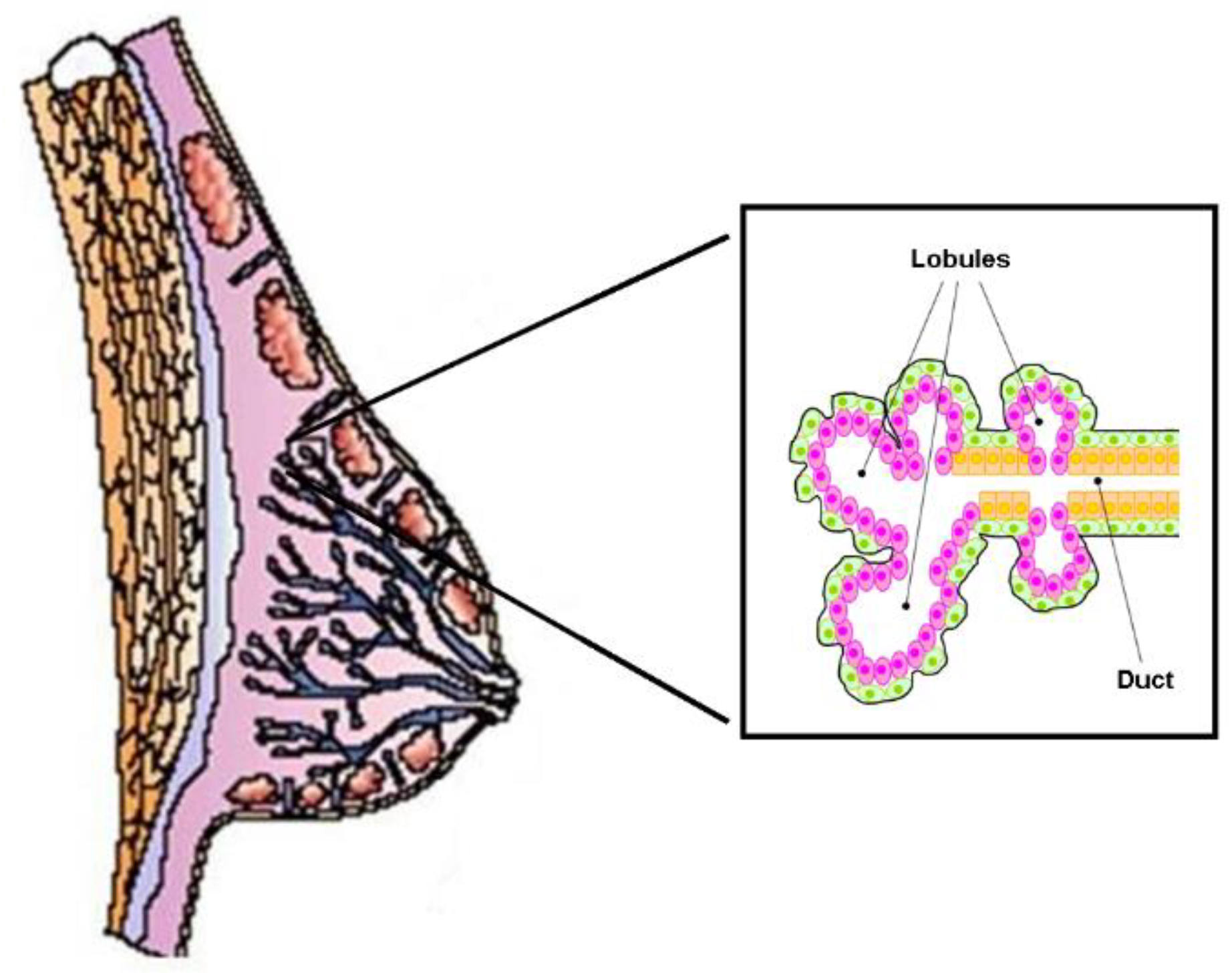
3.2. Cholesterol Biosynthesis, Mammary Stem Cells, and Breast Cancer Stem Cells
3.3. Cholesterol Biosynthesis and Ductal Carcinoma In Situ
3.4. Cholesterol Biosynthesis and Invasive Breast Cancer
4. Cholesterol Biosynthesis and Drug Resistance
5. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- Luo, J.; Yang, H.; Song, B.L. Mechanisms and Regulation of Cholesterol Homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Sugii, S.; Reid, P.C.; Ohgami, N.; Du, H.; Chang, T.Y. Distinct Endosomal Compartments in Early Trafficking of Low Density Lipoprotein-Derived Cholesterol. J. Biol. Chem. 2003, 278, 27180–27189. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.Y.; Chang, C.C.; Ohgami, N.; Yamauchi, Y. Cholesterol Sensing, Trafficking, and Esterification. Annu. Rev. Cell Dev. Biol. 2006, 22, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhou, Y.; Goldstein, J.L.; Brown, M.S.; Radhakrishnan, A. Cholesterol-Induced Conformational Changes in the Sterol-Sensing Domain of the Scap Protein Suggest Feedback Mechanism to Control Cholesterol Synthesis. J. Biol. Chem. 2017, 292, 8729–8737. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.P.; Li, L.; Goldstein, J.L.; Brown, M.S. Insig Required for Sterol-Mediated Inhibition of Scap/SREBP Binding to COPII Proteins in vitro. J. Biol. Chem. 2005, 280, 26483–26490. [Google Scholar] [CrossRef] [PubMed]
- Sever, N.; Yang, T.; Brown, M.S.; Goldstein, J.L.; DeBose-Boyd, R.A. Accelerated Degradation of HMG CoA Reductase Mediated by Binding of Insig-1 to Its Sterol-Sensing Domain. Mol. Cell. 2003, 11, 25–33. [Google Scholar] [CrossRef]
- DeBose-Boyd, R.A. Feedback Regulation of Cholesterol Synthesis: Sterol-Accelerated Ubiquitination and Degradation of HMG CoA Reductase. Cell Res. 2008, 18, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Llaverias, G.; Danilo, C.; Mercier, I.; Daumer, K.; Capozza, F.; Williams, T.M.; Sotgia, F.; Lisanti, M.P.; Frank, P.G. Role of Cholesterol in the Development and Progression of Breast Cancer. Am. J. Pathol. 2011, 178, 402–412. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The Role of Cholesterol Metabolism in Cancer. Am. J. Cancer Res. 2019, 9, 219–227. [Google Scholar]
- Mok, E.H.K.; Lee, T.K.W. The Pivotal Role of the Dysregulation of Cholesterol Homeostasis in Cancer: Implications for Therapeutic Targets. Cancers 2020, 12, 1410. [Google Scholar] [CrossRef]
- Giacomini, I.; Gianfanti, F.; Desbats, M.A.; Orso, G.; Berretta, M.; Prayer-Galetti, T.; Ragazzi, E.; Cocetta, V. Cholesterol Metabolic Reprogramming in Cancer and Its Pharmacological Modulation as Therapeutic Strategy. Front. Oncol. 2021, 11, 682911. [Google Scholar]
- González-Ortiz, A.; Galindo-Hernández, O.; Hernández-Acevedo, G.N.; Hurtado-Ureta, G.; García-González, V. Impact of Cholesterol-Pathways on Breast Cancer Development, a Metabolic Landscape. J. Cancer 2021, 12, 4307–4321. [Google Scholar] [CrossRef] [PubMed]
- Mayengbam, S.S.; Ajay, A.S.; Bhat, M.K. Influence of Cholesterol on Cancer Progression and Therapy. Transl. Oncol. 2021, 14, 101043. [Google Scholar] [CrossRef] [PubMed]
- Mouritsen, O.G.; Zuckermann, M.J. What’s so Special about Cholesterol? Lipids 2004, 39, 1101–1113. [Google Scholar] [CrossRef] [PubMed]
- Dufourc, E.J. Sterols and Membrane Dynamics. J. Chem. Biol. 2008, 1, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J. Lipid Rafts: Bringing Order to Chaos. J. Lipid Res. 2003, 44, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Lingwood, D.; Simons, K. Lipid Rafts as a Membrane-Organizing Principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef]
- Simons, K.; Toomre, D. Lipid Rafts and Signal Transduction. Nat. Rev. Mol. Cell. Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef]
- Hanzal-Bayer, M.F.; Hancock, J.F. Lipid Rafts and Membrane Traffic. FEBS Lett. 2007, 581, 2098–2104. [Google Scholar] [CrossRef]
- Head, B.P.; Patel, H.H.; Insel, P.A. Interaction of Membrane/Lipid Rafts with the Cytoskeleton: Impact on Signaling and Function: Membrane/Lipid Rafts, Mediators of Cytoskeletal Arrangement and Cell Signaling. Biochim. Biophys. Acta 2014, 1838, 532–545. [Google Scholar] [CrossRef]
- Varshney, P.; Yadav, V.; Saini, N. Lipid Rafts in Immune Signalling: Current Progress and Future Perspective. Immunology 2016, 149, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F.; Gajate, C. Lipid Rafts as Signaling Hubs in Cancer Cell Survival/Death and Invasion: Implications in Tumor Progression and Therapy: Thematic Review Series: Biology of Lipid Rafts. J. Lipid Res. 2020, 61, 611–635. [Google Scholar] [CrossRef] [PubMed]
- Vona, R.; Iessi, E.; Matarrese, P. Role of Cholesterol and Lipid Rafts in Cancer Signaling: A Promising Therapeutic Opportunity? Front. Cell. Dev. Biol. 2021, 9, 622908. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, F.; Mader, S.; Philip, A. Cholesterol as an Endogenous Ligand of ERRα Promotes ERRα-Mediated Cellular Proliferation and Metabolic Target Gene Expression in Breast Cancer Cells. Cells 2020, 9, 1765. [Google Scholar] [CrossRef] [PubMed]
- Deblois, G.; Giguère, V. Functional and Physiological Genomics of Estrogen-Related Receptors (ERRs) in Health and Disease. Biochim. Biophys. Acta 2011, 1812, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Deblois, G.; St-Pierre, J.; Giguère, V. The PGC-1/ERR Signaling Axis in Cancer. Oncogene 2013, 32, 3483–3490. [Google Scholar] [CrossRef]
- Wei, W.; Schwaid, A.G.; Wang, X.; Wang, X.; Chen, S.; Chu, Q.; Saghatelian, A.; Wan, Y. Ligand Activation of ERRα by Cholesterol Mediates Statin and Bisphosphonate Effects. Cell Metab. 2016, 23, 479–491. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Tripathi, M.; Singh, B.K. Metabolic Switching of Estrogen-Related Receptor alpha in Breast Cancer Aggression. FEBS J. 2023, 290, 1473–1476. [Google Scholar] [CrossRef]
- Ghanbari, F.; Fortier, A.M.; Park, M.; Philip, A. Cholesterol-Induced Metabolic Reprogramming in Breast Cancer Cells Is Mediated via the ERRα Pathway. Cancers 2021, 13, 2605. [Google Scholar] [CrossRef]
- Brindisi, M.; Frattaruolo, L.; Fiorillo, M.; Dolce, V.; Sotgia, F.; Lisanti, M.P.; Cappello, A.R. New Insights into Cholesterol-Mediated ERRα Activation in Breast Cancer Progression and Pro-Tumoral Microenvironment Orchestration. FEBS J. 2023, 290, 1481–1501. [Google Scholar] [CrossRef] [PubMed]
- Muduli, K.; Prusty, M.; Pradhan, J.; Samal, A.P.; Sahu, B.; Roy, D.S.; Reddy, K.S.; Elangovan, S. Estrogen-Related Receptor Alpha (ERRα) Promotes Cancer Stem Cell-Like Characteristics in Breast Cancer. Stem Cell Rev. Rep. 2023, 19, 2807–2819. [Google Scholar] [CrossRef] [PubMed]
- Brindisi, M.; Fiorillo, M.; Frattaruolo, L.; Sotgia, F.; Lisanti, M.P.; Cappello, A.R. Cholesterol and Mevalonate: Two Metabolites Involved in Breast Cancer Progression and Drug Resistance through the ERRα Pathway. Cells 2020, 9, 1819. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y. Bile Acid Metabolism and Signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [PubMed]
- DuSell, C.D.; Umetani, M.; Shaul, P.W.; Mangelsdorf, D.J.; McDonnell, D.P. 27-Hydroxycholesterol is an Endogenous Selective Estrogen Receptor Modulator. Mol. Endocrinol. 2008, 22, 65–77. [Google Scholar] [CrossRef]
- Nelson, E.R.; Wardell, S.E.; Jasper, J.S.; Park, S.; Suchindran, S.; Howe, M.K.; Carver, N.J.; Pillai, R.V.; Sullivan, P.M.; Sondhi, V.; et al. 27-Hydroxycholesterol Links Hypercholesterolemia and Breast Cancer Pathophysiology. Science 2013, 342, 1094–1098. [Google Scholar] [CrossRef]
- Kimbung, S.; Chang, C.Y.; Bendahl, P.O.; Dubois, L.; Thompson, J.W.; McDonnell, D.P.; Borgquist, S. Impact of 27-Hydroxylase (CYP27A1) and 27-Hydroxycholesterol in Breast Cancer. Endocr. Relat. Cancer 2017, 24, 339–349. [Google Scholar] [CrossRef]
- Wu, Q.; Ishikawa, T.; Sirianni, R.; Tang, H.; McDonald, J.G.; Yuhanna, I.S.; Thompson, B.; Girard, L.; Mineo, C.; Brekken, R.A.; et al. 27-Hydroxycholesterol Promotes Cell-Autonomous, ER-Positive Breast Cancer Growth. Cell Rep. 2013, 5, 637–645. [Google Scholar] [CrossRef]
- Abdalkareem Jasim, S.; Kzar, H.H.; Haider Hamad, M.; Ahmad, I.; Al-Gazally, M.E.; Ziyadullaev, S.; Sivaraman, R.; Abed Jawad, M.; Thaeer Hammid, A.; Oudaha, K.H.; et al. The Emerging Role of 27-Hydroxycholesterol in Cancer Development and Progression: An Update. Int. Immunopharmacol. 2022, 110, 109074. [Google Scholar] [CrossRef]
- Biasi, F.; Leoni, V.; Gamba, P.; Sassi, K.; Lizard, G.; Poli, G. Role of 27-Hydroxycholesterol and Its Metabolism in Cancer Progression: Human Studies. Biochem. Pharmacol. 2022, 196, 114618. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cho, W.; Nelson, E.R. Our Evolving Understanding of How 27-Hydroxycholesterol Influences Cancer. Biochem. Pharmacol. 2021, 196, 114621. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.H.; Hales, D.B. Overview of Steroidogenic Enzymes in the Pathway from Cholesterol to Active Steroid Hormones. Endocr. Rev. 2004, 25, 947–970. [Google Scholar] [CrossRef] [PubMed]
- Whirledge, S.; Cidlowski, J.A. Chapter 5—Steroid Hormone Action. In Yen and Jaffe’s Reproductive Endocrinology, 8th ed.; Strauss, J.F., Barbieri, R.L., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 115–131. ISBN 9780323479127. [Google Scholar] [CrossRef]
- Mahboobifard, F.; Pourgholami, M.H.; Jorjani, M.; Dargahi, L.; Amiri, M.; Sadeghi, S.; Tehrani, F.R. Estrogen as a Key Regulator of Energy Homeostasis and Metabolic Health. Biomed. Pharmacother. 2022, 156, 113808. [Google Scholar] [CrossRef]
- Kim, J.H.; Cho, H.T.; Kim, Y.J. The role of Estrogen in Adipose Tissue Metabolism: Insights into Glucose Homeostasis regulation. Endocr. J. 2014, 61, 1055–1067. [Google Scholar] [CrossRef]
- Miller, W.L.; Auchus, R.J. The Molecular Biology, Biochemistry, and Physiology of Human Steroidogenesis and Its Disorders. Endocr. Rev. 2011, 32, 81–151. [Google Scholar] [CrossRef]
- Miller, W.R.; Langdon, S.P. Steroid Hormones and Cancer: (III) Observations from Human Subjects. Eur. J. Surg. Oncol. 1997, 23, 163–177. [Google Scholar] [CrossRef]
- Bhardwaj, P.; Au, C.C.; Benito-Martin, A.; Ladumor, H.; Oshchepkova, S.; Moges, R.; Brown, K.A. Estrogens and Breast Cancer: Mechanisms Involved in Obesity-Related Development, Growth and Progression. J. Steroid Biochem. Mol. Biol. 2019, 189, 161–170. [Google Scholar] [CrossRef]
- Khan, M.Z.I.; Uzair, M.; Nazli, A.; Chen, J.Z. An Overview on Estrogen Receptors Signaling and Its Ligands in Breast Cancer. Eur. J. Med. Chem. 2022, 241, 114658. [Google Scholar] [CrossRef]
- Hargrove-Wiley, E.; Fingleton, B. Sex Hormones in Breast Cancer Immunity. Cancer Res. 2023, 83, 12–19. [Google Scholar] [CrossRef]
- Walters, M.R.; Nemere, I. Receptors for Steroid hormones: Membrane-associated and nuclear forms. Cell. Mol. Life Sci. 2004, 61, 2309–2321. [Google Scholar] [CrossRef] [PubMed]
- Dahlman-Wright, K.; Cavailles, V.; Fuqua, S.A.; Jordan, V.C.; Katzenellenbogen, J.A.; Korach, K.S.; Maggi, A.; Muramatsu, M.; Parker, M.G.; Gustafsson, J.A. International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol. Rev. 2006, 58, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Soltysik, K.; Czekaj, P. Membrane Estrogen Receptors—Is It an Alternative Way of Estrogen Action? J. Physiol. Pharmacol. 2013, 64, 129–142. [Google Scholar] [PubMed]
- Clark, L.T. Cholesterol and Heart Disease: Current Concepts in Pathogenesis and Treatment. J. Natl. Med. Assoc. 1986, 78, 743–751. [Google Scholar] [PubMed]
- Brown, A.J. Cholesterol, Statins and Cancer. Clin. Exp. Pharmacol. Physiol. 2007, 34, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Kuzu, O.F.; Noory, M.A.; Robertson, G.P. The Role of Cholesterol in Cancer. Cancer Res. 2016, 76, 2063–2070. [Google Scholar] [CrossRef] [PubMed]
- Murdock, D.J.; Sanchez, R.J.; Mohammadi, K.A.; Fazio, S.; Geba, G.P. Serum Cholesterol and the Risk of Developing Hormonally Driven Cancers: A Narrative Review. Cancer Med. 2023, 12, 6722–6767. [Google Scholar] [CrossRef]
- Li, C.; Yang, L.; Zhang, D.; Jiang, W. Systematic Review and Meta-Analysis Suggest that Dietary Cholesterol Intake Increases Risk of Breast Cancer. Nutr. Res. 2016, 36, 627–635. [Google Scholar] [CrossRef]
- Johnson, K.E.; Siewert, K.M.; Klarin, D.; Damrauer, S.M.; Chang, K.M.; Tsao, P.S.; Assimes, T.L.; Maxwell, K.N.; Voight, B.F. The Relationship Between Circulating Lipids and Breast Cancer Risk: A Mendelian randomization study. PLoS Med. 2020, 17, e1003302. [Google Scholar] [CrossRef]
- Touvier, M.; Fassier, P.; His, M.; Norat, T.; Chan, D.S.; Blacher, J.; Hercberg, S.; Galan, P.; Druesne-Pecollo, N.; Latino-Martel, P. Cholesterol and Breast Cancer Risk: A Systematic Review and Meta-Analysis of Prospective Studies. Br. J. Nutr. 2015, 114, 347–357. [Google Scholar] [CrossRef]
- Baek, A.E.; Nelson, E.R. The Contribution of Cholesterol and Its Metabolites to the Pathophysiology of Breast Cancer. Horm. Cancer 2016, 7, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Ben Hassen, C.; Goupille, C.; Vigor, C.; Durand, T.; Guéraud, F.; Silvente-Poirot, S.; Poirot, M.; Frank, P.G. Is Cholesterol a Risk Factor for Breast Cancer Incidence and Outcome? J. Steroid Biochem. Mol. Biol. 2023, 232, 106346. [Google Scholar] [CrossRef] [PubMed]
- Clendening, J.W.; Pandyra, A.; Boutros, P.C.; El Ghamrasni, S.; Khosravi, F.; Trentin, G.A.; Martirosyan, A.; Hakem, A.; Hakem, R.; Jurisica, I.; et al. Dysregulation of the Mevalonate Pathway Promotes Transformation. Proc. Natl. Acad. Sci. USA 2010, 107, 15051–15056. [Google Scholar] [CrossRef]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo Pathway in Organ Size Control, Tissue Regeneration and Stem Cell Self-Renewal. Nat. Cell Biol. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP Oncoprotein by the Hippo Pathway Is Involved in Cell Contact Inhibition and Tissue Growth Control. Genes Dev. 2007, 21, 2747–2761. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.X.; Guan, K.L. The Hippo Pathway: Regulators and Regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in Cell Biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Ridley, A.J.; Hall, A. The Small GTP-Binding Protein Rho Regulates the Assembly of Focal Adhesions and Actin Stress Fibers in Response to Growth Factors. Cell 1992, 70, 389–399. [Google Scholar] [CrossRef]
- Jang, J.W.; Kim, M.K.; Bae, S.C. Reciprocal Regulation of YAP/TAZ by the Hippo Pathway and the Small GTPase Pathway. Small GTPases 2020, 11, 280–288. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Lei, Q.; and Guan, K.L. The Hippo-YAP Pathway in Organ Size Control and Tumorigenesis: An Updated Version. Genes Dev. 2010, 24, 862–874. [Google Scholar] [CrossRef]
- Steinhardt, A.A.; Gayyed, M.F.; Klein, A.P.; Dong, J.; Maitra, A.; Pan, D.; Montgomery, E.A.; Anders, R.A. Expression of Yes-associated Protein in Common Solid Tumors. Hum. Pathol. 2008, 39, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Stingl, J.; Eaves, C.J.; Kuusk, U.; Emerman, J.T. Phenotypic and Functional Characterization In Vitro of a Multipotent Epithelial Cell Present in the Normal Adult Human Breast. Differentiation 1998, 63, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Gudjonsson, T.; Villadsen, R.; Nielsen, H.L.; Rønnov-Jessen, L.; Bissell, M.J.; Petersen, O.W. Isolation, Immortalization, and Characterization of a Human Breast Epithelial Cell Line with Stem Cell Properties. Genes Dev. 2002, 16, 693–706. [Google Scholar] [CrossRef]
- Fu, N.Y.; Nolan, E.; Lindeman, G.J.; Visvader, J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2020, 100, 489–523. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.A.; Jackson, H.W.; Beristain, A.G.; Di Grappa, M.A.; Mote, P.A.; Clarke, C.L.; Stingl, J.; Waterhouse, P.D.; Khokha, R. Progesterone Induces Adult Mammary Stem Cell Expansion. Nature 2010, 465, 803–807. [Google Scholar] [CrossRef]
- Honeth, G.; Schiavinotto, T.; Vaggi, F.; Marlow, R.; Kanno, T.; Shinomiya, I.; Lombardi, S.; Buchupalli, B.; Graham, R.; Gazinska, P.; et al. Models of Breast Morphogenesis Based on Localization of Stem Cells in the Developing Mammary Lobule. Stem Cell Rep. 2015, 4, 699–711. [Google Scholar] [CrossRef]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In Vitro Propagation and Transcriptional Profiling of Human Mammary Stem/Progenitor Cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef]
- Slepicka, P.F.; Somasundara, A.V.H.; Dos Santos, C.O. The Molecular Basis of Mammary Gland Development and Epithelial Differentiation. Semin. Cell Dev. Biol. 2021, 114, 93–112. [Google Scholar] [CrossRef]
- Dontu, G.; El-Ashry, D.; Wicha, M.S. Breast Cancer, Stem/Progenitor Cells and the Estrogen Receptor. Trends Endocrinol. Metab. 2004, 15, 193–197. [Google Scholar] [CrossRef]
- Ercan, C.; van Diest, P.J.; Vooijs, M. Mammary Development and Breast Cancer: The Role of Stem Cells. Curr. Mol. Med. 2011, 11, 270–285. [Google Scholar] [CrossRef]
- Economopoulou, P.; Kaklamani, V.G.; Siziopikou, K. The Role of Cancer Stem Cells in Breast Cancer Initiation and Progression: Potential Cancer Stem Cell-directed Therapies. Oncologist 2012, 17, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Bertucci, F.; Cabaud, O.; Wicinski, J.; Finetti, P.; Josselin, E.; Adelaide, J.; Nguyen, T.T.; Monville, F.; et al. ALDH1-positive Cancer Stem Cells Predict Engraftment of Primary Breast Tumors and Are Governed by a Common Stem Cell Program. Cancer Res. 2013, 73, 7290–7300. [Google Scholar] [CrossRef] [PubMed]
- Ehmsen, S.; Pedersen, M.H.; Wang, G.; Terp, M.G.; Arslanagic, A.; Hood, B.L.; Conrads, T.P.; Leth-Larsen, R.; Ditzel, H.J. Increased Cholesterol Biosynthesis Is a Key Characteristic of Breast Cancer Stem Cells Influencing Patient Outcome. Cell Rep. 2019, 27, 3927–3938.e6. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Monville, F.; Wicinski, J.; Cabaud, O.; Cervera, N.; Josselin, E.; Finetti, P.; Guille, A.; Larderet, G.; Viens, P.; et al. Mevalonate Metabolism Regulates Basal Breast Cancer Stem Cells and Is a Potential Therapeutic Target. Stem Cells 2012, 30, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.A.; Akrap, N.; Garre, E.; Magnusson, Y.; Harrison, H.; Andersson, D.; Jonasson, E.; Rafnsdottir, S.; Choudhry, H.; Buffa, F.; et al. The Mevalonate Precursor Enzyme HMGCS1 Is a Novel Marker and Key Mediator of Cancer Stem Cell Enrichment in Luminal and Basal Models of Breast Cancer. PLoS ONE 2020, 15, e0236187. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Clouthier, S.G.; Wicha, M.S. Role of microRNAs in the Regulation of Breast Cancer Stem Cells. J. Mammary Gland Biol. Neoplasia 2012, 17, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Monchusi, B.; Kaur, M. miRNAs as Modulators of Cholesterol in Breast Cancer Stem Cells: An Approach to Overcome Drug Resistance in Cancer. Curr. Drug Targets 2022, 23, 656–677. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- Sanders, M.E.; Schuyler, P.A.; Simpson, J.F.; Page, D.L.; Dupont, W.D. Continued Observation of the Natural History of Low-grade Ductal Carcinoma in situ Reaffirms Proclivity for Local Recurrence Even after More than 30 Years of Follow-Up. Mod. Pathol. 2015, 28, 662–669. [Google Scholar] [CrossRef]
- Schuetz, C.S.; Bonin, M.; Clare, S.E.; Nieselt, K.; Sotlar, K.; Walter, M.; Fehm, T.; Solomayer, E.; Riess, O.; Wallwiener, D.; et al. Progression-specific Genes Identified by Expression Profiling of Matched Ductal Carcinomas in situ and Invasive Breast Tumors, Combining Laser Capture Microdissection and Oligonucleotide Microarray Analysis. Cancer Res. 2006, 66, 5278–5286. [Google Scholar] [CrossRef]
- Tamimi, R.M.; Baer, H.J.; Marotti, J.; Galan, M.; Galaburda, L.; Fu, Y.; Deitz, A.C.; Connolly, J.L.; Schnitt, S.J.; Colditz, G.A.; et al. Comparison of Molecular Phenotypes of Ductal Carcinoma in situ and Invasive Breast Cancer. Breast Cancer Res. 2008, 10, R67. [Google Scholar] [CrossRef] [PubMed]
- Wiechmann, L.; Kuerer, H.M. The Molecular Journey from Ductal Carcinoma in situ to Invasive Breast Cancer. Cancer 2008, 112, 2130–2142. [Google Scholar] [CrossRef] [PubMed]
- Coradini, D. Interaction of de novo Cholesterol Biosynthesis and Hippo Signaling Pathway in Ductal Carcinoma in situ (DCIS)—Comparison with the Corresponding Normal Breast Epithelium. Transl. Breast. Cancer Res. 2023, 4, 26. [Google Scholar] [CrossRef]
- Butt, S.; Butt, T.; Jirström, K.; Hartman, L.; Amini, R.M.; Zhou, W.; Wärnberg, F.; Borgquist, S. The Target for Statins, HMG-CoA Reductase, is Expressed in Ductal Carcinoma in situ and May Predict Patient Response to Radiotherapy. Ann. Surg. Oncol. 2014, 21, 2911–2919. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.; Stevenson, J.; Kristiana, I.; Brown, A.J. Cholesterol-dependent Degradation of Squalene Monooxygenase, a Control Point in Cholesterol Synthesis Beyond HMG-CoA Reductase. Cell Metab. 2011, 13, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Göbel, A.; Riffel, R.M.; Hofbauer, L.C.; Rachner, T.D. The Mevalonate Pathway in Breast Cancer Biology. Cancer Lett. 2022, 542, 215761. [Google Scholar] [CrossRef]
- Kim, H.Y.; Bae, S.J.; Choi, J.W.; Han, S.; Bae, S.H.; Cheong, J.H.; Jang, H. Cholesterol Synthesis Is Important for Breast Cancer Cell Tumor Sphere Formation and Invasion. Biomedicines 2022, 10, 1908. [Google Scholar] [CrossRef]
- Maja, M.; Mohammed, D.; Dumitru, A.C.; Verstraeten, S.; Lingurski, M.; Mingeot-Leclercq, M.P.; Alsteens, D.; Tyteca, D. Surface Cholesterol-Enriched Domains Specifically Promote Invasion of Breast Cancer Cell Lines by Controlling Invadopodia and Extracellular Matrix Degradation. Cell Mol. Life Sci. 2022, 79, 417. [Google Scholar] [CrossRef]
- Tang, Q.; Liang, B.; Zhang, L.; Li, X.; Li, H.; Jing, W.; Jiang, Y.; Zhou, F.; Zhang, J.; Meng, Y.; et al. Enhanced Cholesterol Biosynthesis Promotes Breast Cancer Metastasis Via Modulating CCDC25 Expression and Neutrophil Extracellular Traps Formation. Sci. Rep. 2022, 12, 17350. [Google Scholar] [CrossRef]
- Coradini, D.; Ambrogi, F.; Infante, G. Cholesterol de novo Biosynthesis in Paired Samples of Breast Cancer and Adjacent Histologically Normal Tissue: Association with Proliferation Index, Tumor Grade, and Recurrence-Free Survival. Arch. Breast Cancer 2023, 10, 187–199. [Google Scholar] [CrossRef]
- Yoon, S.H.; Kim, H.S.; Kim, R.N.; Jung, S.Y.; Hong, B.S.; Kang, E.J.; Moon, H.G.; Noh, D.Y.; Han, W. NAD(P)-dependent Steroid Dehydrogenase-like is Involved in Breast Cancer Cell Growth and Metastasis. BMC Cancer 2020, 20, 375. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene Expression Profiling in Breast Cancer: Understanding the Molecular Basis of Histologic Grade to Improve Prognosis. J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Matikas, A.; Foukakis, T.; Swain, S.; Bergh, J. Avoiding over- and Undertreatment in Patients with Resected Node-Positive Breast Cancer with the Use of Gene Expression Signatures: Are We There Yet? Ann. Oncol. 2019, 30, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Hultsch, S.; Kankainen, M.; Paavolainen, L.; Kovanen, R.M.; Ikonen, E.; Kangaspeska, S.; Pietiäinen, V.; Kallioniemi, O. Association of Tamoxifen Eesistance and Lipid Reprogramming in Breast Cancer. BMC Cancer 2018, 18, 850. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.K.L.; Jose, J.; Wahba, M.; Bernaus-Esqué, M.; Hoy, A.J.; Enrich, C.; Rentero, C.; Grewal, T. Linking Late Endosomal Cholesterol with Cancer Progression and Anticancer Drug Resistance. Int. J. Mol. Sci. 2022, 23, 7206. [Google Scholar] [CrossRef] [PubMed]
- Henriques Palma, G.B.; Kaur, M. Cholesterol Depletion Modulates Drug Resistance Pathways to Sensitize Resistant Breast Cancer Cells to Tamoxifen. Anticancer Res. 2022, 42, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Yue, W.; Wang, J.P.; Aiyar, S.; Li, Y.; Kim, T.H.; Santen, R.J. Mechanisms of Resistance to Structurally Diverse Antiestrogens Differ Under Premenopausal and Postmenopausal Conditions: Evidence from in vitro Breast Cancer Cell Models. Endocrinology 2009, 150, 2036–2045. [Google Scholar] [CrossRef]
- O’Grady, S.; Crown, J.; Duffy, M.J. Statins Inhibit Proliferation and Induce Apoptosis in Triple-Negative Breast Cancer Cells. Med. Oncol. 2022, 39, 142. [Google Scholar] [CrossRef]
- Zipinotti Dos Santos, D.; Santos Guimaraes, I.D.; Hakeem-Sanni, M.F.; Cochran, B.J.; Rye, K.A.; Grewal, T.; Hoy, A.J.; Rangel, L.B.A. Atorvastatin Improves cCisplatin Sensitivity Through Modulation of Cholesteryl Ester Homeostasis in Breast Cancer Cells. Discov. Oncol. 2022, 13, 135. [Google Scholar] [CrossRef]
- Borgquist, S.; Giobbie-Hurder, A.; Ahern, T.P.; Garber, J.E.; Colleoni, M.; Láng, I.; Debled, M.; Ejlertsen, B.; von Moos, R.; Smith, I.; et al. Cholesterol, Cholesterol-Lowering Medication Use, and Breast Cancer Outcome in the BIG 1-98 Study. J. Clin. Oncol. 2017, 35, 1179–1188. [Google Scholar] [CrossRef]
- Marti, J.L.G.; Beckwitt, C.H.; Clark, A.M.; Wells, A. Atorvastatin Facilitates Chemotherapy Effects in Metastatic Triple-Negative Breast Cancer. Br. J. Cancer 2021, 125, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Coradini, D.; Ambrogi, F. Cholesterol de novo Biosynthesis: A Promising Target to Overcome the Resistance to Aromatase Inhibitors in Postmenopausal Patients with ER-positive Breast Cancer. Explor. Med. 2023; in press. [Google Scholar] [CrossRef]
- Zwijsen, R.M.; Wientjens, E.; Klompmaker, R.; van der Sman, J.; Bernards, R.; Michalides, R.J. CDK-Independent Activation of Estrogen Receptor by Cyclin D1. Cell 1997, 88, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Zelivianski, S.; Cooley, A.; Kall, R.; Jeruss, J.S. Cyclin-Dependent Kinase 4-Mediated Phosphorylation Inhibits Smad3 Activity in Cyclin D-Overexpressing Breast Cancer Cells. Mol. Cancer Res. 2010, 8, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; McKenna, C.E. Farnesyl Pyrophosphate Synthase Modulators: A Patent Review (2006–2010). Expert Opin. Ther. Pat. 2011, 21, 1433–1451. [Google Scholar] [CrossRef] [PubMed]
- Drake, M.T.; Clarke, B.L.; Khosla, S. Bisphosphonates: Mechanism of Action and Role in Clinical Practice. Mayo Clin. Proc. 2008, 83, 1032–1045. [Google Scholar] [CrossRef]
- Iannelli, F.; Lombardi, R.; Milone, M.R.; Pucci, B.; De Rienzo, S.; Budillon, A.; Bruzzese, F. Targeting Mevalonate Pathway in Cancer Treatment: Repurposing of Statins. Recent Pat. Anticancer Drug Discov. 2018, 13, 184–200. [Google Scholar] [CrossRef]
- Bjarnadottir, O.; Romero, Q.; Bendahl, P.O.; Jirström, K.; Rydén, L.; Loman, N.; Uhlén, M.; Johannesson, H.; Rose, C.; Grabau, D.; et al. Targeting HMG-CoA Reductase with Statins in a Window-of-Opportunity Breast Cancer Trial. Breast Cancer Res. Treat. 2013, 138, 499–508. [Google Scholar] [CrossRef]
- Beckwitt, C.H.; Brufsky, A.; Oltvai, Z.N.; Wells, A. Statin Drugs to Reduce Breast Cancer Recurrence and Mortality. Breast Cancer Res. 2018, 20, 144. [Google Scholar] [CrossRef]
- Lluch, A.; Cueva, J.; Ruiz-Borrego, M.; Ponce, J.; Pérez-Fidalgo, J.A. Zoledronic Acid in the Treatment of Metastatic Breast Cancer. Anticancer Drugs 2014, 25, 1–7. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Embury, M.D.; Ju, Z.; Wang, J.; Bedrosian, I. Gene Signature Associated with Resistance to Fluvastatin Chemoprevention for Breast Cancer. BMC Cancer 2022, 22, 282. [Google Scholar] [CrossRef] [PubMed]
- Haney, S.L.; Varney, M.L.; Chhonker, Y.; Talmon, G.; Smith, L.M.; Murry, D.J.; Holstein, S.A. In vivo Evaluation of Combination Therapy Targeting the Isoprenoid Biosynthetic Pathway. Pharmacol. Res. 2021, 167, 105528. [Google Scholar] [CrossRef] [PubMed]
- Undela, K.; Srikanth, V.; Bansal, D. Statin Use and Risk of Breast Cancer: A Meta-Analysis of Observational Studies. Breast Cancer Res. Treat. 2012, 135, 261–269. [Google Scholar] [CrossRef] [PubMed]
- El-Refai, S.M.; Brown, J.D.; Arnold, S.M.; Black, E.P.; Leggas, M.; Talbert, J.C. Epidemiologic Analysis Along the Mevalonate Pathway Reveals Improved Cancer Survival in Patients Who Receive Statins Alone and in Combination With Bisphosphonates. JCO Clin. Cancer Inform. 2017, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Bandyopadhayaya, S.; Chowdhury, K.; Sharma, T.; Maheshwari, R.; Das, A.; Chakrabarti, G.; Kumar, V.; Mandal, C.C. Metformin Exhibited Anticancer Activity by Lowering Cellular Cholesterol Content in Breast Cancer Cells. PLoS ONE 2019, 14, e0209435. [Google Scholar] [CrossRef]
- Stein, E.A.; Bays, H.; O’Brien, D.; Pedicano, J.; Piper, E.; Spezzi, A. Lapaquistat Acetate: Development of a Squalene Synthase Inhibitor for the Treatment of Hypercholesterolemia. Circulation 2011, 123, 1974–1985. [Google Scholar] [CrossRef]
- Maione, F.; Oliaro-Bosso, S.; Meda, C.; Di Nicolantonio, F.; Bussolino, F.; Balliano, G.; Viola, F.; Giraudo, E. The Cholesterol Biosynthesis Enzyme Oxidosqualene Cyclase is a New Target to Impair Tumour Angiogenesis and Metastasis Dissemination. Sci. Rep. 2015, 5, 9054. [Google Scholar] [CrossRef]
- Liang, Y.; Besch-Williford, C.; Aebi, J.D.; Mafuvadze, B.; Cook, M.T.; Zou, X.; Hyder, S.M. Cholesterol Biosynthesis Inhibitors as Potent Novel Anti-cancer Agents: Suppression of Hormone-dependent Breast Cancer by the Oxidosqualene Cyclase Inhibitor RO 48-8071. Breast Cancer Res. Treat. 2014, 146, 51–62. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, S.; Tang, Q.; Xia, H.; Bi, F. Cholesterol Metabolism: New Functions and Therapeutic Approaches in Cancer. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188394. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coradini, D. Impact of De Novo Cholesterol Biosynthesis on the Initiation and Progression of Breast Cancer. Biomolecules 2024, 14, 64. https://doi.org/10.3390/biom14010064
Coradini D. Impact of De Novo Cholesterol Biosynthesis on the Initiation and Progression of Breast Cancer. Biomolecules. 2024; 14(1):64. https://doi.org/10.3390/biom14010064
Chicago/Turabian StyleCoradini, Danila. 2024. "Impact of De Novo Cholesterol Biosynthesis on the Initiation and Progression of Breast Cancer" Biomolecules 14, no. 1: 64. https://doi.org/10.3390/biom14010064
APA StyleCoradini, D. (2024). Impact of De Novo Cholesterol Biosynthesis on the Initiation and Progression of Breast Cancer. Biomolecules, 14(1), 64. https://doi.org/10.3390/biom14010064





