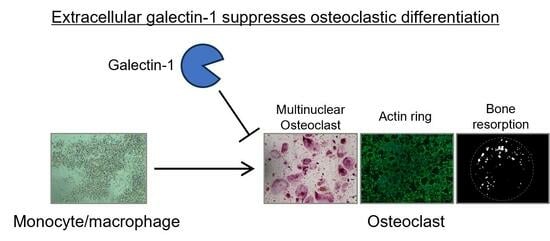
Reduced form of Galectin-1 Suppresses Osteoclastic Differentiation of Human Peripheral Blood Mononuclear Cells and Murine RAW264 Cells In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of Recombinant Gal-1 Proteins
2.3. Osteoclastic Differentiation
2.4. RNA Interference (RNAi)
2.5. Cell Viability Assay
2.6. TRAP Staining
2.7. Actin Staining
2.8. Real-Time Reverse Transcription PCR
2.9. Resorption Assay
2.10. Oxidation of Recombinant Gal-1 Protein
2.11. Hemagglutination Assay
2.12. Redox State Monitoring of Cys Residues
2.13. Statistical Analysis
3. Results
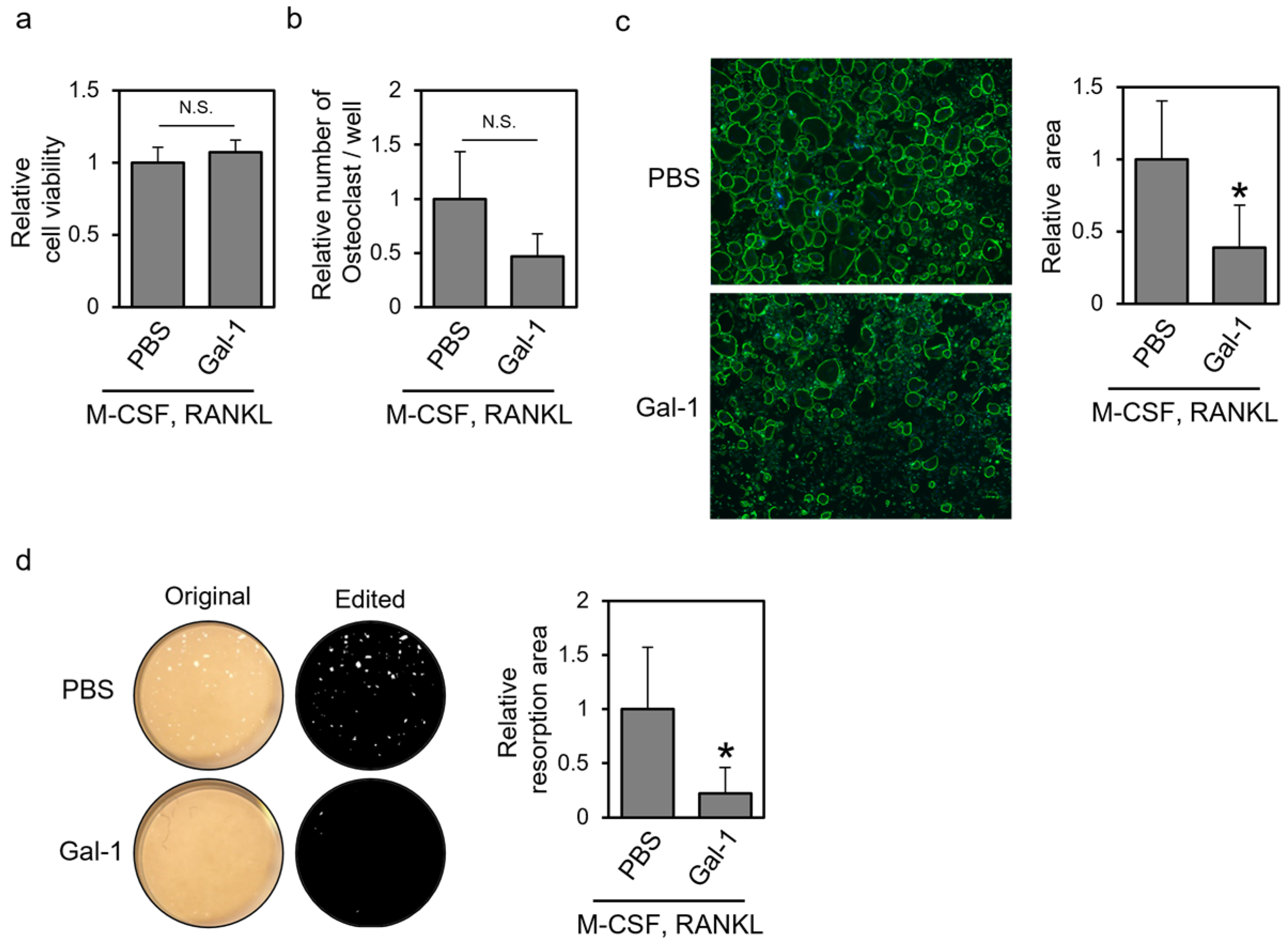
3.1. Recombinant Gal-1 Protein Suppresses Osteoclastic Differentiation of Human PBMCs
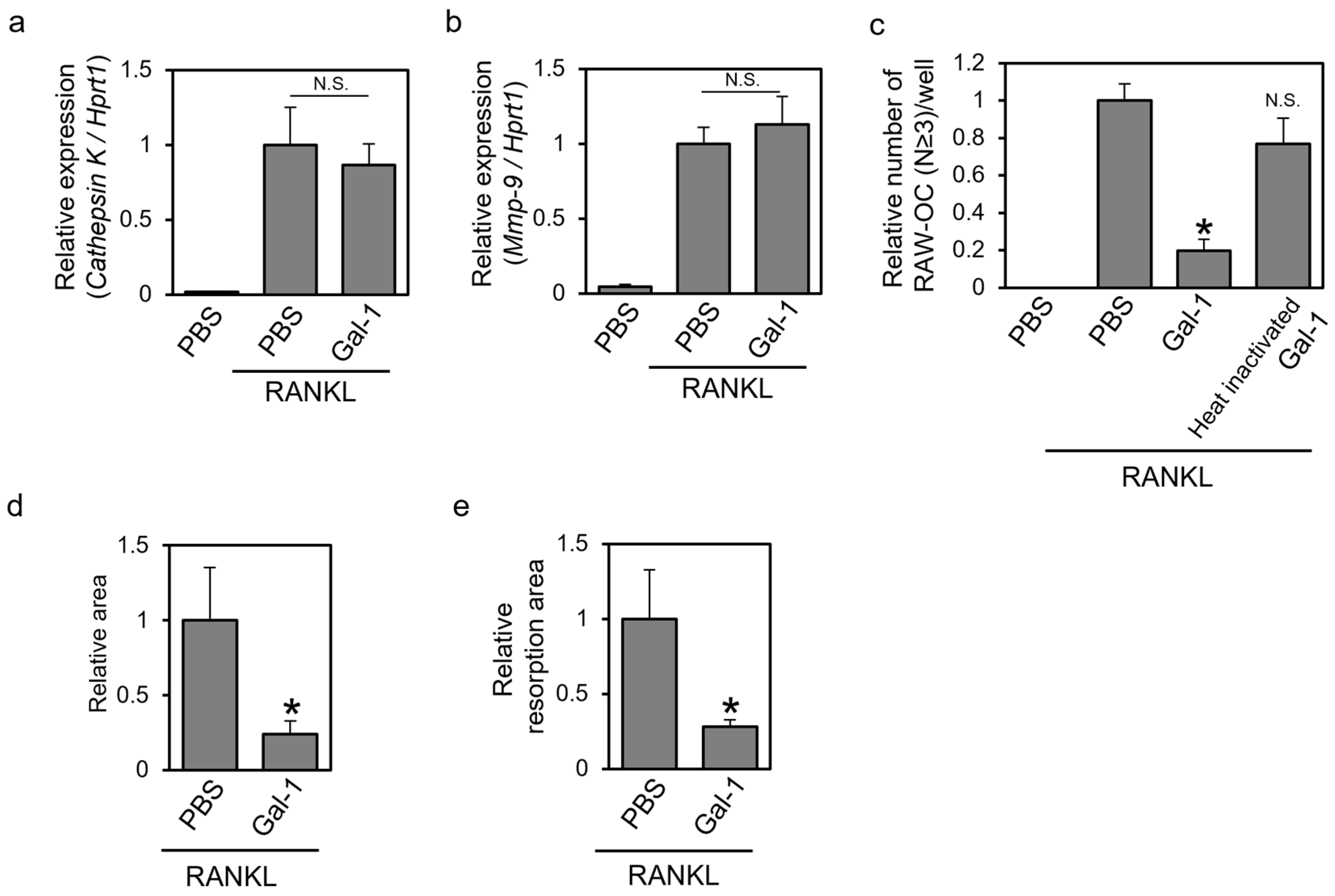
3.2. Recombinant Gal-1 Protein Suppresses the Osteoclastic Differentiation of Murine RAW264 Cells
3.3. Knockdown of Gal-1 Increases Formed TRAP-Positive Multinuclear Cells
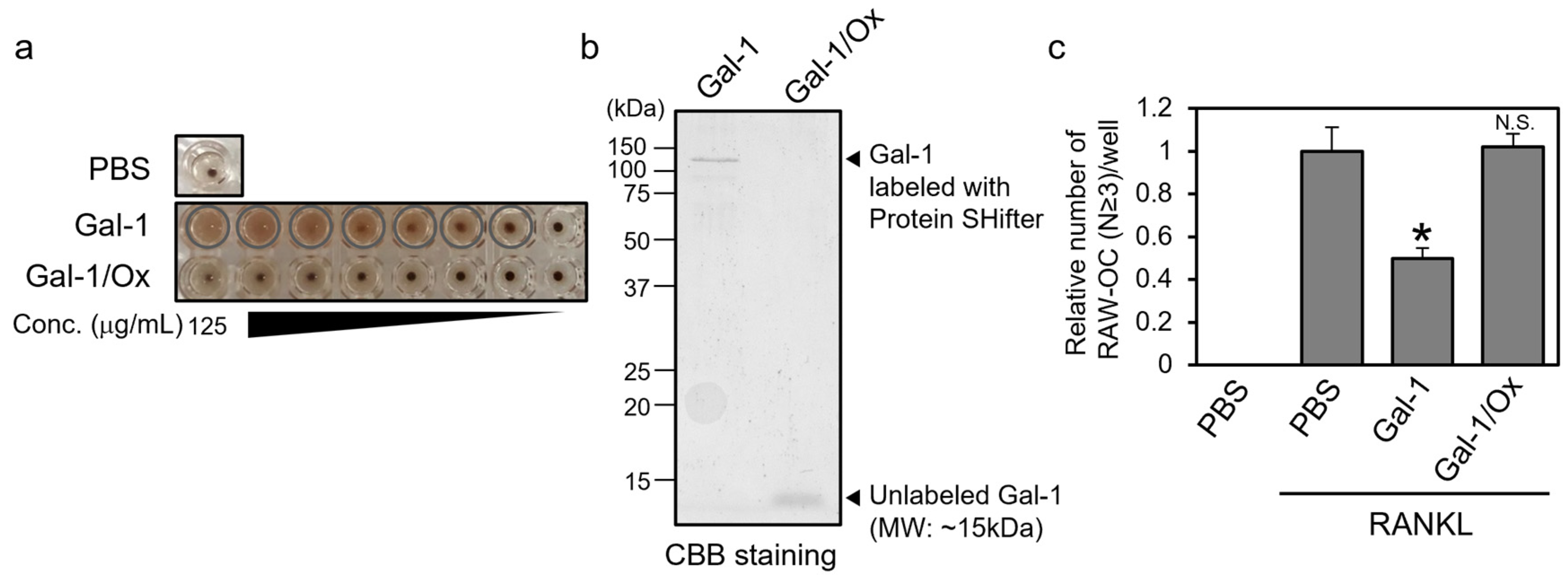
3.4. Reduced Form, Not Oxidized Form, of Gal-1 Suppresses the Formation of TRAP-Positive Multinuclear Cells
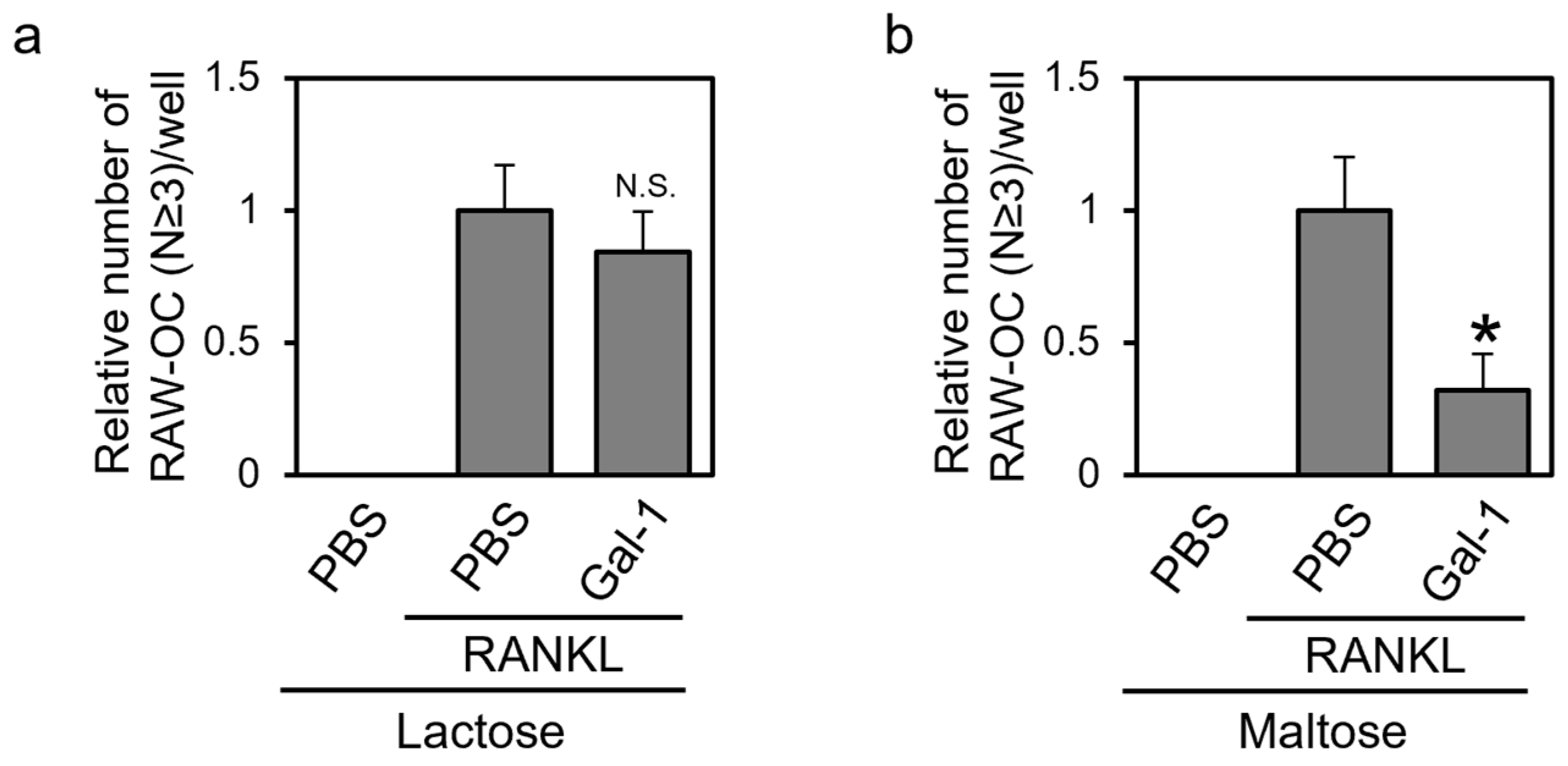
3.5. Gal-1 Suppresses the Formation of TRAP-Positive Multinuclear Cells in a β-Galactoside-dependent Manner
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikeda, K.; Takeshita, S. The role of osteoclast differentiation and function in skeletal homeostasis. J. Biochem. 2016, 159, 1–8. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Ren, F.; Ye, Y.; Wang, F.; Zheng, C.; Qian, Y.; Zhang, M. The Macrophage-Osteoclast Axis in Osteoimmunity and Osteo-Related Diseases. Front. Immunol. 2021, 12, 664871. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Nakashima, T. Recent advances in osteoclast biology. Histochem. Cell Biol. 2018, 149, 325–341. [Google Scholar] [CrossRef]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J. Cell Sci. 2018, 131, jcs208884. [Google Scholar] [CrossRef]
- Nehme, R.; St-Pierre, Y. Targeting intracellular galectins for cancer treatment. Front. Immunol. 2023, 14, 1269391. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.J.; Vasudevan, S.O.; Mendez-Huergo, S.P.; Kumari, P.; Menoret, A.; Duduskar, S.; Wang, C.; Perez Saez, J.M.; Fettis, M.M.; Li, C.; et al. Intracellular immune sensing promotes inflammation via gasdermin D-driven release of a lectin alarmin. Nat. Immunol. 2021, 22, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Sundblad, V.; Morosi, L.G.; Geffner, J.R.; Rabinovich, G.A. Galectin-1: A Jack-of-All-Trades in the Resolution of Acute and Chronic Inflammation. J. Immunol. 2017, 199, 3721–3730. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Kasai, K. Effect of amino acid substitution by sited-directed mutagenesis on the carbohydrate recognition and stability of human 14-kDa beta-galactoside-binding lectin. J. Biol. Chem. 1991, 266, 23648–23653. [Google Scholar] [CrossRef]
- Stowell, S.R.; Cho, M.; Feasley, C.L.; Arthur, C.M.; Song, X.; Colucci, J.K.; Karmakar, S.; Mehta, P.; Dias-Baruffi, M.; McEver, R.P.; et al. Ligand reduces galectin-1 sensitivity to oxidative inactivation by enhancing dimer formation. J. Biol. Chem. 2009, 284, 4989–4999. [Google Scholar] [CrossRef]
- Guardia, C.M.; Caramelo, J.J.; Trujillo, M.; Mendez-Huergo, S.P.; Radi, R.; Estrin, D.A.; Rabinovich, G.A. Structural basis of redox-dependent modulation of galectin-1 dynamics and function. Glycobiology 2014, 24, 428–441. [Google Scholar] [CrossRef]
- Robinson, B.S.; Arthur, C.M.; Evavold, B.; Roback, E.; Kamili, N.A.; Stowell, C.S.; Vallecillo-Zuniga, M.L.; Van Ry, P.M.; Dias-Baruffi, M.; Cummings, R.D.; et al. The Sweet-Side of Leukocytes: Galectins as Master Regulators of Neutrophil Function. Front. Immunol. 2019, 10, 1762. [Google Scholar] [CrossRef]
- Inagaki, Y.; Sohma, Y.; Horie, H.; Nozawa, R.; Kadoya, T. Oxidized galectin-1 promotes axonal regeneration in peripheral nerves but does not possess lectin properties. Eur. J. Biochem. 2000, 267, 2955–2964. [Google Scholar] [PubMed]
- Echigo, Y.; Sugiki, H.; Koizumi, Y.; Hikitsuchi, S.; Inoue, H. Activation of RAW264.7 macrophages by oxidized galectin-1. Immunol. Lett. 2010, 131, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Andersen, H.; Jensen, O.N.; Moiseeva, E.P.; Eriksen, E.F. A proteome study of secreted prostatic factors affecting osteoblastic activity: Galectin-1 is involved in differentiation of human bone marrow stromal cells. J. Bone Miner. Res. 2003, 18, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Zhang, Y.; Yu, J.; Tang, L. Galectin-1 deletion in mice causes bone loss via impaired osteogenic differentiation potential of BMSCs. FASEB J. 2022, 36, e22516. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.; Duray, E.; Lejeune, M.; Dubois, S.; Plougonven, E.; Leonard, A.; Storti, P.; Giuliani, N.; Cohen-Solal, M.; Hempel, U.; et al. Loss of Stromal Galectin-1 Enhances Multiple Myeloma Development: Emphasis on a Role in Osteoclasts. Cancers 2019, 11, 261. [Google Scholar] [CrossRef]
- Xu, W.; Ni, C.; Wang, Y.; Zheng, G.; Zhang, J.; Xu, Y. Age-related trabecular bone loss is associated with a decline in serum Galectin-1 level. BMC Musculoskelet. Disord. 2021, 22, 394. [Google Scholar] [CrossRef]
- Kong, L.; Smith, W.; Hao, D. Overview of RAW264.7 for osteoclastogensis study: Phenotype and stimuli. J. Cell Mol. Med. 2019, 23, 3077–3087. [Google Scholar] [CrossRef]
- Lampiasi, N.; Russo, R.; Kireev, I.; Strelkova, O.; Zhironkina, O.; Zito, F. Osteoclasts Differentiation from Murine RAW 264.7 Cells Stimulated by RANKL: Timing and Behavior. Biology 2021, 10, 117. [Google Scholar] [CrossRef]
- Raschke, W.C.; Baird, S.; Ralph, P.; Nakoinz, I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 1978, 15, 261–267. [Google Scholar] [CrossRef]
- Takeuchi, T.; Tamura, M.; Nishiyama, K.; Iwaki, J.; Hirabayashi, J.; Takahashi, H.; Natsugari, H.; Arata, Y.; Kasai, K. Mammalian galectins bind galactosebeta1-4fucose disaccharide, a unique structural component of protostomial N-type glycoproteins. Biochem. Biophys. Res. Commun. 2013, 436, 509–513. [Google Scholar] [CrossRef]
- Takeuchi, T. Preparation of Recombinant Galectin Protein: Expression, Affinity Purification, and Removal of Lipopolysaccharide. In Glycoscience Protocols (GlycoPODv2); Nishihara, S., Angata, K., Aoki-Kinoshita, K.F., Hirabayashi, J., Eds.; Japan Consortium for Glycobiology and Glycotechnology: Saitama, Japan, 2021. [Google Scholar]
- Takeuchi, T.; Sugimoto, A.; Imazato, N.; Tamura, M.; Nakatani, S.; Kobata, K.; Arata, Y. Glucosamine Suppresses Osteoclast Differentiation through the Modulation of Glycosylation Including O-GlcNAcylation. Biol. Pharm. Bull. 2017, 40, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Nagasaka, M.; Shimizu, M.; Tamura, M.; Arata, Y. N-acetylglucosamine suppresses osteoclastogenesis in part through the promotion of O-GlcNAcylation. Bone Rep. 2016, 5, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Nairn, A.V.; York, W.S.; Harris, K.; Hall, E.M.; Pierce, J.M.; Moremen, K.W. Regulation of glycan structures in animal tissues: Transcript profiling of glycan-related genes. J. Biol. Chem. 2008, 283, 17298–17313. [Google Scholar] [CrossRef]
- Takeuchi, T.; Nakamura, R.; Hamasaki, M.; Oyama, M.; Hamano, S.; Hatanaka, T. In vitro evaluation of the effect of galectins on Schistosoma mansoni motility. BMC Res. Notes 2023, 16, 266. [Google Scholar] [CrossRef]
- Gauthier, L.; Rossi, B.; Roux, F.; Termine, E.; Schiff, C. Galectin-1 is a stromal cell ligand of the pre-B cell receptor (BCR) implicated in synapse formation between pre-B and stromal cells and in pre-BCR triggering. Proc. Natl. Acad. Sci. USA 2002, 99, 13014–13019. [Google Scholar] [CrossRef] [PubMed]
- Elantak, L.; Espeli, M.; Boned, A.; Bornet, O.; Bonzi, J.; Gauthier, L.; Feracci, M.; Roche, P.; Guerlesquin, F.; Schiff, C. Structural basis for galectin-1-dependent pre-B cell receptor (pre-BCR) activation. J. Biol. Chem. 2012, 287, 44703–44713. [Google Scholar] [CrossRef]
- Toudic, C.; Vargas, A.; Xiao, Y.; St-Pierre, G.; Bannert, N.; Lafond, J.; Rassart, E.; Sato, S.; Barbeau, B. Galectin-1 interacts with the human endogenous retroviral envelope protein syncytin-2 and potentiates trophoblast fusion in humans. FASEB J. 2019, 33, 12873–12887. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, H.; Yang, X.; Fan, L.; Tian, S.; Niu, R.; Yan, M.; Zheng, M.; Zhang, S. Cell Fusion-Related Proteins and Signaling Pathways, and Their Roles in the Development and Progression of Cancer. Front. Cell Dev. Biol. 2021, 9, 809668. [Google Scholar] [CrossRef]
- Soe, K.; Andersen, T.L.; Hobolt-Pedersen, A.S.; Bjerregaard, B.; Larsson, L.I.; Delaisse, J.M. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone 2011, 48, 837–846. [Google Scholar] [CrossRef]
- Kodama, J.; Kaito, T. Osteoclast Multinucleation: Review of Current Literature. Int. J. Mol. Sci. 2020, 21, 5685. [Google Scholar] [CrossRef] [PubMed]
- Avni, O.; Pur, Z.; Yefenof, E.; Baniyash, M. Complement receptor 3 of macrophages is associated with galectin-1-like protein. J. Immunol. 1998, 160, 6151–6158. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Nakahama, K.; Sato, T.; Tuchiya, T.; Asakawa, Y.; Maemura, T.; Tanaka, M.; Morita, M.; Morita, I. The role of Mac-1 (CD11b/CD18) in osteoclast differentiation induced by receptor activator of nuclear factor-kappaB ligand. FEBS Lett. 2008, 582, 3243–3248. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, M.A.; Croci, D.O.; Lapponi, M.J.; Tribulatti, M.V.; Negrotto, S.; Poirier, F.; Campetella, O.; Rabinovich, G.A.; Schattner, M. Binding of galectin-1 to alphaIIbbeta(3) integrin triggers "outside-in" signals, stimulates platelet activation, and controls primary hemostasis. FASEB J. 2012, 26, 2788–2798. [Google Scholar] [CrossRef]
- Vilen, Z.; Joeh, E.; Critcher, M.; Parker, C.G.; Huang, M.L. Proximity Tagging Identifies the Glycan-Mediated Glycoprotein Interactors of Galectin-1 in Muscle Stem Cells. ACS Chem. Biol. 2021, 16, 1994–2003. [Google Scholar] [CrossRef]
- McHugh, K.P.; Hodivala-Dilke, K.; Zheng, M.H.; Namba, N.; Lam, J.; Novack, D.; Feng, X.; Ross, F.P.; Hynes, R.O.; Teitelbaum, S.L. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J. Clin. Investig. 2000, 105, 433–440. [Google Scholar] [CrossRef]
- Georgiadis, V.; Stewart, H.J.; Pollard, H.J.; Tavsanoglu, Y.; Prasad, R.; Horwood, J.; Deltour, L.; Goldring, K.; Poirier, F.; Lawrence-Watt, D.J. Lack of galectin-1 results in defects in myoblast fusion and muscle regeneration. Dev. Dyn. 2007, 236, 1014–1024. [Google Scholar] [CrossRef]
- Blazev, R.; Ashwood, C.; Abrahams, J.L.; Chung, L.H.; Francis, D.; Yang, P.; Watt, K.I.; Qian, H.; Quaife-Ryan, G.A.; Hudson, J.E.; et al. Integrated Glycoproteomics Identifies a Role of N-Glycosylation and Galectin-1 on Myogenesis and Muscle Development. Mol. Cell. Proteom. 2021, 20, 100030. [Google Scholar] [CrossRef]
- Cerri, D.G.; Rodrigues, L.C.; Stowell, S.R.; Araujo, D.D.; Coelho, M.C.; Oliveira, S.R.; Bizario, J.C.; Cummings, R.D.; Dias-Baruffi, M.; Costa, M.C. Degeneration of dystrophic or injured skeletal muscles induces high expression of Galectin-1. Glycobiology 2008, 18, 842–850. [Google Scholar] [CrossRef]
- Kaji, H. Effects of myokines on bone. BoneKey Rep. 2016, 5, 826. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Xiao, W.; Xie, W.; Fu, X.; Pan, L.; Jin, H.; Yu, Y.; Zhang, Y.; Li, Y. The Role of Osteokines in Sarcopenia: Therapeutic Directions and Application Prospects. Front. Cell Dev. Biol. 2021, 9, 735374. [Google Scholar] [CrossRef] [PubMed]
- Lecourt, S.; Lepelletier, Y.; Vanneaux, V.; Jarray, R.; Domet, T.; Raynaud, F.; Hadj-Slimane, R.; Cras, A.; Hermine, O.; Marolleau, J.P.; et al. Human Muscle Progenitor Cells Displayed Immunosuppressive Effect through Galectin-1 and Semaphorin-3A. Stem. Cells Int. 2012, 2012, 412610. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeuchi, T.; Oyama, M.; Tamura, M.; Arata, Y.; Hatanaka, T. Reduced form of Galectin-1 Suppresses Osteoclastic Differentiation of Human Peripheral Blood Mononuclear Cells and Murine RAW264 Cells In Vitro. Biomolecules 2024, 14, 121. https://doi.org/10.3390/biom14010121
Takeuchi T, Oyama M, Tamura M, Arata Y, Hatanaka T. Reduced form of Galectin-1 Suppresses Osteoclastic Differentiation of Human Peripheral Blood Mononuclear Cells and Murine RAW264 Cells In Vitro. Biomolecules. 2024; 14(1):121. https://doi.org/10.3390/biom14010121
Chicago/Turabian StyleTakeuchi, Tomoharu, Midori Oyama, Mayumi Tamura, Yoichiro Arata, and Tomomi Hatanaka. 2024. "Reduced form of Galectin-1 Suppresses Osteoclastic Differentiation of Human Peripheral Blood Mononuclear Cells and Murine RAW264 Cells In Vitro" Biomolecules 14, no. 1: 121. https://doi.org/10.3390/biom14010121
APA StyleTakeuchi, T., Oyama, M., Tamura, M., Arata, Y., & Hatanaka, T. (2024). Reduced form of Galectin-1 Suppresses Osteoclastic Differentiation of Human Peripheral Blood Mononuclear Cells and Murine RAW264 Cells In Vitro. Biomolecules, 14(1), 121. https://doi.org/10.3390/biom14010121