The History and Science of the Major Birch Pollen Allergen Bet v 1
Abstract
1. Events That Led to the Discovery of the Major Birch Pollen Allergen Bet v 1
2. The PR-10-like Family of Allergenic Proteins
3. Natural Ligands of PR-10-like Allergenic Proteins and Their Biological Functions
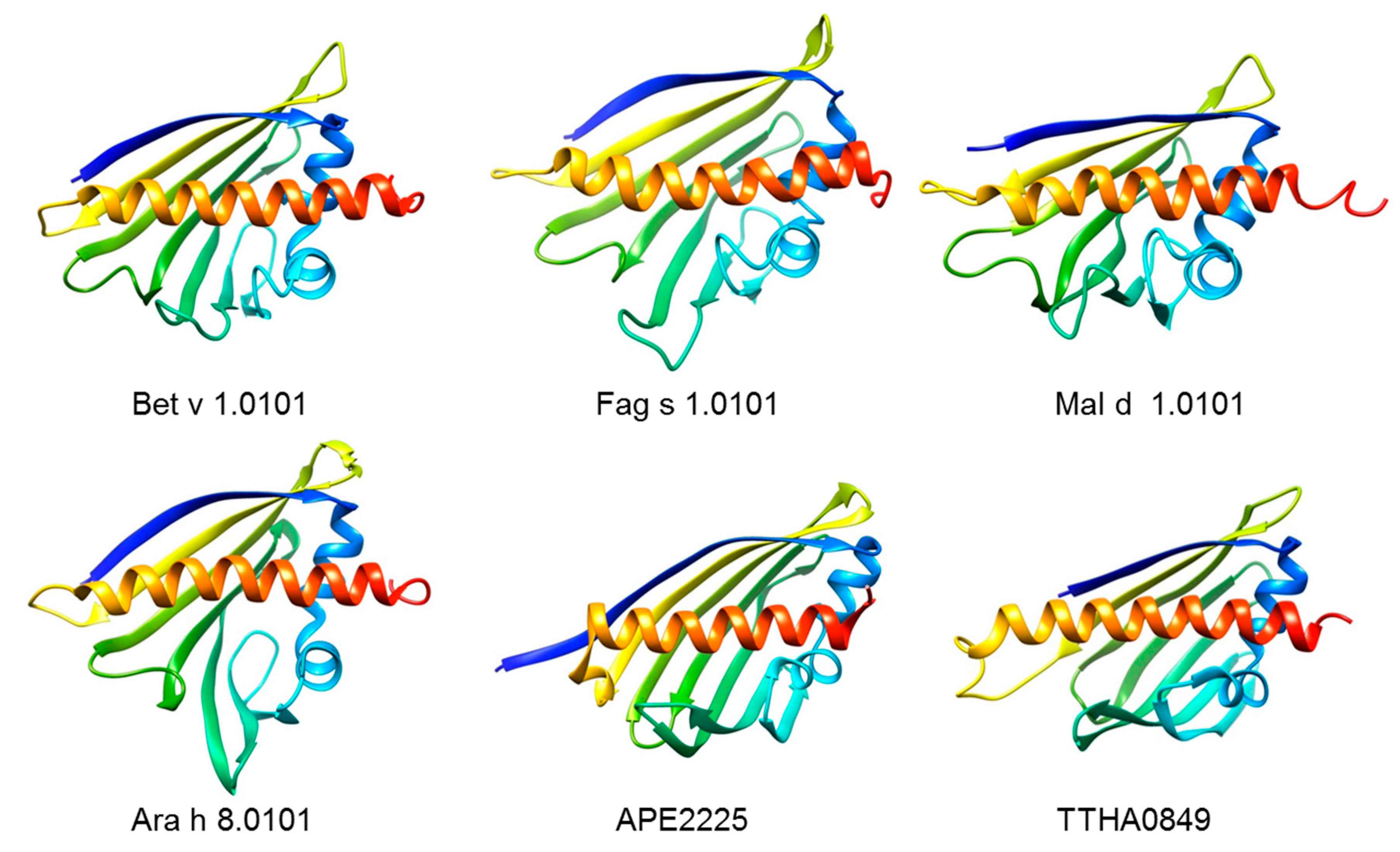
4. The Bet v 1-like Superfamily of Proteins
5. The Yeast Connection
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Breiteneder, H.; Hendler, P.N.; Kraft, D. Legends of allergy and immunology: Clemens von Pirquet. Allergy 2020, 75, 1276–1277. [Google Scholar] [CrossRef]
- Breiteneder, H. Legends of allergy/immunology: Dietrich Kraft. Allergy 2019, 74, 1591–1593. [Google Scholar] [CrossRef]
- King, T.P.; Hoffman, D.; Lowenstein, H.; Marsh, D.G.; Platts-Mills, T.A.; Thomas, W. Allergen nomenclature. Allergy 1995, 50, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Seiser, C.; Loffelhardt, W.; Michalowski, C.; Bohnert, H.J. Physical map and protein gene map of cyanelle DNA from the second known isolate of Cyanophora paradoxa (Kies-strain). Curr. Genet. 1988, 13, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Hassfeld, W.; Pettenburger, K.; Jarolim, E.; Breitenbach, M.; Rumpold, H.; Kraft, D.; Scheiner, O. Isolation and characterization of messenger RNA from male inflorescences and pollen of the white birch (Betula verrucosa). Int. Arch. Allergy Appl. Immunol. 1988, 87, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Pettenburger, K.; Bito, A.; Valenta, R.; Kraft, D.; Rumpold, H.; Scheiner, O.; Breitenbach, M. The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J. 1989, 8, 1935–1938. [Google Scholar] [CrossRef] [PubMed]
- Valenta, R.; Breiteneder, H.; Petternburger, K.; Breitenbach, M.; Rumpold, H.; Kraft, D.; Scheiner, O. Homology of the major birch-pollen allergen, Bet v I, with the major pollen allergens of alder, hazel, and hornbeam at the nucleic acid level as determined by cross-hybridization. J. Allergy Clin. Immunol. 1991, 87, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Ferreira, F.; Reikerstorfer, A.; Duchene, M.; Valenta, R.; Hoffmann-Sommergruber, K.; Ebner, C.; Breitenbach, M.; Kraft, D.; Scheiner, O. Complementary DNA cloning and expression in Escherichia coli of Aln g I, the major allergen in pollen of alder (Alnus glutinosa). J. Allergy Clin. Immunol. 1992, 90, 909–917. [Google Scholar] [CrossRef]
- Breiteneder, H.; Ferreira, F.; Hoffmann-Sommergruber, K.; Ebner, C.; Breitenbach, M.; Rumpold, H.; Kraft, D.; Scheiner, O. Four recombinant isoforms of Cor a I, the major allergen of hazel pollen, show different IgE-binding properties. Eur. J. Biochem. 1993, 212, 355–362. [Google Scholar] [CrossRef]
- Vanek-Krebitz, M.; Hoffmann-Sommergruber, K.; Laimer da Camara Machado, M.; Susani, M.; Ebner, C.; Kraft, D.; Scheiner, O.; Breiteneder, H. Cloning and sequencing of Mal d 1, the major allergen from apple (Malus domestica), and its immunological relationship to Bet v 1, the major birch pollen allergen. Biochem. Biophys. Res. Commun. 1995, 214, 538–551. [Google Scholar] [CrossRef]
- Breiteneder, H.; Hoffmann-Sommergruber, K.; O’Riordain, G.; Susani, M.; Ahorn, H.; Ebner, C.; Kraft, D.; Scheiner, O. Molecular characterization of Api g 1, the major allergen of celery (Apium graveolens), and its immunological and structural relationships to a group of 17-kDa tree pollen allergens. Eur. J. Biochem. 1995, 233, 484–489. [Google Scholar] [CrossRef]
- Antoniw, J.F.; Ritter, C.E.; Pierpoint, W.S.; Vanloon, L.C. Comparison of 3 pathogenesis-related proteins from plants of 2 cultivars of tobacco infected with TMV. J. Gen. Virol. 1980, 47, 79–87. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Sancho, A.I.; Wangorsch, A.; Jensen, B.M.; Watson, A.; Alexeev, Y.; Johnson, P.E.; Mackie, A.R.; Neubauer, A.; Reese, G.; Ballmer-Weber, B.; et al. Responsiveness of the major birch allergen Bet v 1 scaffold to the gastric environment: Impact on structure and allergenic activity. Mol. Nutr. Food Res. 2011, 55, 1690–1699. [Google Scholar] [CrossRef] [PubMed]
- Rib-Schmidt, C.; Riedl, P.; Meisinger, V.; Schwaben, L.; Schulenborg, T.; Reuter, A.; Schiller, D.; Seutter von Loetzen, C.; Rosch, P. pH and heat resistance of the major celery allergen Api g 1. Mol. Nutr. Food Res. 2018, 62, e1700886. [Google Scholar] [CrossRef]
- Jacob, T.; Vogel, L.; Reuter, A.; Wangorsch, A.; Kring, C.; Mahler, V.; Wohrl, B.M. Food processing does not abolish the allergenicity of the carrot allergen Dau c 1: Influence of pH, temperature, and the food matrix. Mol. Nutr. Food Res. 2020, 64, e2000334. [Google Scholar] [CrossRef] [PubMed]
- Dramburg, S.; Hilger, C.; Santos, A.F.; de Las Vecillas, L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; Altmann, F.; Arruda, K.L.; Asero, R.; et al. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34 (Suppl. S28), e13854. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.; Asam, C.; Himly, M.; Palazzo, P.; Voltolini, S.; Montanari, C.; Briza, P.; Bernardi, M.L.; Mari, A.; Ferreira, F.; et al. Bet v 1-like pollen allergens of multiple Fagales species can sensitize atopic individuals. Clin. Exp. Allergy 2011, 41, 1804–1814. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.N.; Stroman, P.; Ipsen, H. PCR based cloning and sequencing of isogenes encoding the tree pollen major allergen Car b I from Carpinus betulus, hornbeam. Mol. Immunol. 1992, 29, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Wallner, M.; Erler, A.; Hauser, M.; Klinglmayr, E.; Gadermaier, G.; Vogel, L.; Mari, A.; Bohle, B.; Briza, P.; Ferreira, F. Immunologic characterization of isoforms of Car b 1 and Que a 1, the major hornbeam and oak pollen allergens. Allergy 2009, 64, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.Y.; Lee, J.; Sang, M.K.; Lee, Y.S.; Park, K.H.; Lee, J.H.; Park, J.W. Sensitization profile to sawtooth oak component allergens and their clinical implications. J. Clin. Lab. Anal. 2021, 35, e23825. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, M.; Guerrero-Sanchez, V.M.; Canales-Bueno, N.; Loli-Ausejo, D.; Castillejo, M.A.; Quirce, S.; Jorrin-Novo, J.V.; Rodriguez-Perez, R. Quercus ilex pollen allergen, Que i 1, responsible for pollen food allergy syndrome caused by fruits in Spanish allergic patients. Clin. Exp. Allergy 2020, 50, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Yang, M.; Jeong, K.Y.; Sim, D.W.; Park, J.H.; Park, K.H.; Lee, J.H.; Park, J.W. Characterization of a major allergen from Mongolian oak, Quercus mongolica, a dominant species of oak in Korea. Int. Arch. Allergy Immunol. 2017, 174, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Vieths, S.; Scheurer, S.; Ballmer-Weber, B. Current understanding of cross-reactivity of food allergens and pollen. Ann. N. Y. Acad. Sci. 2002, 964, 47–68. [Google Scholar] [CrossRef]
- Oberhuber, C.; Bulley, S.M.; Ballmer-Weber, B.K.; Bublin, M.; Gaier, S.; DeWitt, A.M.; Briza, P.; Hofstetter, G.; Lidholm, J.; Vieths, S.; et al. Characterization of Bet v 1-related allergens from kiwifruit relevant for patients with combined kiwifruit and birch pollen allergy. Mol. Nutr. Food Res. 2008, 52 (Suppl. S2), S230–S240. [Google Scholar] [CrossRef]
- D’Avino, R.; Bernardi, M.L.; Wallner, M.; Palazzo, P.; Camardella, L.; Tuppo, L.; Alessandri, C.; Breiteneder, H.; Ferreira, F.; Ciardiello, M.A.; et al. Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy 2011, 66, 870–877. [Google Scholar] [CrossRef]
- Zhao, L.; Xie, H.; Wang, X.; Liu, M.; Ma, T.; Fu, W.; Wang, Y.; Wu, D.; Feng, Y.; Liu, Y.; et al. Molecular characterization of allergens and component-resolved diagnosis of IgE-mediated mango fruit allergy. Allergy 2023. [Google Scholar] [CrossRef]
- Hoffmann-Sommergruber, K.; O’Riordain, G.; Ahorn, H.; Ebner, C.; Laimer Da Camara Machado, M.; Puhringer, H.; Scheiner, O.; Breiteneder, H. Molecular characterization of Dau c 1, the Bet v 1 homologous protein from carrot and its cross-reactivity with Bet v 1 and Api g 1. Clin. Exp. Allergy 1999, 29, 840–847. [Google Scholar] [CrossRef]
- Ebo, D.G.; Decuyper, I.I.; Rihs, H.P.; Mertens, C.; Van Gasse, A.L.; van der Poorten, M.M.; De Puysseleyr, L.; Faber, M.A.; Hagendorens, M.M.; Bridts, C.H.; et al. IgE-binding and mast cell-activating capacity of the homologue of the major birch pollen allergen and profilin from Cannabis sativa. J. Allergy Clin. Immunol. Pract. 2021, 9, 2509–2512.e3. [Google Scholar] [CrossRef]
- Luttkopf, D.; Muller, U.; Skov, P.S.; Ballmer-Weber, B.K.; Wuthrich, B.; Skamstrup Hansen, K.; Poulsen, L.K.; Kastner, M.; Haustein, D.; Vieths, S. Comparison of four variants of a major allergen in hazelnut (Corylus avellana) Cor a 1.04 with the major hazel pollen allergen Cor a 1.01. Mol. Immunol. 2002, 38, 515–525. [Google Scholar] [CrossRef]
- Mittag, D.; Akkerdaas, J.; Ballmer-Weber, B.K.; Vogel, L.; Wensing, M.; Becker, W.M.; Koppelman, S.J.; Knulst, A.C.; Helbling, A.; Hefle, S.L.; et al. Ara h 8, a Bet v 1-homologous allergen from peanut, is a major allergen in patients with combined birch pollen and peanut allergy. J. Allergy Clin. Immunol. 2004, 114, 1410–1417. [Google Scholar] [CrossRef]
- Crowell, D.N.; John, M.E.; Russell, D.; Amasino, R.M. Characterization of a stress-induced, developmentally regulated gene family from soybean. Plant Mol. Biol. 1992, 18, 459–466. [Google Scholar] [CrossRef]
- Mittag, D.; Vieths, S.; Vogel, L.; Wagner-Loew, D.; Starke, A.; Hunziker, P.; Becker, W.M.; Ballmer-Weber, B.K. Birch pollen-related food allergy to legumes: Identification and characterization of the Bet v 1 homologue in mungbean (Vigna radiata), Vig r 1. Clin. Exp. Allergy 2005, 35, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Guhsl, E.E.; Hofstetter, G.; Hemmer, W.; Ebner, C.; Vieths, S.; Vogel, L.; Breiteneder, H.; Radauer, C. Vig r 6, the cytokinin-specific binding protein from mung bean (Vigna radiata) sprouts, cross-reacts with Bet v 1-related allergens and binds IgE from birch pollen allergic patients’ sera. Mol. Nutr. Food Res. 2014, 58, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Wangorsch, A.; Jamin, A.; Lidholm, J.; Grani, N.; Lang, C.; Ballmer-Weber, B.; Vieths, S.; Scheurer, S. Identification and implication of an allergenic PR-10 protein from walnut in birch pollen associated walnut allergy. Mol. Nutr. Food Res. 2017, 61, 1600902. [Google Scholar] [CrossRef] [PubMed]
- Musidlowska-Persson, A.; Alm, R.; Emanuelsson, C. Cloning and sequencing of the Bet v 1-homologous allergen Fra a 1 in strawberry (Fragaria ananassa) shows the presence of an intron and little variability in amino acid sequence. Mol. Immunol. 2007, 44, 1245–1252. [Google Scholar] [CrossRef]
- Scheurer, S.; Pastorello, E.A.; Wangorsch, A.; Kastner, M.; Haustein, D.; Vieths, S. Recombinant allergens Pru av 1 and Pru av 4 and a newly identified lipid transfer protein in the in vitro diagnosis of cherry allergy. J. Allergy Clin. Immunol. 2001, 107, 724–731. [Google Scholar] [CrossRef]
- Kabasser, S.; Crvenjak, N.; Schmalz, S.; Kalic, T.; Hafner, C.; Dubiela, P.; Kucharczyk, A.; Bazan-Socha, S.; Lukaszyk, M.; Breiteneder, H.; et al. Pru du 1, the Bet v 1-homologue from almond, is a major allergen in patients with birch pollen associated almond allergy. Clin. Transl. Allergy 2022, 12, e12177. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.; Bassett, C.; Arora, R. Distribution and partial characterization of seasonally expressed proteins in different aged shoots and roots of ‘Loring’ peach (Prunus persica). Tree Physiol. 2004, 24, 339–345. [Google Scholar] [CrossRef]
- Karamloo, F.; Scheurer, S.; Wangorsch, A.; May, S.; Haustein, D.; Vieths, S. Pyr c 1, the major allergen from pear (Pyrus communis), is a new member of the Bet v 1 allergen family. J. Chromatogr. B Biomed. Sci. Appl. 2001, 756, 281–293. [Google Scholar] [CrossRef]
- Marzban, G.; Herndl, A.; Kolarich, D.; Maghuly, F.; Mansfeld, A.; Hemmer, W.; Katinger, H.; Laimer, M. Identification of four IgE-reactive proteins in raspberry (Rubus ideaeus L.). Mol. Nutr. Food Res. 2008, 52, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Wangorsch, A.; Jamin, A.; Foetisch, K.; Malczyk, A.; Reuter, A.; Vierecke, S.; Schulke, S.; Bartel, D.; Mahler, V.; Lidholm, J.; et al. Identification of Sola l 4 as Bet v 1 homologous pathogenesis related-10 allergen in tomato fruits. Mol. Nutr. Food Res. 2015, 59, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Tebbe, J.; Vogel, L.; Crowell, D.N.; Haustein, U.F.; Vieths, S. Severe oral allergy syndrome and anaphylactic reactions caused by a Bet v 1- related PR-10 protein in soybean, SAM22. J. Allergy Clin. Immunol. 2002, 110, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Gajhede, M.; Osmark, P.; Poulsen, F.M.; Ipsen, H.; Larsen, J.N.; Joost van Neerven, R.J.; Schou, C.; Lowenstein, H.; Spangfort, M.D. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat. Struct. Biol. 1996, 3, 1040–1045. [Google Scholar] [CrossRef]
- Mogensen, J.E.; Wimmer, R.; Larsen, J.N.; Spangfort, M.D.; Otzen, D.E. The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 2002, 277, 23684–23692. [Google Scholar] [CrossRef]
- Kofler, S.; Asam, C.; Eckhard, U.; Wallner, M.; Ferreira, F.; Brandstetter, H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen bet v 1. J. Mol. Biol. 2012, 422, 109–123. [Google Scholar] [CrossRef]
- Aglas, L.; Soh, W.T.; Kraiem, A.; Wenger, M.; Brandstetter, H.; Ferreira, F. Ligand Binding of PR-10 Proteins with a Particular Focus on the Bet v 1 Allergen Family. Curr. Allergy Asthma Rep. 2020, 20, 25. [Google Scholar] [CrossRef]
- McBride, J.K.; Cheng, H.; Maleki, S.J.; Hurlburt, B.K. Purification and characterization of pathogenesis related class 10 panallergens. Foods 2019, 8, 609. [Google Scholar] [CrossRef]
- Seutter von Loetzen, C.; Hoffmann, T.; Hartl, M.J.; Schweimer, K.; Schwab, W.; Rosch, P.; Hartl-Spiegelhauer, O. Secret of the major birch pollen allergen Bet v 1: Identification of the physiological ligand. Biochem. J. 2014, 457, 379–390. [Google Scholar] [CrossRef]
- Brunetti, C.; Fini, A.; Sebastiani, F.; Gori, A.; Tattini, M. Modulation of Phytohormone Signaling: A Primary Function of Flavonoids in Plant-Environment Interactions. Front. Plant Sci. 2018, 9, 1042. [Google Scholar] [CrossRef]
- Agati, G.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Pollastri, S.; Tattini, M. Functional roles of flavonoids in photoprotection: New evidence, lessons from the past. Plant Physiol. Biochem. 2013, 72, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Grutsch, S.; Fuchs, J.E.; Freier, R.; Kofler, S.; Bibi, M.; Asam, C.; Wallner, M.; Ferreira, F.; Brandstetter, H.; Liedl, K.R.; et al. Ligand binding modulates the structural dynamics and compactness of the major birch pollen allergen. Biophys. J. 2014, 107, 2972–2981. [Google Scholar] [CrossRef] [PubMed]
- Asam, C.; Batista, A.L.; Moraes, A.H.; de Paula, V.S.; Almeida, F.C.; Aglas, L.; Kitzmuller, C.; Bohle, B.; Ebner, C.; Ferreira, F.; et al. Bet v 1—A Trojan horse for small ligands boosting allergic sensitization? Clin. Exp. Allergy 2014, 44, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Soh, W.T.; Aglas, L.; Mueller, G.A.; Gilles, S.; Weiss, R.; Scheiblhofer, S.; Huber, S.; Scheidt, T.; Thompson, P.M.; Briza, P.; et al. Multiple roles of Bet v 1 ligands in allergen stabilization and modulation of endosomal protease activity. Allergy 2019, 74, 2382–2393. [Google Scholar] [CrossRef]
- Freier, R.; Dall, E.; Brandstetter, H. Protease recognition sites in Bet v 1a are cryptic, explaining its slow processing relevant to its allergenicity. Sci. Rep. 2015, 5, 12707. [Google Scholar] [CrossRef]
- Seutter von Loetzen, C.; Jacob, T.; Hartl-Spiegelhauer, O.; Vogel, L.; Schiller, D.; Sporlein-Guttler, C.; Schobert, R.; Vieths, S.; Hartl, M.J.; Rosch, P. Ligand recognition of the major birch pollen allergen Bet v 1 is isoform dependent. PLoS ONE 2015, 10, e0128677. [Google Scholar] [CrossRef]
- Jacob, T.; von Loetzen, C.S.; Reuter, A.; Lacher, U.; Schiller, D.; Schobert, R.; Mahler, V.; Vieths, S.; Rosch, P.; Schweimer, K.; et al. Identification of a natural ligand of the hazel allergen Cor a 1. Sci. Rep. 2019, 9, 8714. [Google Scholar] [CrossRef] [PubMed]
- Hjerno, K.; Alm, R.; Canback, B.; Matthiesen, R.; Trajkovski, K.; Bjork, L.; Roepstorff, P.; Emanuelsson, C. Down-regulation of the strawberry Bet v 1-homologous allergen in concert with the flavonoid biosynthesis pathway in colorless strawberry mutant. Proteomics 2006, 6, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Casanal, A.; Zander, U.; Munoz, C.; Dupeux, F.; Luque, I.; Botella, M.A.; Schwab, W.; Valpuesta, V.; Marquez, J.A. The strawberry pathogenesis-related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J. Biol. Chem. 2013, 288, 35322–35332. [Google Scholar] [CrossRef]
- Lackner, P.; Koppensteiner, W.A.; Sippl, M.J.; Domingues, F.S. ProSup: A refined tool for protein structure alignment. Protein Eng. 2000, 13, 745–752. [Google Scholar] [CrossRef]
- Radauer, C.; Lackner, P.; Breiteneder, H. The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 2008, 8, 286. [Google Scholar] [CrossRef] [PubMed]
- Sako, Y.; Nomura, N.; Uchida, A.; Ishida, Y.; Morii, H.; Koga, Y.; Hoaki, T.; Maruyama, T. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int. J. Syst. Bacteriol. 1996, 46, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, M.; Shibata, N.; Komori, H.; Ueda, Y.; Iino, H.; Ebihara, A.; Kuramitsu, S.; Higuchi, Y. Structure of a conserved hypothetical protein, TTHA0849 from Thermus thermophilus HB8, at 2.4 A resolution: A putative member of the StAR-related lipid-transfer (START) domain superfamily. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Schmalz, S.; Mayr, V.; Shosherova, A.; Gepp, B.; Ackerbauer, D.; Sturm, G.; Bohle, B.; Breiteneder, H.; Radauer, C. Isotype-specific binding patterns of serum antibodies to multiple conformational epitopes of Bet v 1. J. Allergy Clin. Immunol. 2022, 149, 1786–1794.e1712. [Google Scholar] [CrossRef]
- Berkner, H.; Seutter von Loetzen, C.; Hartl, M.; Randow, S.; Gubesch, M.; Vogel, L.; Husslik, F.; Reuter, A.; Lidholm, J.; Ballmer-Weber, B.; et al. Enlarging the toolbox for allergen epitope definition with an allergen-type model protein. PLoS ONE 2014, 9, e111691. [Google Scholar] [CrossRef]
- Holthuis, J.C.; Menon, A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature 2014, 510, 48–57. [Google Scholar] [CrossRef]
- Gatta, A.T.; Wong, L.H.; Sere, Y.Y.; Calderon-Norena, D.M.; Cockcroft, S.; Menon, A.K.; Levine, T.P. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife 2015, 4, e07253. [Google Scholar] [CrossRef]
- Elbaz-Alon, Y.; Eisenberg-Bord, M.; Shinder, V.; Stiller, S.B.; Shimoni, E.; Wiedemann, N.; Geiger, T.; Schuldiner, M. Lam6 regulates the extent of contacts between organelles. Cell Rep. 2015, 12, 7–14. [Google Scholar] [CrossRef]
- Murley, A.; Sarsam, R.D.; Toulmay, A.; Yamada, J.; Prinz, W.A.; Nunnari, J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 2015, 209, 539–548. [Google Scholar] [CrossRef]
- Dresden, C.E.; Ashraf, Q.; Husbands, A.Y. Diverse regulatory mechanisms of StARkin domains in land plants and mammals. Curr. Opin. Plant Biol. 2021, 64, 102148. [Google Scholar] [CrossRef]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 2001, 43, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Levine, T.P. Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem. Soc. Trans. 2016, 44, 517–527. [Google Scholar] [CrossRef]
- Jentsch, J.A.; Kiburu, I.; Pandey, K.; Timme, M.; Ramlall, T.; Levkau, B.; Wu, J.; Eliezer, D.; Boudker, O.; Menon, A.K. Structural basis of sterol binding and transport by a yeast StARkin domain. J. Biol. Chem. 2018, 293, 5522–5531. [Google Scholar] [CrossRef]
- Tong, J.; Manik, M.K.; Im, Y.J. Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites. Proc. Natl. Acad. Sci. USA 2018, 115, E856–E865. [Google Scholar] [CrossRef] [PubMed]
- Gibrat, J.F.; Madej, T.; Bryant, S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996, 6, 377–385. [Google Scholar] [CrossRef]
- Madej, T.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Bryant, S.H. Biological assembly comparison with VAST. Methods Mol. Biol. 2020, 2112, 175–186. [Google Scholar] [CrossRef] [PubMed]


| Plant Family | Subfamily | Allergen Source | Allergen | References | UniProt/PDB |
|---|---|---|---|---|---|
| Betulaceae | Betuloideae | Birch (Betula pendula) | Bet v 1.0101 | [6] | P15494/4A88 |
| Alder (Alnus glutinosa) | Aln g 1.0101 | [8] | P38948 | ||
| Coryloideae | Hazel (Corylus avellana) | Cor a 1.0101 | [9] | Q09407 | |
| Hornbeam (Carpinus betulus) | Car b 1.0101 | [19] | P38949 | ||
| Hop-hornbeam (Ostrya carpinifolia) | Ost c 1.0101 | [18] | E2GL17 | ||
| Fagaceae | Fagoideae | Beech (Fagus silvatica) | Fag s 1.0101 | [18] | B7TWE6/6ALK |
| Quercoideae | Oak (Quercus alba) | Que a 1.0201 | [20] | B6RQS1 | |
| Sawtooth oak (Quercus acutissima) | Que ac 1.0101 | [21] | GenBank QOL10866.1 | ||
| Holly oak (Quercus ilex) | Que i 1.0101 | [22] | A0A7D0TA82 | ||
| Mongolian oak (Quercus mongolica) | Que m 1.0101 | [23] | GenBank AUH28179 | ||
| Castanoideae | Chestnut (Castanea sativa) | Cas s 1.0101 | [18] | B7TWE3 |
| Plant Family | Allergen Source | Allergen | References | UniProt/PDB |
|---|---|---|---|---|
| Actinidiaceae | Golden kiwifruit (Actinidia chinensis) | Act c 8.0101 | [25] | D1YSM4 |
| Green kiwifruit (Actinidia deliciosa) | Act d 8.0101 Act d 11.0101 | [25] [26] | D1YSM5 P85524/4IGV | |
| Anacardiaceae | Mango (Mangifera indica) | Man i 2.0101 | [27] | GenBank UYO79702.1 |
| Apiaceae | Celery (Apium graveolens) | Api g 1.0101 | [11] | P49372/2BK0 |
| Carrot (Daucus carota) | Dau c 1.0103 | [28] | O04298/2WQL | |
| Cannabaceae | Indian hemp (Cannabis sativa) | Can s 5.0101 | [29] | I6XT51 |
| Corylaceae | Hazelnut (Corylus avellana) | Cor a 1.0401 | [30] | Q9SWR4/6GQ9, 6Y3H |
| Fabaceae | Peanut (Arachis hypogaea) | Ara h 8.0101 | [31] | Q6VT83/4M9B |
| Soybean (Glycine max) | Gly m 4.0101 | [32] | P26987/2K7H | |
| Mung bean (Vigna radiata) | Vig r 1.0101 Vig r 6.0101 | [33] [34] | Q2VU97 A0A1S3THR8/2FLH, 3C0V | |
| Juglandaceae | English walnut (Juglans regia) | Jug r 5.0101 | [35] | GenBank Acc. No. KX034087.1 |
| Rosaceae | Strawberry (Fragaria x ananassa) | Fra a 1.0101 | [36] | Q5ULZ4/6ST8 |
| Apple (Malus domestica) | Mal d 1.0101 | [10] | P43211/5MMU | |
| Apricot (Prunus armeniaca) | Pru ar 1.0101 | unpublished | O50001 | |
| Cherry (Prunus avium) | Pru av 1.0101 | [37] | O24248/1E09, 1H20 | |
| Almond (Prunus dulcis) | Pru du 1.0101 | [38] | B6CQS9 | |
| Peach (Prunus persica) | Pru p 1.0101 | [39] | Q2I6V8/6Z98 | |
| Pear (Pyrus communis) | Pyr c 1.0101 | [40] | O65200 | |
| Raspberry (Rubus idaeus) | Rub 1 1.0101 | [41] | Q0Z8U9 | |
| Solanaceae | Tomato (Solanum lycopersicum) | Sola l 4.0101 | [42] | K4CWC5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Breiteneder, H.; Kraft, D. The History and Science of the Major Birch Pollen Allergen Bet v 1. Biomolecules 2023, 13, 1151. https://doi.org/10.3390/biom13071151
Breiteneder H, Kraft D. The History and Science of the Major Birch Pollen Allergen Bet v 1. Biomolecules. 2023; 13(7):1151. https://doi.org/10.3390/biom13071151
Chicago/Turabian StyleBreiteneder, Heimo, and Dietrich Kraft. 2023. "The History and Science of the Major Birch Pollen Allergen Bet v 1" Biomolecules 13, no. 7: 1151. https://doi.org/10.3390/biom13071151
APA StyleBreiteneder, H., & Kraft, D. (2023). The History and Science of the Major Birch Pollen Allergen Bet v 1. Biomolecules, 13(7), 1151. https://doi.org/10.3390/biom13071151






